Molecular Characterization of Watermelon Chlorotic Stunt Virus (WmCSV) from Palestine
Abstract
:1. Introduction
2. Results and Discussion
2.1. Field Survey

| Season/Year | Area | Crop | Fields | +ve by deg. primers * | +ve for WmCSV (PCR) | +ve for WmCSV (RCA) | Total no. of samples +ve for WmCSV (%) | No. of Samples mixed Infected with SLCV (% ϕ) |
|---|---|---|---|---|---|---|---|---|
| Summer/2010 | Tulkarm | Watermelon | 3 | 58/62 | 0 | 0 | 0 (0) | 50 (80.6) |
| Jenin | Watermelon | 4 | 65/110 | 43 | 37 | 43 (39.1) | 27 (62.7) | |
| Summer/2011 | Jenin | Watermelon | 4 | ND (120) | 0 | 0 | 0 (0) | 100 (83.3) |
| Arifa S. Khan | Jericho | Watermelon | 7 | 59/84 | 0 | 0 | 0 (0) | 54 (64.3) |
| Total | 23 | 288/501 | 53 | 73 | 79 (15.8) | 261 (52.1) | ||
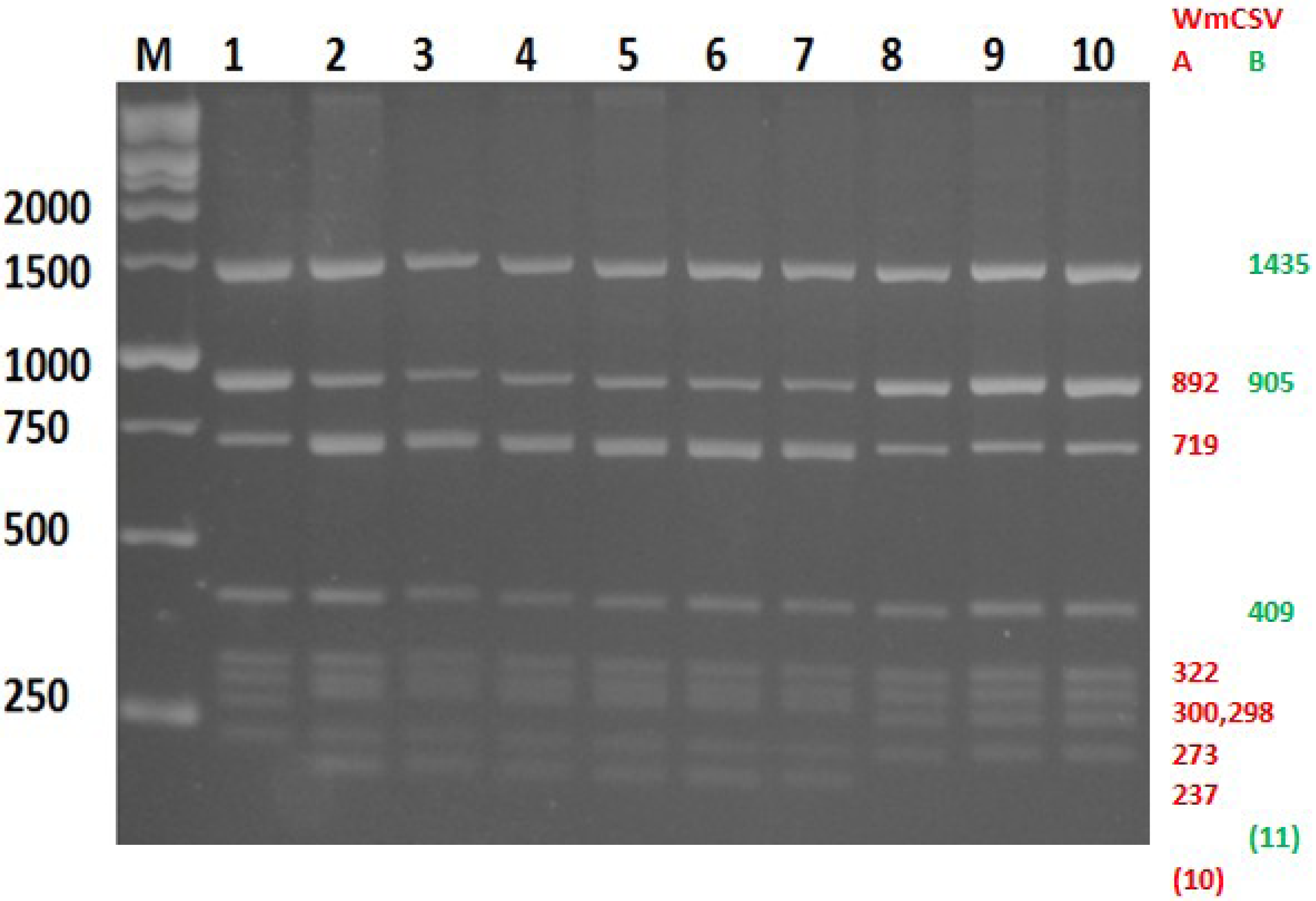
2.2. Cloning and Sequencing of WmCSV-[PAL]
| Primer Name | 5'–3' Sequence | Target | Fragment Size |
|---|---|---|---|
| WmA150F 1 | GTCAGTATGTGGGATCCATTGC | DNA-A | 1201 bp |
| WmA 1350R 1 | GCAAATACGATTCAACCACAACC | ||
| WmA170R 1 | GCAATGGATCCCACATACTGAC | DNA-A | 1597 bp |
| WmA1325F 1 | GGTTGTGGTTGAATCGTATTTGC | ||
| WmB672F | CGCCGTTGCCTGGAGGATGTTCAC | DNA-B | 1329 bp |
| WmB2000R | GCAGCACAGGCTGCCTTCACCTTC | ||
| WmB1977F | GAAGGTGAAGGCAGCCTGTGCTGC | DNA-B | 1479 bp |
| WmB695R | GTGAACATCCTCCAGGCAACGGCG |
| DNA-A | DNA-B | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Position of Genes (Coordinates of Start/Stop Codons [Predicted Coding Capacity in kDa] | Position of Genes (Coordinates of Start/Stop Codons [Predicted Coding Capacity in kDa] | ||||||||||||
| AV2 | AV1 | AC1 | AC2 | AC3 | AC4 | AC5 | BC1 | BV1 | |||||
