Ion Channel Activity of Vpu Proteins Is Conserved throughout Evolution of HIV-1 and SIV
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bioinformatics:
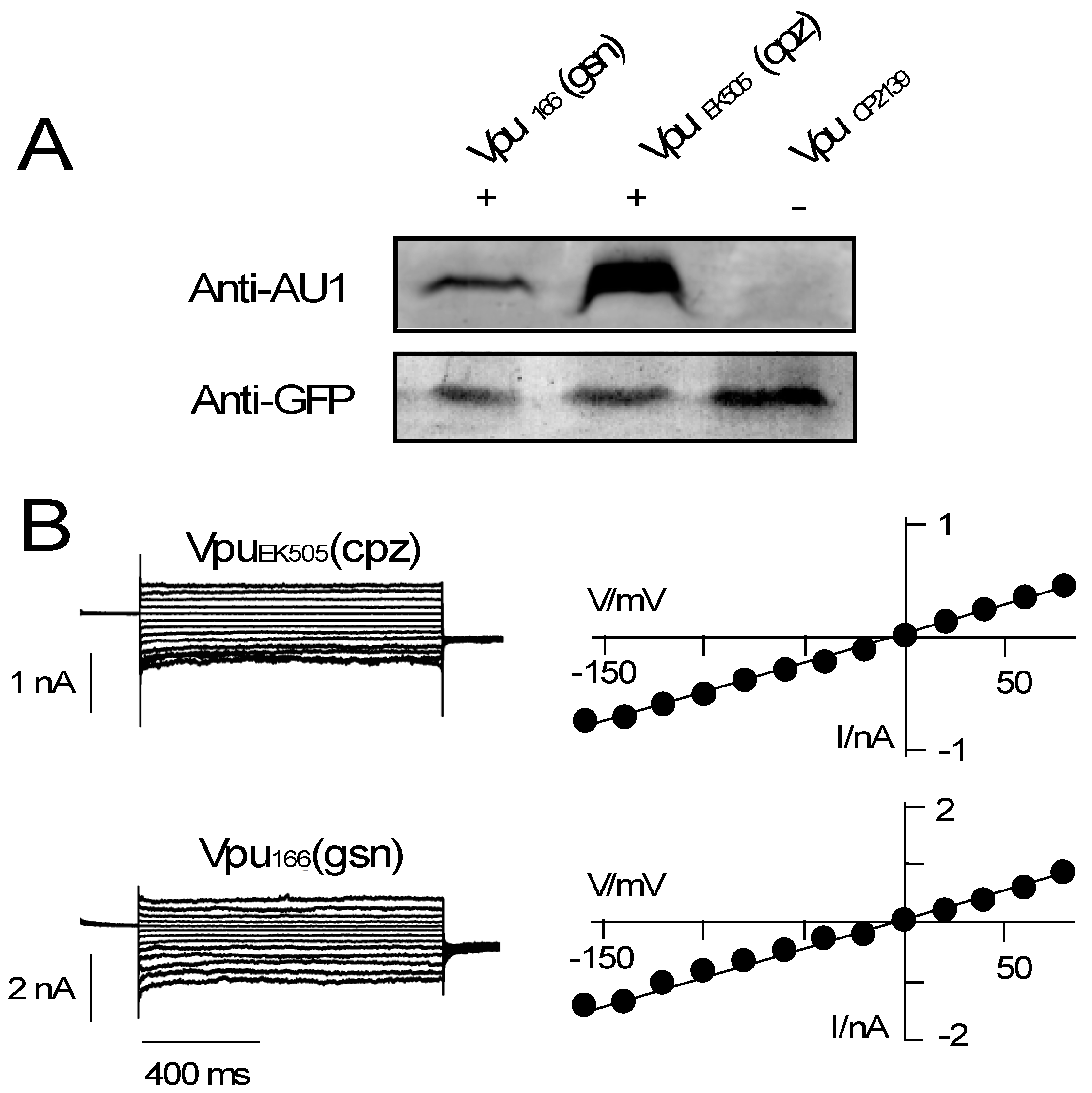
2.2. Heterologous Expression of Vpus:
2.3. Electrophysiological Characterization:
3. Results
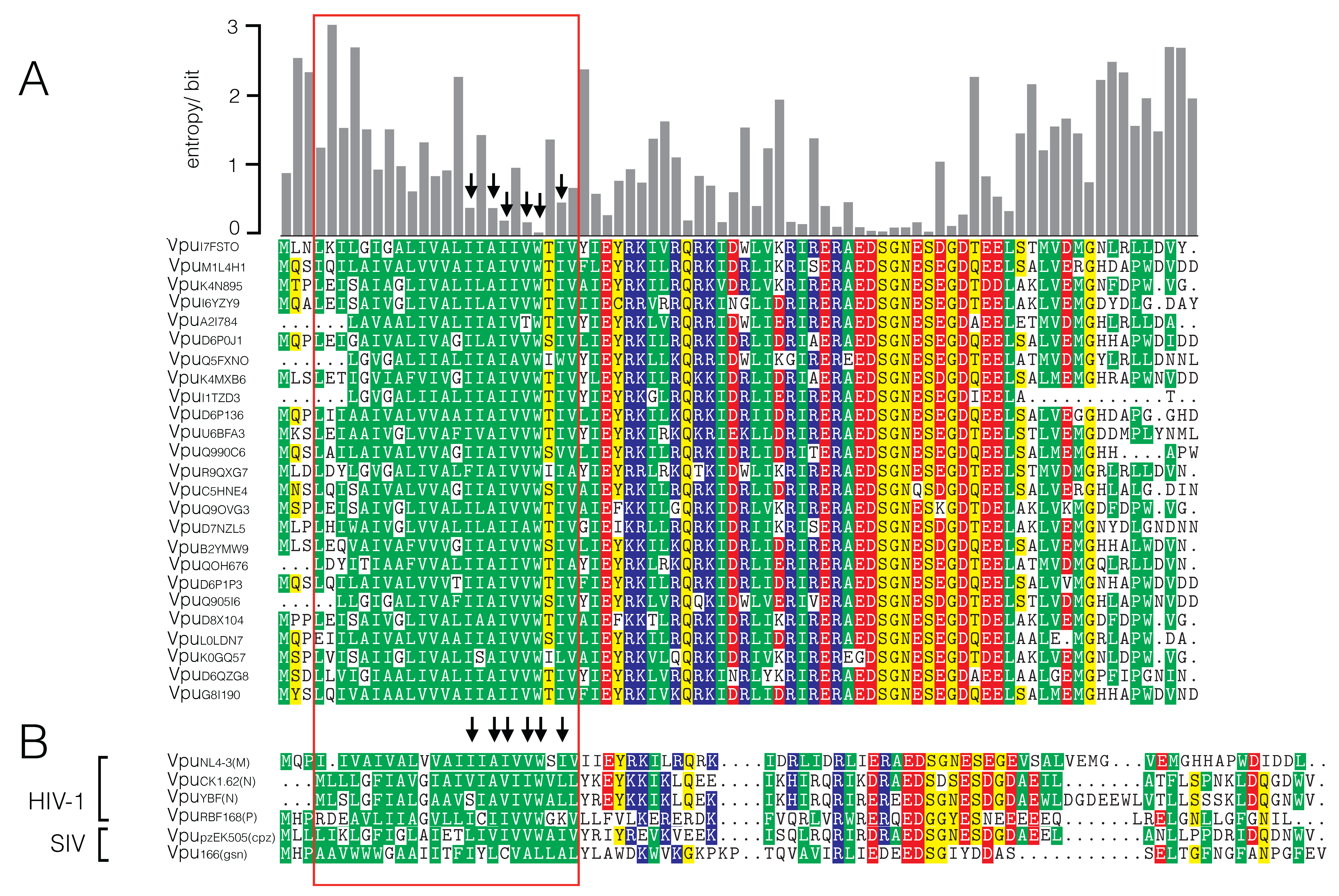
3.1. Sequence Variability of Vpu Proteins
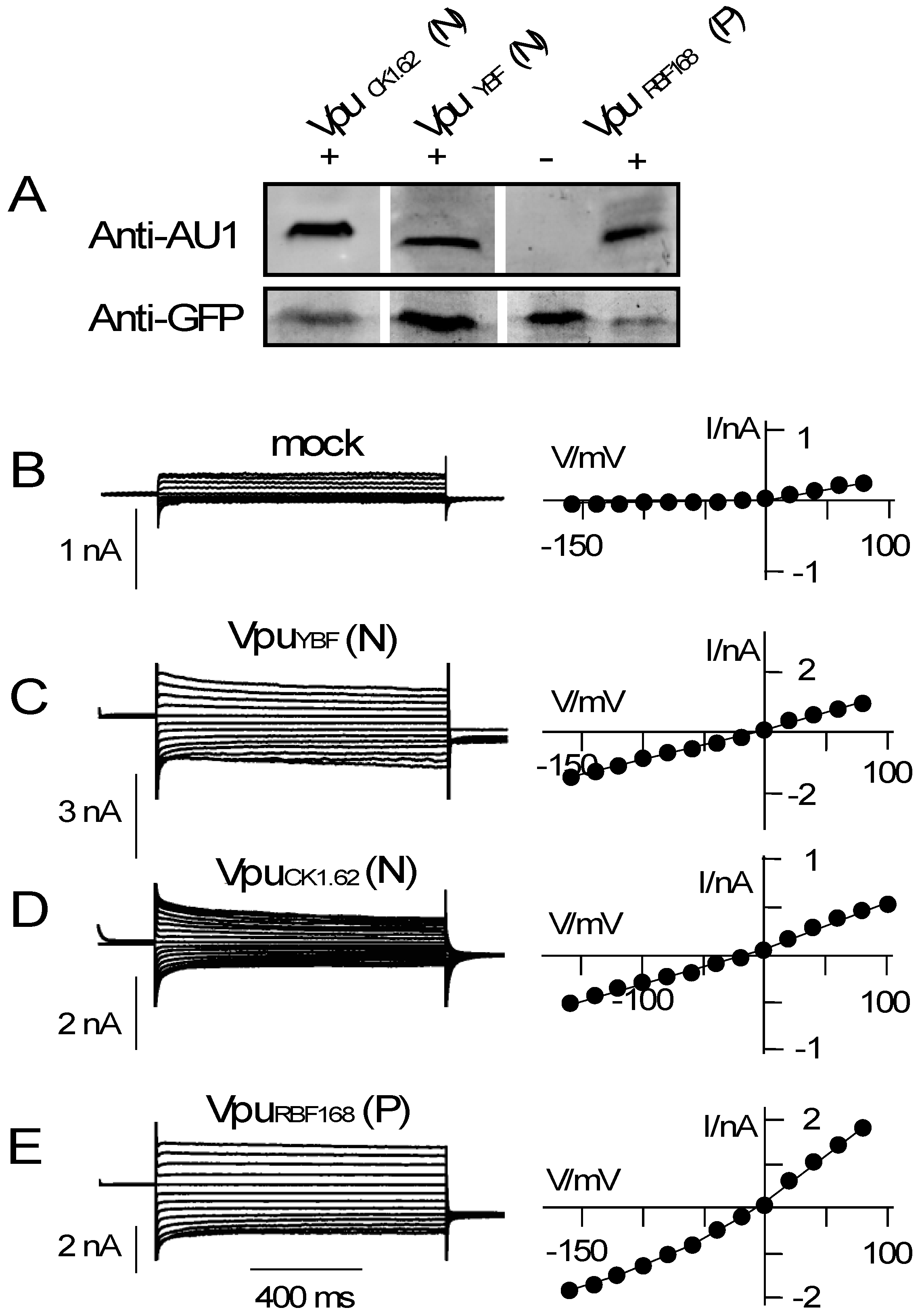
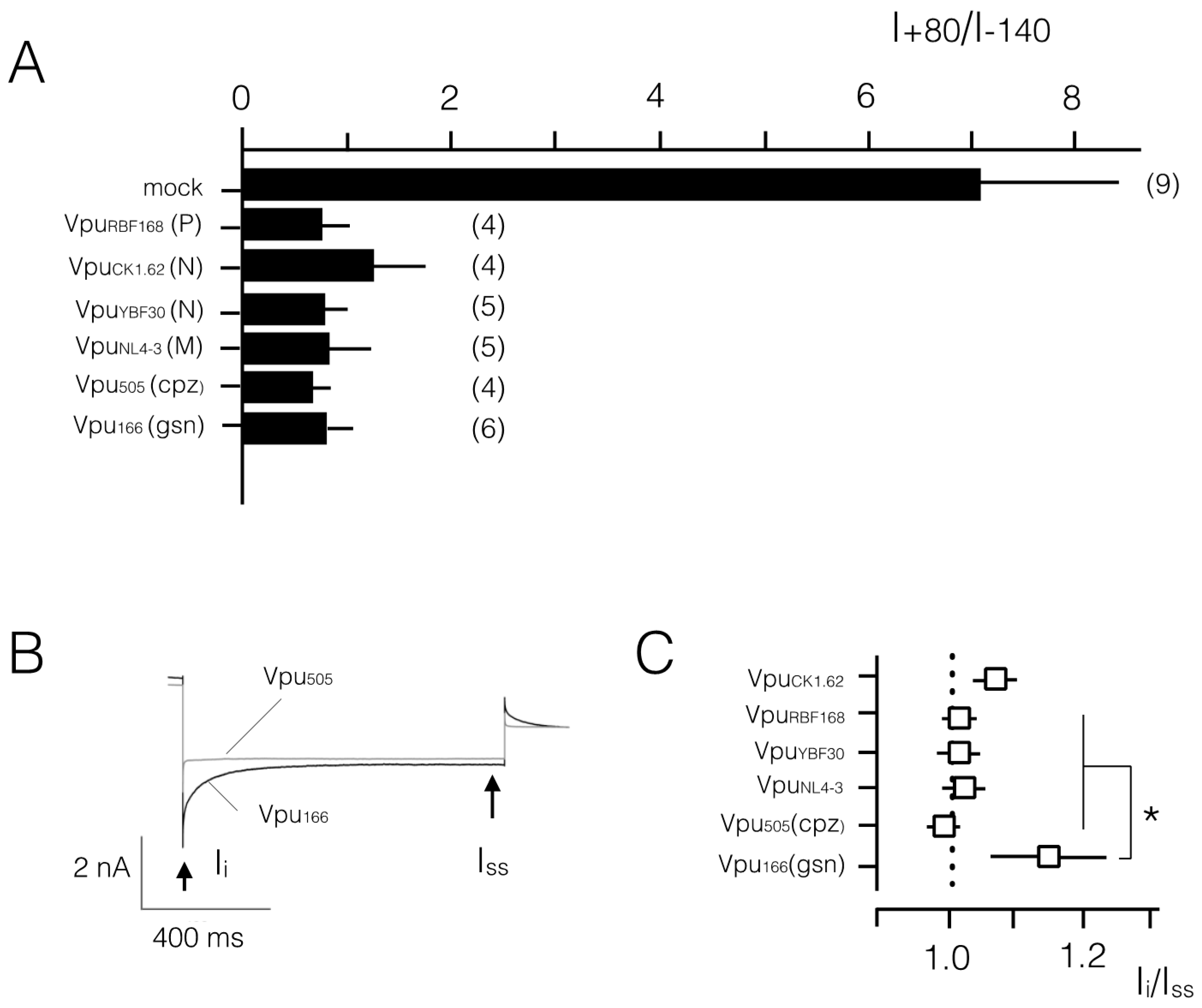
3.2. Various Vpu Proteins from Human Immunodeficiency Virus (HIV) and Simian Immunodeficiency Virus (SIV) Generate Channel Function
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cohen, E.A.; Terwilliger, E.F.; Sodroski, J.G.; Haseltine, W.A. Identification of a protein encoded by the vpu gene of HIV-1. Nature 1988, 334, 532–534. [Google Scholar] [CrossRef] [PubMed]
- Maldarelli, F.; Chen, M.Y.; Willey, R.L.; Strebel, K. Human immunodeficiency virus type 1 Vpu protein is an oligomeric type I integral membrane protein. J. Virol. 1993, 67, 5056–5061. [Google Scholar] [PubMed]
- Strebel, K.; Klimkait, T.; Martin, M.A. A novel gene of HIV-1 vpu and its 16-kilodalton product. Science 1988, 241, 1221–1223. [Google Scholar] [CrossRef] [PubMed]
- Neil, S.J.D.; Zang, T.; Bieniasz, P.D. Tetherin inhibits retrovirus release and is antagonized by HIV-1 vpu. Nature 2008, 451, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, N.; Goff, D.; Katsura, C.; Jorgenson, R.L.; Mitchell, R.; Johnson, M.C.; Stephens, E.B.; Guatelli, J. The interferon-induced protein BST-2 restricts HIV-1 release and is down regulated