Transspecies Transmission of Gammaretroviruses and the Origin of the Gibbon Ape Leukaemia Virus (GaLV) and the Koala Retrovirus (KoRV)
Abstract
:1. Introduction
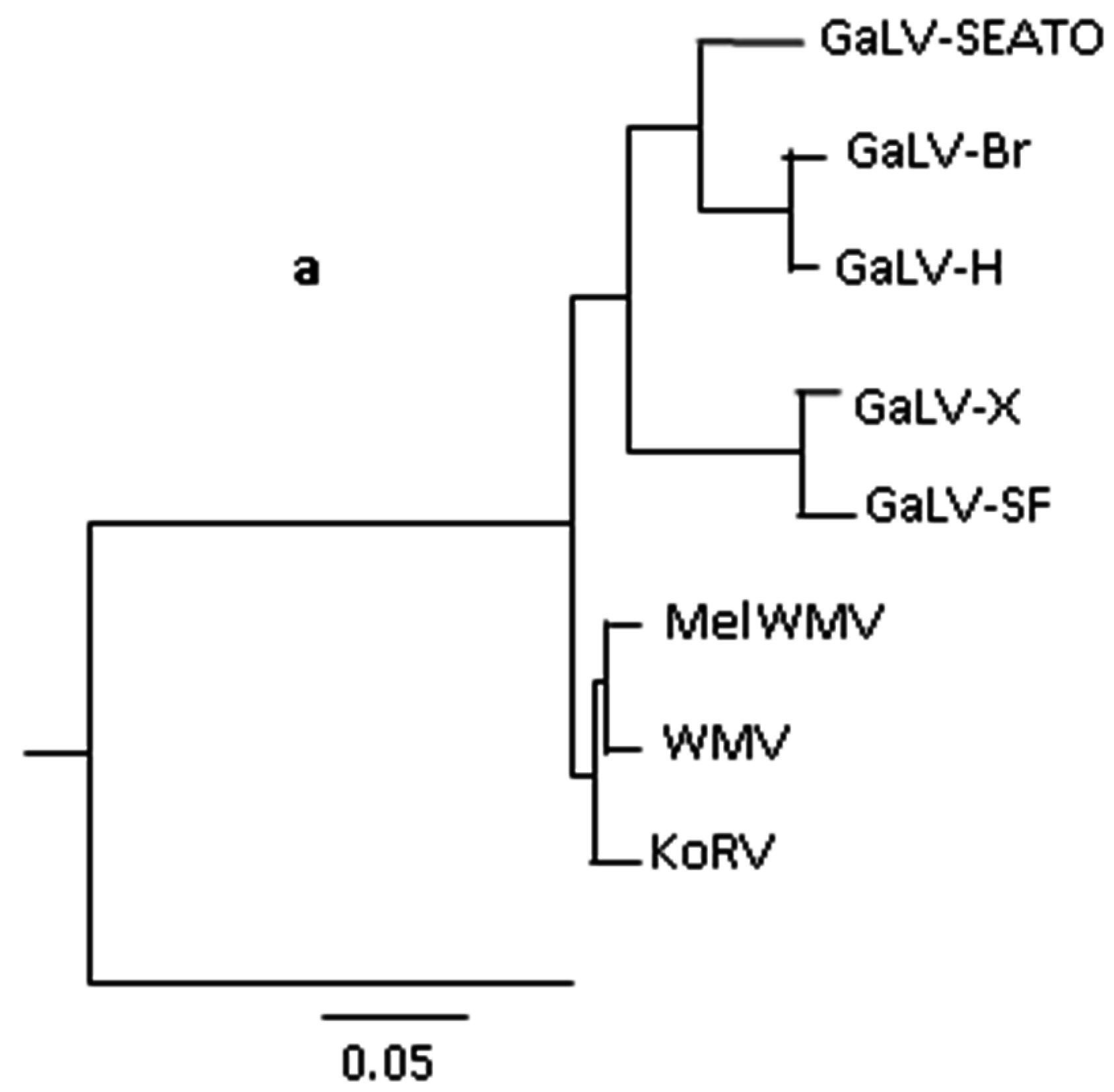
2. The Gibbon Ape Leukaemia Virus (GaLV)
3. The Koala Retrovirus (KoRV)
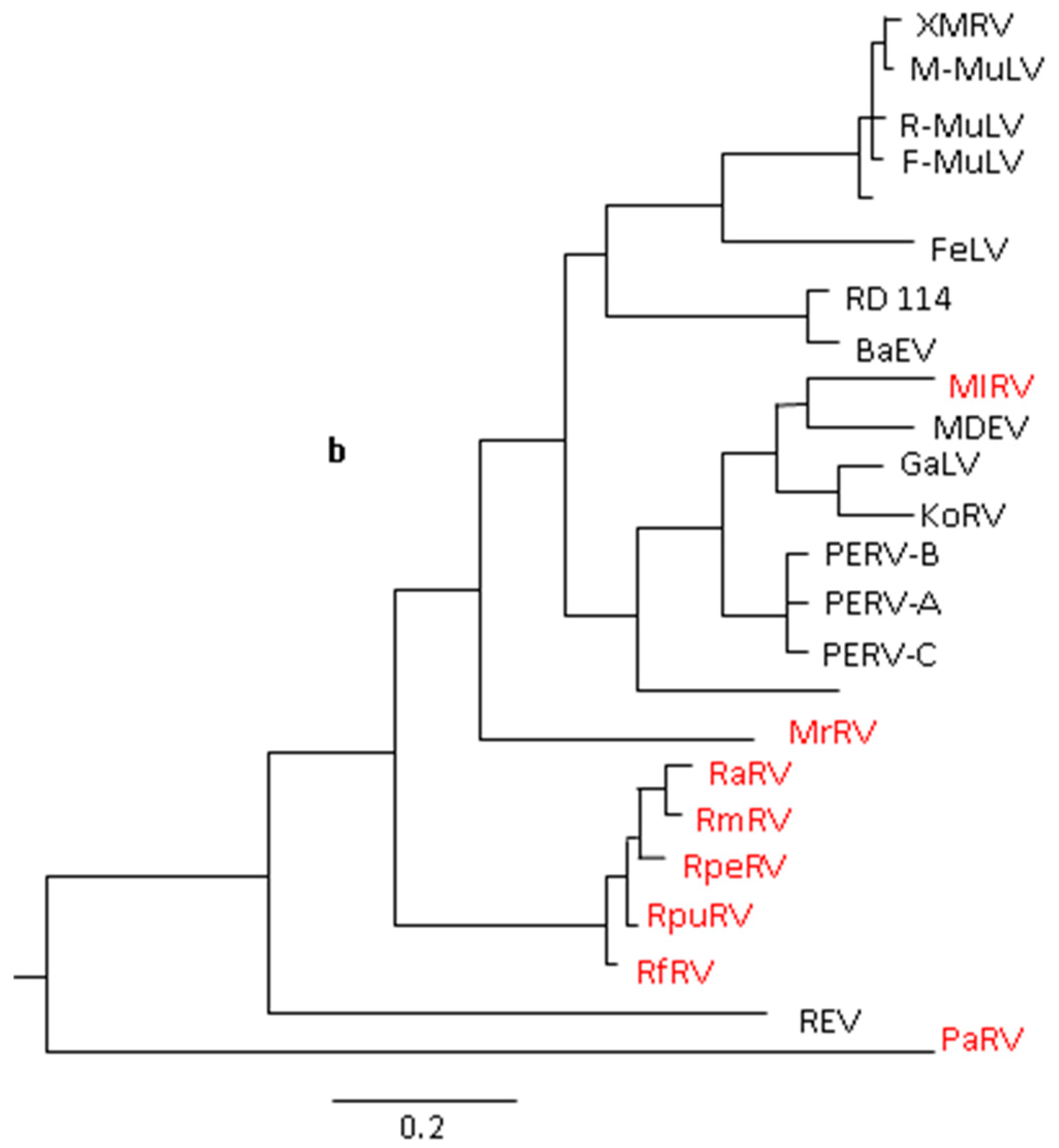
4. Related Rodent Viruses
5. Related Bat Retroviruses and Their Possible Transmission
6. Open Questions
7. Gammaretroviruses and Xenotransplantation
8. Conclusions
Acknowledgments
Conflicts of Interest
References
- Peeters, M.; Jung, M.; Ayouba, A. The origin and molecular epidemiology of HIV. Expert Rev. Anti-Infect. Ther. 2013, 11, 885–896. [Google Scholar] [CrossRef] [PubMed]
- Denner, J. Transspecies transmissions of retroviruses: New cases. Virology 2007, 369, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Beneventiste, R.E.; Todaro, G.J. Evolution of type C viral genes preservation of ancestral murine type C viral sequences in pig cellular DNA. Proc. Nat. Acad. Sci. USA 1975, 72, 4090–4094. [Google Scholar] [CrossRef]
- Tönjes, R.R.; Niebert, M. Relative age of proviral porcine endogenous retrovirus sequences in Sus scrofa based on the molecular clock hypothesis. J. Virol. 2003, 77, 12363–12368. [Google Scholar] [CrossRef] [PubMed]
- Delassus, S.; Sonigo, P.; Wain-Hobson, S. Genetic organization of gibbon ape leukemia virus. Virology 1989, 173, 205–213. [Google Scholar] [CrossRef]
- Denner, J.; Young, P.R. Koala retroviruses: Characterization and impact on the life of koalas. Retrovirology 2013, 10, 108. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, L.H.; Hishes, S.; Suyanto, A.; Newbound, C.N.; Kitchener, D.J.; How, R.A. Crossing the line: The impact of contemporary and historical sea barriers on the population structure of bats in Southern Wallacea. In Island Bats: Evolution, Ecology, and Conservation; Flemming, T.H., Racey, P.A., Eds.; The University of Chicago Press: Chicago, IL, USA, 2009; pp. 59–96. [Google Scholar]
- Ferguson, B.; Smith, D.G. Evolutionary History and Genetic Variation of Macaca mulatta and Macaca fascicularis. In The Nonhuman Primate in Nonclinical Drug Development and Safety Assessment; Bluemel, J., Korte, S., Schenck, E., Weinbauer, G.F., Eds.; Academic Press: Cambridge, MA, USA, 2015; pp. 17–35. [Google Scholar]