| Isolate | Accession Number | Size (nt) | nt | nt | Nt | nt | Nt | nt | nt | Accession Number | Size (nt) | nt | nt |
| PAL-1-J40 | KC462552 | 2751 | 154–513 (13.2) | 314–1090 (28.7) | 1539–2624 (40.2) | 1234–1641 (15.1) | 1087–1491 (14.9) | 2327–2470 (5.3) | 218–985 (28.4) | KC462553 | 2760 | 1326–2321 (36.5) | 522–1286 (28.3) |
| PAL-2-J44 | KJ854912 | 2753 | 156–515 (13.2) | 316–1092 (28.7) | 1541–2626 (40.2) | 1234–1641 (15.1) | 1089–1493 (14.9) | 2329–2472 (5.3) | 220–987 (28.4) | - | - | ||
| PAL-3-J45 | KJ854913 | 2752 | 155–514 (13.2) | 315–1091 (28.7) | 1540–2625 (40.2) | 1233–1640 (15.1) | 1088–1492 (14.9) | 2328–2471 (5.3) | 219–986 (28.4) | - | - | ||
| PAL-4-J56 | KJ854914 | 2752 | 155–514 (13.2) | 315–1091 (28.7) | 1540–2625 (40.2) | 1233–1640 (15.1) | 1088–1492 (14.9) | 2328–2471 (5.3) | 219–986 (28.4) | - | - | ||
| PAL-5-J67 | KJ854915 | 2752 | 155–514 (13.2) | 315–1091 (28.7) | 1540–2625 (40.2) | 1233–1640 (15.1) | 1088–1492 (14.9) | 2328–2471 (5.3) | 219–986 (28.4) | - | - | ||
| PAL-6-Q13 | KJ854916 | 2753 | 156–515 (13.2) | 316–1092 (28.7) | 1541–2626 (40.2) | 1234–1641 (15.1) | 1089–1493 (14.9) | 2329–2472 (5.3) | 220–987 (28.4) | - | - | ||
| PAL-7-Q17 | KJ854917 | 2752 | 156–515 (13.2) | 316–1092 (28.7) | 1541–2626 (40.2) | 1233–1640 (15.1) | 1088–1492 (14.9) | 2328–1471 (5.3) | 219–986 (28.4) | - | - | ||
| PAL-8-Q20 | KJ854918 | 2753 | 156–515 (13.2) | 316–1092 (28.7) | 1541–2626 (40.2) | 1234–1641 (15.1) | 1089–1493 (14.9) | 2329–2472 (5.3) | 220–987 (28.4) | - | - | ||
| PAL-9-Q23 | KJ854919 | 2753 | 156–515 (13.2) | 316–1092 (28.7) | 1541–2626 (40.2) | 1234–1641 (15.1) | 1089–1493 (14.9) | 2329–2472 (5.3) | 220–987 (28.4) | - | - | ||