from the cell surface by the viral vpu protein. Cell Host Microbe 2008, 3, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Binette, J.; Bubé, M.; Mercier, J.; Halawani, D.; Latterich, M.; Cohen, E.A. Requirements for the selective degradation of CD4 receptor molecules by the human immunodeficiency virus type 1 Vpu protein in the endoplasmic reticulum. Retrovirology 2007, 4, 75. [Google Scholar] [CrossRef] [PubMed]
- Magadán, J.G.; Pérez-Victoria, F.J.; Sougrat, R.; Ye, Y.; Strebel, K.; Bonifacino, J.S. Multilayered mechanism of CD4 downregulation by HIV-1 Vpu involving distinct ER retention and ERAD targeting steps. PLoS Pathog. 2010, 6, e1000869. [Google Scholar] [CrossRef] [PubMed]
- Schubert, U.; Antón, L.C.; Bacík, I.; Cox, J.H.; Bour, S.; Bennink, J.R.; Orlowski, M.; Strebel, K.; Yewdell, J.W. CD4 glycoprotein degradation induced by human immunodeficient virus type 1 Vpu protein requires the function of proteasomes and the ubiquitin pathway. J. Virol. 1998, 72, 2280–2288. [Google Scholar] [PubMed]
- Willey, R.L.; Maldarelli, F.; Martin, M.A.; Strebel, K. Human immunodeficiency virus type 1 Vpu protein induces rapid degradation of CD4. J. Virol. 1992, 66, 7193–7200. [Google Scholar] [PubMed]
- Schubert, U.; Henklein, P.; Boldyreff, B.; Wingender, E.; Strebel, K.; Porstmann, T. The human immunodeficiency virus type 1 encoded Vpu protein is phosphorylated by casein kinase-2 (CK-2) at positions Ser52 and Ser56 within a predicted alpha–helix–turn–alpha–helix–motif. J. Mol. Biol. 1994, 236, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Friborg, J.; Ladha, A.; Gottlinger, H.; Haseltine, W.A.; Cohen, E.A. Functional analysis of the phosphorylation sites on the human immunodeficiency virus type 1 Vpu protein. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 1995, 8, 10–22. [Google Scholar] [CrossRef] [PubMed]
- Schubert, U.; Schneider, T.; Henklein, P.; Hoffmann, K.; Berthold, E.; Hauser, H.; Pauli, G.; Porstmann, T. Human-immunodeficiency-virus-type-1-encoded Vpu protein is phosphorylated by casein kinase II. Eur. J. Biochem. 1992, 204, 875–883. [Google Scholar] [CrossRef] [PubMed]
- Schubert, U.; Strebel, K. Differential activities of the human immunodeficiency virus type 1-encoded Vpu protein are regulated by phosphorylation and occur in different cellular compartments. J. Virol. 1994, 68, 2260–2271. [Google Scholar] [PubMed]
- Lin, M.-H.; Chen, C.-P.; Fischer, B. Patch formation of a viral channel forming protein within a lipid membrane—Vpu of HIV-1. Mol. BioSyst. 2016, 12, 1118–1127. [Google Scholar] [CrossRef] [PubMed]
- Ewart, G.D.; Sutherland, T.; Gage, P.W.; Cox, G.B. The Vpu protein of human immunodeficiency virus type 1 forms cation-selective ion channels. J. Virol. 1996, 70, 7108–7115. [Google Scholar] [PubMed]
- Schubert, U.; Ferrer-Montiel, A.V.; Oblatt-Montal, M.; Henklein, P.; Strebel, K.; Montal, M. Identification of an ion channel activity of the Vpu transmembrane domain and its involvement in the regulation of virus release from HIV-1-infected cells. FEBS Lett. 1996, 398, 12–18. [Google Scholar] [CrossRef]