- Todaro, G.J.; Benveniste, R.E.; Sherr, C.J.; Lieber, M.M.; Callahan, R. Infectious primate type C virus group: Evidence for an origin from an endogenous virus of the rodent, Mus caroli. Bibl. Haematol. 1975, 43, 115–120. [Google Scholar]
- Lieber, M.M.; Sherr, C.J.; Todaro, G.J.; Benveniste, R.E.; Callahan, R.; Coon, H.G. Isolation from the asian mouse Mus caroli of an endogenous type C virus related to infectious primate type C viruses. Proc. Natl. Acad. Sci. USA 1975, 72, 2315–2319. [Google Scholar] [CrossRef] [PubMed]
- Wolgamot, G.; Miller, A.D. Replication of Mus dunni endogenous retrovirus depends on promoter activation followed by enhancer multimerization. J. Virol. 1999, 73, 9803–9809. [Google Scholar] [PubMed]
- Simmons, G.; Clarke, D.; McKee, J.; Young, P.; Meers, J. Discovery of a novel retrovirus sequence in an Australian native rodent (Melomys burtoni): A putative link between gibbon ape leukemia virus and koala retrovirus. PLoS ONE 2014, 9, e106954. [Google Scholar] [CrossRef] [PubMed]
- Alfano, N.; Michaux, J.; Morand, S.; Aplin, K.; Tsangaras, K.; Löber, U.; Fabre, P.H.; Fitriana, Y.; Semiadi, G.; Ishida, Y.; et al. Greenwood AD9. Endogenous Gibbon Ape Leukemia Virus Identified in a Rodent (Melomys burtoni subsp.) from Wallacea (Indonesia). J. Virol. 2016, 90, 8169–8180. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Tachedjian, G.; Tachedjian, M.; Holmes, E.C.; Zhang, S.; Wang, L.F. Identification of diverse groups of endogenous gammaretroviruses in mega- and microbats. J. Gen. Virol. 2012, 93, 2037–2045. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Tachedjian, M.; Wang, L.; Tachedjian, G.; Wang, L.F.; Zhang, S. Discovery of retroviral homologs in bats: Implications for the origin of mammalian gammaretroviruses. J. Virol. 2012, 86, 4288–4293. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, T.G.; Kollias, G.V., Jr.; Holmberg, C. Oncogenicity of gibbon type-C myelogenous leukemia virus. Int. J. Cancer 1980, 25, 641–646. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, T.G.; Huff, S.D.; Buckley, P.M.; Dungworth, D.L.; Synder, S.P.; Gilden, R.V. C-type virus associated with gibbon lymphosarcoma. Nat. New Biol. 1972, 235, 170–171. [Google Scholar] [CrossRef] [PubMed]