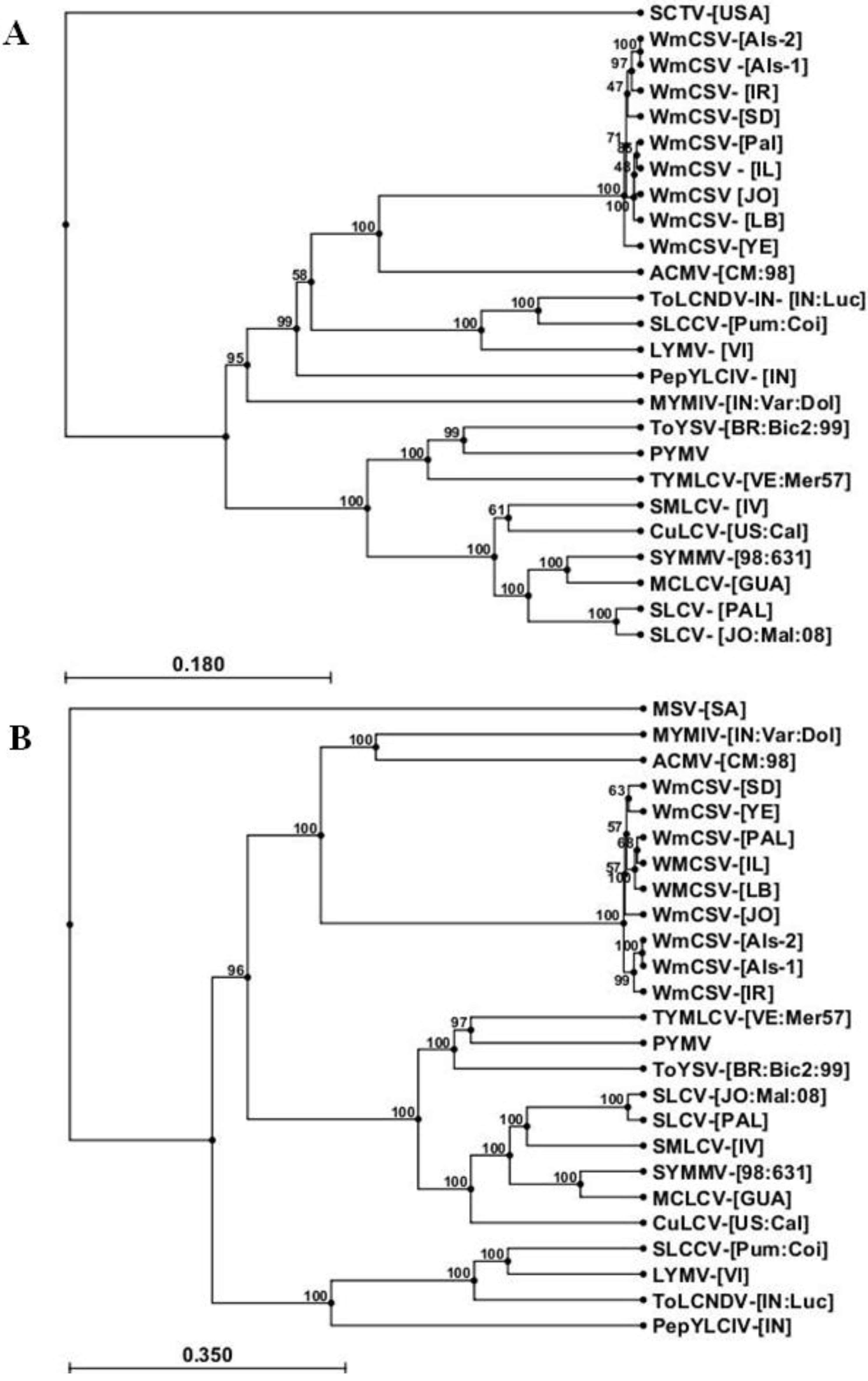
2.3. Sequence Analysis of DNA-A and DNA-B of WmCSV-[PA:Pal:10]
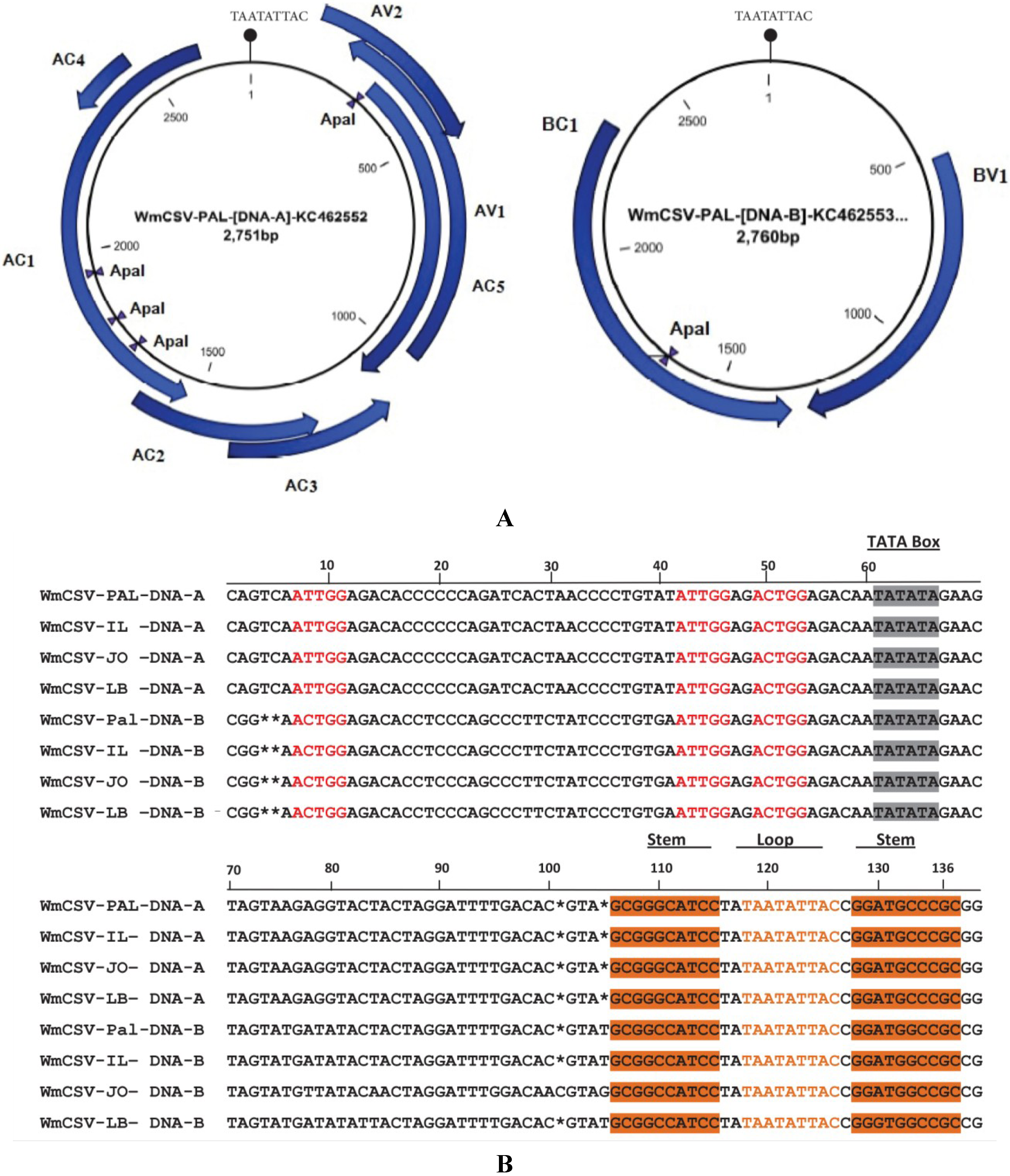
| Virus | Total nt | DNA-A | DNA-B | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AV2 | AV1 | AC1 | AC2 | AC3 | AC4 | AC5 | BC1 | BV1 | ||||||||||||
| DNA-A | DNA-B | Nt | aa | nt | Aa | Nt | Aa | nt | Aa | Nt | Aa | nt | aa | nt | aa | nt | aa | nt | aa | |
| WmCSV [YE] | 97.60 | 95.80 | 99.44 | 98.32 | 97.68 | 95.35 | 97.88 | 97.50 | 97.55 | 95.00 | 97.28 | 94.78 | 98.61 | 95.74 | 98.57 | 96.86 | 89.86 | 91.56 | 96.60 | 93.65 |
| WmCSV [SD] | 98.22 | 95.36 | 98.89 | 98.32 | 98.97 | 100.00 | 98.25 | 98.61 | 97.30 | 94.17 | 97.78 | 95.52 | 100.00 | 100.00 | 99.09 | 97.65 | 89.96 | 91.87 | 95.03 | 92.11 |
| WmCSV [JO] | 99.20 | 95.80 | 99.72 | 100.00 | 99.61 | 99.61 | 99.45 | 99.17 | 98.28 | 95.83 | 98.27 | 96.27 | 100.00 | 100.00 | 99.61 | 99.22 | 90.16 | 91.46 | 96.60 | 92.59 |
| WmCSV [IL] | 99.42 | 98.26 | 100.00 | 100.00 | 99.49 | 99.61 | 99.63 | 99.17 | 98.77 | 96.67 | 98.77 | 97.76 | 100.00 | 100.00 | 99.61 | 99.22 | 91.57 | 99.59 | 97.78 | 94.71 |
| WmCSV [IR] | 98.26 | 94.46 | 99.44 | 99.16 | 98.97 | 100.00 | 98.16 | 98.06 | 96.57 | 92.50 | 97.04 | 95.52 | 100.00 | 100.00 | 99.22 | 97.65 | 89.66 | 91.06 | 96.56 | 92.67 |
| WmCSV [LB] | 99.06 | 98.26 | 100.00 | 100.00 | 98.58 | 97.29 | 99.45 | 99.17 | 98.77 | 96.67 | 99.01 | 98.51 | 100.00 | 100.00 | 98.83 | 97.25 | 98.69 | 99.59 | 98.04 | 94.74 |
| WmCSV [OM:1] | 97.86 | 93.16 | 99.44 | 99.16 | 98.46 | 98.84 | 97.61 | 97.50 | 96.57 | 92.50 | 97.78 | 95.52 | 99.31 | 97.87 | 98.96 | 96.86 | 98.16 | 89.84 | 94.77 | 93.72 |