- Bolduan, S.; Votteler, J.; Lodermeyer, V.; Greiner, T.; Koppensteiner, H.; Schindler, M.; Thiel, G.; Schubert, U. Ion channel activity of HIV-1 Vpu is dispensible for counteraction of CD317. Virology 2011, 416, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Strebel, K. HIV-1 Vpu- an ion channel in search of a job. Biochim. Biophys. Acta 2014, 1838, 1074–1081. [Google Scholar] [CrossRef] [PubMed]
- Ewart, G.D.; Nasr, N.; Naif, H.; Cox, G.B.; Cunningham, A.L.; Gage, P.W. Potential new anti-human immunodeficiency virus type 1 compounds depress virus replication in cultured human macrophages. Antimicrob. Agents Chemother. 2004, 48, 2325–2330. [Google Scholar] [CrossRef] [PubMed]
- Khoury, G.; Ewart, G.; Luscombe, C.; Miller, M.; Wilkinson, J. Antiviral efficacy of novel compound BIT225 against HIV-1 release from human macrophages. Antimicrob. Agents Chemother. 2010, 54, 835–845. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, J.; Ewart, G.; Luscombe, C.; McBride, K.; Ratanasuwan, W.; Miller, M.; Murphy, R.L. A Phase 1b/2a study of the safety, pharmacokinetics and antiviral activity of BIT225 in patients with HIV-1 infection. J. Antimicrob. Chemother. 2016, 71, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Khoury, G.; Ewart, G.; Luscombe, C.; Miller, M.; Wilkinson, J. The antiviral compound BIT225 inhibits HIV-1 replication in myeloid dendritic cells. AIDS Res. Ther. 2016, 13, 7. [Google Scholar] [CrossRef] [PubMed]
- González, M.E. Vpu protein: The viroporin encoded by HIV-1. Viruses 2015, 7, 4352–4368. [Google Scholar] [CrossRef] [PubMed]