- Snyder, S.P.; Dungworth, D.L.; Kawakami, T.G.; Callaway, E.; Lau, D.T. Lymphosarcomas in two gibbons (Hylobates lar) with associated C-type virus. J. Natl. Cancer Inst. 1973, 51, 89–94. [Google Scholar] [PubMed]
- Gallo, R.C.; Gallagher, R.E.; Wong-Staal, F.; Aoki, T.; Markham, P.D.; Schetters, H.; Ruscetti, F.; Valerio, M.; Walling, M.J.; O’Keeffe, R.T.; et al. Isolation and tissue distribution of type-C virus and viral components from a gibbon ape (Hylobates lar) with lymphocytic leukemia. Virology 1978, 84, 359–373. [Google Scholar] [CrossRef]
- Reitz, M.S., Jr.; Wong-Staal, F.; Haseltine, W.A.; Kleid, D.G.; Trainor, C.D.; Gallagher, R.E.; Gallo, R.C. Gibbon ape leukemia virus-Hall’s Island: New strain of gibbon ape leukemia virus. J. Virol. 1979, 29, 395–400. [Google Scholar] [PubMed]
- Todaro, G.J.; Lieber, M.M.; Benveniste, R.E.; Sherr, C.J. Infectious primate type C viruses: Three isolates belonging to a new subgroup from the brains of normal gibbons. Virology 1975, 67, 335–343. [Google Scholar] [CrossRef]
- Brown, J.; Tarlinton, E. Is gibbon ape leukaemia virus still a threat? Mamm. Rev. 2016, 47, 53–61. [Google Scholar] [CrossRef]
- Theilen, G.H.; Gould, D.; Fowler, M.; Dungworth, D.L. C-type virus in tumor tissue of a woolly monkey (Lagothrix spp.) with fibrosarcoma. J. Natl. Cancer Inst. 1971, 47, 881–889. [Google Scholar] [PubMed]
- Wolfe, L.G.; Smith, R.K.; Deinhardt, F. Simian sarcoma virus, type 1 (Lagothrix): Focus assay and demonstration of nontransforming associated virus. J. Natl. Cancer Inst. 1972, 48, 1905–1908. [Google Scholar] [PubMed]
- Hino, S.; Stephenson, J.R.; Aaronson, S.A. Antigenic determinants of the 70,000 molecular weight glycoprotein of woolly monkey type C RNA virus. J. Immunol. 1975, 115, 922–927. [Google Scholar] [PubMed]
- Rangan, S.R. Antigenic relatedness of simian C-type viruses. Int. J. Cancer 1974, 13, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Krakower, J.M.; Tronick, S.R.; Gallagher, R.E.; Gallo, R.C.; Aaronson, S.A. Antigenic characterization of a new gibbon ape leukemia virus isolate: Seroepidemiologic assessment of an outbreak of gibbon leukemia. Int. J. Cancer 1978, 22, 715–720. [Google Scholar] [CrossRef] [PubMed]