| WmCSV [OM:2] | 97.79 | 93.77 | 99.44 | 99.16 | 98.46 | 98.84 | 97.43 | 97.23 | 96.53 | 93.33 | 97.78 | 95.52 | 99.31 | 97.87 | 98.96 | 96.86 | 88.96 | 89.02 | 92.96 | 92.15 |
| Virus Name | Abbreviation | DNA-A | DNA-B |
|---|---|---|---|
| Watermelon chlorotic stunt virus-[Yemen] | WmCSV-[YE] | AJ012081 | AJ012082 |
| Watermelon chlorotic stunt virus-[Sudan] | WmCSV-[SD] | AJ245650 | AJ245651 |
| Watermelon chlorotic stunt virus-[Oman]-Als-2 | WmCSV-[Als-2] | JN618982 | HE800539 |
| Watermelon chlorotic stunt virus-[Oman]-Als-1 | WmCSV-[Als-1] | JN618981 | JN618980 |
| Watermelon chlorotic stunt virus-[Lebanon] | WmCSV-[LB] | HM368371 | HM368372 |
| Watermelon chlorotic stunt virus-[Jordan] | WmCSV-[JO] | EU561237 | EU561236 |
| Watermelon chlorotic stunt virus-[Israel] | WmCSV-[IL] | EF201809.1 | EF201810.1 |
| Watermelon chlorotic stunt virus-[Iran] | WmCSV-[IR] | AJ245652 | AJ245653 |
| Tomato yellow spot virus[Brazil:Bicas 2:1999] | ToYSV-[BR:Bic2:99] | DQ336350 | DQ336351 |
| Tomato yellow margin leaf curl virus-[Venezuela] | TYMLCV-[VE:Mer57] | AY508993 | AY508994 |
| Tomato leaf curl New Delhi virus India-[India] | ToLCNDV-[INLuc] | Y16421 | X89653 |
| Squash yellow mild mottle virus-[Costa Rica:1998] | SYMMV-[98:631] | AY064391 | AF440790 |
| Squash mild leaf curl virus-[Imperial Valley] | SMLCV-[IV] | NC_004645 | NC_004646 |
| Squash leaf curl virus-[Palestine] | SLCV-[PA: Pal:10] | KC441465 | KC441466 |
| Squash leaf curl virus-[Jordan] | SLCV-[JO:Mal:08] | EF532620 | EF532621 |
| Squash leaf curl China virus -[Pumpkin:Coimbatore] | SLCCV-[Pum:Coi] | AY184487 | AY184488 |