- Kirchhoff, F. Is the high virulence of HIV-1 an unfortunate coincidence of primate lentiviral evolution? Nat. Rev. Microbiol. 2009, 7, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Sauter, D.; Schindler, M.; Specht, A.; Landford, W.N.; Münch, J.; Kim, K.A.; Votteler, J.; Schubert, U.; Bibollet-Ruche, F.; Keele, B.F.; et al. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 2009, 6, 409–421. [Google Scholar] [CrossRef] [PubMed]
- Sauter, D.; Hue, S.; Petit, S.J.; Plantier, J.C.; Toers, G.J.; Kirchhoff, F.; Gupta, R.K. HIV-1 Group P is unable to antagonize human Tetherin by Vpu Env or Nef. Retrovirology 2011, 8, 103. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acid Res. 2013, 42, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Hoffgaard, F.; Weil, P.; Hamacher, K. BioPhysConnectoR: Connecting sequence information and biophysical models. BMC Bioinform. 2010, 11, 199. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. Clustal, W. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting position-specific gap penalties and weight matrix choice. Nucleic Acid Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Warterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Hamill, O.; Marty, A.; Neher, E.; Sakmann, B.; Sigworth, F. Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflügers Arch. 1981, 391, 85–100. [Google Scholar] [CrossRef] [PubMed]
- Adachi, A.; Gendelman, H.E.; Koenig, S.; Folks, T.; Willey, R.; Rabson, A.; Martin, M.A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 1986, 59, 284–291. [Google Scholar] [PubMed]