| Potato yellow mosaic virus-[Puerto Rico] | PYMV | AY965897 | AY965898 |
| Pepper yellow leaf curl virus-[Indonesia] | PepYLCIV-[IN] | AB267834 | AB267839 |
| Mungbean yellow mosaic virus-[India] | MYMIV-[IN:Var:Dol] | AY547317 | DQ061273 |
| Melon chlorotic leaf curl virus-[Guatemala] | MCLCV-[GUA] | AF325497 | AF325498 |
| Loofa yellow mosaic virus-[Vietnam] | LYMV-[VI:02] | AF509739 | AF509740 |
| Cucurbit leaf curl virus-[California] | CuLCV-[US:Cal:00] | AF224760 | AF224761 |
| African cassava mosaic virus-[Cameroon] | ACMV-[CM:98] | AF112352 | AF112353 |
| Spinach curly top virus-[USA] | SCTV-[USA] | NC_005860 | - |
| Maize streak virus - [South Africa: Sasri_s:2007] | MSV-[SA:Sa:07] | - | EU152254 |
2.4. Discrimination between WmCSV-[PAL], WmCSV-[JO] and WmCSV-[IL] Isolates
2.5. Inoculation of Watermelon Plants by Particle Bombardment (Biolistic Inoculation)
3. Experimental
3.1. Samples Collection
3.2. Nucleic Acid Extraction
3.3. Detection of WmCSV-[PAL] by Polymerase Chain Reaction Using Degenerate Primers
3.3.1. Rolling Circle Amplification (RCA)
3.3.2. Restriction Fragment Length Polymorphism (RFLP)
3.3.3. Amplification of DNA-A and DNA-B of WmCSV-[PA]
3.4. Cloning and Sequence Analysis
3.5. Discrimination between WmCSV-[PAL] and WmCSV-[IL] and WmCSV-[JO] Isolates Using in Silico Enzymatic Digestion
3.6. Inoculation of Plants by Particle Bombardment (Biolistic Inoculation)
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References and Notes
- International Committee on Taxonomy of Viruses (ICTV). Available online: http://ictvonline.org/virusTaxonomy.asp?taxnode_id=20120007/ (accessed on 21 September 2013).
- Hanley-Bowdoin, L.; Bejarano, E.R.; Robertson, D.; Mansoor, S. Geminiviruses: Masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 2013. [Google Scholar] [CrossRef]
- Nawaz-ul-Rehman, M.S.; Fauquet, C.M. Evolution of geminiviruses and their satellites. FEBS Lett. 2009, 583, 1825–1832. [Google Scholar] [CrossRef]
- Lapidot, M.; Friedmann, M. Breeding for resistance to whitefly-transmitted geminiviruses. Annals Appl. Biol. 2002, 140, 109–127. [Google Scholar] [CrossRef]
- Mansoor, S.; Briddon, R.W.; Bull, S.E.; Bedford, I.D.; Bashir, A.; Hussain, M.; Saeed, M.; Zafar, Y.; Malik, K.A.; Fauquet, C.; Markham, P.G. Cotton leaf curl disease is associated with multiple monopartite begomoviruses supported by single DNAb. Arch. Virol. 2003, 148, 1969–1986. [Google Scholar] [CrossRef]
- Abudy, A.; Sufrin-Ringwald, T.; Dayan-Glick, C.; Guenoune-Gelbert, D.; Livneh, O.; Zaccai, M.; Lapidot, M. Watermelon chlorotic stunt and Squash leaf curl begomoviruses—New threats to cucurbit crops in the Middle East. Isr. J. Plant Sci. 2010, 58, 33–42. [Google Scholar]
- Gutierrez, C. Strategies for geminivirus DNA replication and cell cycle interference. Physiol. Mol. Plant Pathol. 2002, 60, 219–230. [Google Scholar] [CrossRef]
- Hanley-Bowdoin, L.; Settlage, S.B.; Robertson, D. Reprogramming plant gene expression: A prerequisite to geminivirus DNA replication. Mol. Plant Pathol. 2004, 5, 149–156. [Google Scholar] [CrossRef]
- Cohen, S.; Duffus, J.E.; Larsen, R.C.; Liu, H.Y.; Flock, R.A. Purification, serology, and vector relationships of squash leaf curl virus, a whitefly-transmitted geminivirus. Phytopathology 1983, 73, 1669–1673. [Google Scholar] [CrossRef]
- Al-Musa, A.; Anfoka, G.; Misbeh, S.; Abhary, M.; Ahmad, F.H. Detection and molecular characterization of Squash leaf curl virus (SLCV) in Jordan. J. Phytopathol. 2008, 156, 311–316. [Google Scholar] [CrossRef]
- Ali-Shtayeh, M.S.; Jamous, R.M.; Hussein, E.H.; Mallah, O.B.; Abu-Zeitoun, S.Y. Squash leaf curl virus (SLCV): A serious disease threatening cucurbits production in Palestine. Virus Genes 2013. [Google Scholar] [CrossRef]
- Brown, J.K.; Idris, A.M.; Alteri, C.; Stenger, D.C. Emergence of a new cucurbit-infecting begomovirus species capable of forming viable reassortments with related viruses in the squash leaf curl virus cluster. Phytopathology 2002, 92, 734–742. [Google Scholar] [CrossRef]
- Brown, J.K.; Idris, A.M.; Rogan, D.; Hussein, M.H.; Palmieri, M. Melon chlorotic leaf curl virus, a new begomovirus associated with Bemisia tabaci infestations in Guatemala. Plant Dis. 2001, 85. [Google Scholar] [CrossRef]
- Idris, A.M.; Mills-Lujan, K.; Martin, K.; Brown, J.K. Melon chlorotic leaf curl virus: Characterization and differential reassortment with closest relatives reveal adaptive virulence in the Squash leaf curl virus clade and host shifting by the host-restricted Bean Calico Mosaic Virus. J. Virol. 2008, 82, 1959–1967. [Google Scholar] [CrossRef]
- Kheyr-Pour, A.; Bananej, K.; Dafalla, G.A.; Caciagli, P.; Noris, E.; Ahoonmanesh, A.; Lecoq, H.; Gronenborn, B. Watermelon chlorotic stunt virus from the Sudan and Iran: Sequence comparisons and identification of a whitefly-transmission determinant. Phytopathology 2000, 90, 629–635. [Google Scholar] [CrossRef]
- Al-Musa, A.; Anfoka, G.; Al-Abdulat, A.; Misbeh, S.; Haj Ahmed, F.; Otri, I. Watermelon chlorotic stunt virus (WmCSV): A serious disease threatening watermelon production in Jordan. Virus Genes 2011, 43, 79–89. [Google Scholar] [CrossRef]
- Bananej, K.; Dafalla, G.A.; Ahoonmanesh, A.; Kheyr-Pour, A. Host range of an Iranian isolate of Watermelon chlorotic stunt virus as determined by whitefly-mediated inoculation and agroinfection, and its geographical distribution. J. Phytopathol. 2002, 150, 423–430. [Google Scholar] [CrossRef]
- Yousif, M.T.; Kheyr-Pour, A.; Gronenborn, B.; Pitrat, M.; Dogimont, C. Sources of resistance to Watermelon Chlorotic Stunt Virus in melon. Plant Breed. 2007, 126, 422–427. [Google Scholar] [CrossRef]
- Rojas, M.R.; Gilbertson, R.L.; Russell, D.R.; Maxwell, D.P. Use of degenerate primers in the polymerase chain reaction to detect whitefly-transmitted geminiviruses. Plant Dis. 1993, 77, 340–347. [Google Scholar] [CrossRef]
- Ali-Shtayeh, M.S.; Jamous, R.M.; Hussein, E.Y.; Mallah, O.B.; Abu-Zaitoun, S.Y. First report of Watermelon chlorotic stunt virus in watermelon in the Palestinian Authority. Plant Dis. 2012, 4. [Google Scholar] [CrossRef]
- Jones, P.; Sattar, M.H.A.; al Kaff, N. The incidence of virus disease in watermelon and sweetmelon crops in the Peoples Democratic Republic of Yemen and its impact on cropping policy. Ann. Appl. Biol. 1988, 17, 203–207. [Google Scholar]