- Marassi, F.M.; Ma, C.; Gratkowski, H.; Straus, S.K.; Strebel, K.; Oblatt-Montal, M.; Montal, M.; Opella, S.J. Correlation of the structural and functional domains in the membrane protein Vpu from HIV-1. Proc. Natl. Acad. Sci. USA 1999, 96, 14336–14341. [Google Scholar] [CrossRef] [PubMed]
- Mehnert, T.; Lam, Y.H.; Judge, P.J.; Routh, A.; Fischer, D.; Watts, A.; Fischer, W. Towards a mechanism of function of the viral ion channel Vpu from HIV-1. J. Biomol. Struct. Dyn. 2007, 24, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.; Smart, T.G. HEK293 cell line: A vehicle for the expression of recombinant proteins. J. Pharmacol. Toxicol. Methods 2005, 51, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Varghese, A.; TenBroek, E.M.; Coles, J.; Sigg, D.C. Endogenous channels in HEK cells and potential roles in HCN ionic current measurements. Prog. Biophys. Mol. Biol. 2006, 90, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Hsu, K.; Seharaseyon, J.; Dong, P.; Bour, S.; Marban, E. Mutualfunctionaldestruction of HIV-1 Vpu and host TASK-1 channel. Mol. Cell 2004, 14, 259–267. [Google Scholar] [CrossRef]
- Schewe, M.; Nematian-Ardestani, E.; Sun, H.; Musinszki, M.; Cordeiro, S.; Bucci, G.; de Groot, B.L.; Tucker, S.J.; Rapedius, M.; Baukrowitz, T. A non-canonical voltage-sensing mechanism controls gating in K2P K+ channels. Cell 2016, 164, 937–949. [Google Scholar] [CrossRef] [PubMed]
- Sauter, D.; Schwarz, S.; Wang, K.; Zhang, R.; Sun, B.; Schwarz, W. Genistein as antiviral drug against HIV ion channel. Planta Med. 2014, 80, 682–687. [Google Scholar] [CrossRef] [PubMed]