- Walkey, D.G.A.; Alhubaishi, A.A.; Webb, M.J.W. Plant virus diseases in the Yemen Arab Republic. Trop. Pest Manag. 1990, 36, 195–206. [Google Scholar] [CrossRef]
- Bedford, I.D.; Briddon, R.W.; Jones, P.; Al-Kaff, N.; Markham, P.G. Differentiation of three whitefly-transmitted geminiviruses from the Republic of Yemen. Eur. J. Plant Pathol. 1994, 100, 243–257. [Google Scholar] [CrossRef]
- Abou-Jawdah, Y.; Sobh, H.; Haidar, A.; Samsatly, J. First report in Lebanon on detection of two whitefly transmitted cucurbit viruses and their molecular characterization. In Petria, Giornale di Patologia delle Piante 20, Proceedings of the 13th Congress of the Mediterranean Phytopathological Union, MPU, Plant Pathology Research Centre in Rome, Rome, Italy, 20–25 June 2010; Barba, M., Motta, E., Tomassoli, L., Riccioni, L., Eds.; CRA-Centro di Ricerca per la Patologia Vegetale: Rome, Italy, 2010; p. 268. [Google Scholar]
- Khan, A.J.; Akhtar, S.; Briddon, R.W.; Ammara, U.; Al-Matrooshi, A.M.; Mansoor, S. Complete nucleotide sequence of watermelon chlorotic stunt virus originating from Oman. Viruses 2012, 4, 1169–1181. [Google Scholar] [CrossRef]
- Food and Agriculture Organization (FAO). The FAO Statistical Database-Agriculture 2012. Available online: http://faostat.fao.org/site/567/DesktopDefault.aspx?PageID=567#ancor/ (accessed on 30 December 2013).
- Haible, D.; Kober, S.; Jeske, H. Rolling circle amplification revolutionizes diagnosis and genomics of geminiviruses. J. Virol. Methods 2006, 135, 9–16. [Google Scholar] [CrossRef]
- Givord, L.; Fargette, D.; Kounounguissa, B.; Thouvenel, J.C.; Walter, B.; van Regenmorte, M.H.V. Detection of geminiviruses from tropical countries by a double monoclonal antibody ELISA using antibodies to African cassava mosaic virus. Plant Pathol. 1994, 14, 327–333. [Google Scholar]
- Harrison, B.D.; Swanson, M.M.; Fargette, D. Begomovirus coat protein: Serology, variation and functions. Physiol. Mol. Plant Pathol. 2002, 60, 257–271. [Google Scholar] [CrossRef]
- Perveen, R.; Fani, I.; Rasheed, I.; Chohan, S.; Rehman, A.; Haider, S. Identification of cotton leaf curl begomovirus in Pakistan in different symptomatic and asymptomatic plants though enzyme-linked immunosorbent assay (ELISA). Eur. J. Soc. Sci. 2010, 14, 502–206. [Google Scholar]
- Briddon, R.W.; Markham, P.G. Universal primers for the PCR amplification of dicot-infecting geminiviruses. Mol. Biotechnol. 1994, 1, 202–205. [Google Scholar] [CrossRef]
- Li, R.; Salih, S.; Hurtt, S. Detection of geminiviruses in sweetpotato by polymerase chain reaction. Plant Dis. 2004, 88, 1347–1351. [Google Scholar] [CrossRef]
- Rouhibakhsh, A.; Priya, J.; Periasamy, M.; Haq, Q.M.I.; Malathi, V.G. An improved DNA isolation method and PCR protocol for efficient detection of multicomponents of begomovirus in legumes. J. Virol. Methods 2008, 147, 37–42. [Google Scholar] [CrossRef]
- Jeske, H.; Gotthardt, D.; Kober, S. In planta cloning of geminiviral DNA: The true Sida micrantha mosaic virus. J. Virol. Methods 2010, 163, 301–308. [Google Scholar] [CrossRef]
- Inoue-Nagata, A.K.; Albuquerque, L.C.; Rocha, W.B.; Nagata, T. A simple method for cloning the complete begomovirus genome using the bacteriophage phi 29 DNA polymerase. J. Virol. Methods 2004, 116, 209–211. [Google Scholar] [CrossRef]
- Kumar, Y.; Hallan, V.; Zaidi, A.A. Molecular characterization of a distinct bipartite begomovirus species infecting tomato in India. Virus Genes 2008, 37, 425–431. [Google Scholar] [CrossRef]
- Paprotka, T.; Metzler, V.; Jeske, H. The first DNA 1-like alpha satellites in association with New World begomoviruses in natural infections. Virology 2010, 404, 148–157. [Google Scholar] [CrossRef]
- Wyant, P.S. The use of rolling circle amplification (RCA) for diagnosis and characterization of geminiviruses. Ph.D. Thesis, University of Stuttgart, Stuttgart, Germany, 2011. [Google Scholar]
- Sufrin-Ringwald, T. Molecular identification and characterization of the interaction between two Geminiviruses: Squash Leaf Curl Virus (SLCV) and Watermelon Chlorotic Stunt Virus (WmCSV). Master’s Thesis, Hebrew University, Jerusalem, Pakistan, 2008. [Google Scholar]
- Ali-Shtayeh, M.S.; Jamous, R.M.; Husein, E.Y.; Alkhader, M.Y. First report of Squash leaf curl virus in squash (Cucurbita pepo), melon (Cucumis melo), and cucumber (Cucumis sativa) in the northern West Bank of the Palestinian Authority. Plant Dis. 2010, 94. [Google Scholar] [CrossRef]
- Al-Shahwan, I.M.; Abdalla, O.A.; Al-Saleh, M.A. Squash Leaf Curl Virus (SqLCV) and other begomoviruses in Saudi Arabia. Dirasa Agric. Sci. 2002, 29, 28–36. [Google Scholar]
- CLC Main Workbench program, version 6.9; CLC bio: Aarhus, Denmark, 2013.