- Weber, W.M. Endogenous ion channels in oocytes of Xenopus laevis: Recent developments. J. Membr. Biol. 1999, 170, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gifford, R.J.; Katzourakis, A.; Tristem, M.; Pybus, O.G.; Winters, M.; Shafer, R.W. A translational endogenous lentivirus from the genome of a basal primate and implications for lentivirus evolution. Proc. Natl. Acad. Sci. USA 2008, 105, 20362–20367. [Google Scholar] [CrossRef] [PubMed]
| Position | 17 | 19 | 20 | 22 | 23 | 25 | |||||||||||||
| HIV | I | A | I | V | W | I | |||||||||||||
| 94.8 | 94.4 | 97.3 | 97.9 | 99.6 | 93.5 | ||||||||||||||
| SIV | I | L | A | V | A, N | T | I | V | V | I | A | W | K | I | |||||
| 36.4 | 22.7 | 22.7 | 27.3 | 22.7 | 18.2 | 63.6 | 27.3 | 40.9 | 27.3 | 18.2 | 95.5 | 40.9 | 27.3 | ||||||
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Greiner, T.; Bolduan, S.; Hertel, B.; Groß, C.; Hamacher, K.; Schubert, U.; Moroni, A.; Thiel, G. Ion Channel Activity of Vpu Proteins Is Conserved throughout Evolution of HIV-1 and SIV. Viruses 2016, 8, 325. https://doi.org/10.3390/v8120325
Greiner T, Bolduan S, Hertel B, Groß C, Hamacher K, Schubert U, Moroni A, Thiel G. Ion Channel Activity of Vpu Proteins Is Conserved throughout Evolution of HIV-1 and SIV. Viruses. 2016; 8(12):325. https://doi.org/10.3390/v8120325
Chicago/Turabian StyleGreiner, Timo, Sebastian Bolduan, Brigitte Hertel, Christine Groß, Kay Hamacher, Ulrich Schubert, Anna Moroni, and Gerhard Thiel. 2016. "Ion Channel Activity of Vpu Proteins Is Conserved throughout Evolution of HIV-1 and SIV" Viruses 8, no. 12: 325. https://doi.org/10.3390/v8120325