- Fauquet, C.M.; Briddon, R.W.; Brown, J.K.; Moriones, E.; Stanley, J.; Zerbini, M.; Zhou, X. Geminivirus strain demarcation and nomenclature. Arch. Virol. 2008, 153, 783–821. [Google Scholar] [CrossRef]
- Settlage, S.B.; See, R.G.; Hanley-Bowdoin, L. Geminivirus C3 protein: Replication enhancement and protein interactions. J. Virol. 2005, 79, 9885–9895. [Google Scholar] [CrossRef]
- Sunter, G.; Bisaro, D.M. Identification of a minimal sequence required for activation of the Tomato golden mosaic virus coat protein promoter in protoplasts. Virology 2003, 305, 452–462. [Google Scholar] [CrossRef]
- Hanley-Bowdoin, L.; Settlage, S.B.; Orozco, B.M.; Nagar, S.; Robertson, D. Geminviruses: Models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Plant Sci. 1999, 18, 71–106. [Google Scholar] [CrossRef]
- Bisaro, D.M. Silencing suppression by geminivirus proteins. Virology 2006, 344, 158–168. [Google Scholar] [CrossRef]
- Albrechtsen, S.E. Testing Methods for Seed Transmitted Viruses: Principles and Protocols; CABL Publishing Oxfordshire: Wallingford, UK, 2006; p. 259. [Google Scholar]
- Guenoune-Gelbart, D.; Sufrin-Ringwald, T.; Capobianco, H.; Gaba, V.; Polston, J.E.; Lapidot, M. Inoculation of plants with begomoviruses by particle bombardment without cloning: Using rolling-circle amplification of total DNA from infected plants. J. Virol. Methods 2010, 168, 87–93. [Google Scholar] [CrossRef]
- Finer, J.J.; Vain, P.; Jones, M.W.; McMullen, D. Development of the particle inflow gun for DNA delivery to plant cells. Plant Cell Rep. 1992, 11, 323–328. [Google Scholar]
- Lapidot, M.; Weil, G.; Cohen, L.; Segev, I.; Gaba, V. Biolistic inoculation of plants with Tomato yellow leaf curl virus DNA. J. Virol. Methods 2007, 144, 143–148. [Google Scholar] [CrossRef]
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. Maize DNA miniprep. Plant Mol. Biol. Report. 1983, 1, 19–21. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor: New York, NY, USA, 2001. [Google Scholar]
- ORF Finder (Open Reading Frame Finder). Available online: http://www.ncbi.nlm.nih.gov/projects/gorf/ (accessed on 1 January 2014).
- Clone manager Software, version 7; Sci-Ed Software: Cary, NC, USA, 2007.
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ali-Shtayeh, M.S.; Jamous, R.M.; Mallah, O.B.; Abu-Zeitoun, S.Y. Molecular Characterization of Watermelon Chlorotic Stunt Virus (WmCSV) from Palestine. Viruses 2014, 6, 2444-2462. https://doi.org/10.3390/v6062444
Ali-Shtayeh MS, Jamous RM, Mallah OB, Abu-Zeitoun SY. Molecular Characterization of Watermelon Chlorotic Stunt Virus (WmCSV) from Palestine. Viruses. 2014; 6(6):2444-2462. https://doi.org/10.3390/v6062444
Chicago/Turabian StyleAli-Shtayeh, Mohammed S., Rana M. Jamous, Omar B. Mallah, and Salam Y. Abu-Zeitoun. 2014. "Molecular Characterization of Watermelon Chlorotic Stunt Virus (WmCSV) from Palestine" Viruses 6, no. 6: 2444-2462. https://doi.org/10.3390/v6062444