- Burtonboy, G.; Delferriere, N.; Mousset, B.; Heusterspreute, M. Isolation of a C-type retrovirus from an HIV infected cell line. Arch. Virol. 1933, 130, 289–300. [Google Scholar] [CrossRef]
- Parent, I.; Qin, Y.; Vandenbroucke, A.T.; Walon, C.; Delferriere, N.; Godfroid, E.; Burtonboy, G. Characterization of a C-type retrovirus isolated from an HIV infected cell line: Complete nucleotide sequence. Arch. Virol. 1998, 143, 1077–1092. [Google Scholar] [CrossRef] [PubMed]
- Arias, M.; Fan, H. The saga of XMRV: A virus that infects human cells but is not a human virus. Emerg. Microbes. Infect. 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Siegal-Willott, J.L.; Jensen, N.; Kimi, D.; Taliaferro, D.; Blankenship, T.; Malinsky, B.; Murray, S.; Eiden, M.V.; Xu, W. Evaluation of captive gibbons (Hylobates spp., Nomascus spp., Symphalangus spp.) in North American Zoological Institutions for Gibbon Ape Leukemia Virus (GALV). J. Zoo Wildl. Med. 2015, 46, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Kinney, M.E.; Pye, G.W. Koala retrovirus: A review. J. Zoo Wildl. Med. 2016, 47, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Tarlinton, R.; Meers, J.; Young, P. Biology and evolution of the endogenous koala retrovirus. Cell Mol. Life Sci. 2008, 65, 3413–3421. [Google Scholar] [CrossRef] [PubMed]
- Canfield, P.J.; Sabine, J.M.; Love, D.N. Virus particles associated with leukaemia in a koala. Aust. Vet. J. 1988, 65, 327–328. [Google Scholar] [CrossRef] [PubMed]
- Hanger, J.J.; Bromham, L.D.; McKee, J.J.; O’Brien, T.M.; Robinson, W.F. The nucleotide sequence of koala (Phascolarctos cinereus) retrovirus: A novel type C endogenous virus related to gibbon ape leukemia virus. J. Virol. 2000, 74, 4264–4272. [Google Scholar] [CrossRef] [PubMed]
- Tarlinton, R.E.; Meers, J.; Young, P.R. Retroviral invasion of the koala genome. Nature 2006, 442, 79–81. [Google Scholar] [CrossRef] [PubMed]
- Miyazawa, T.; Shojima, T.; Yoshikawa, R.; Ohata, T. Isolation of koala retroviruses from koalas in Japan. J. Vet. Med. Sci. 2011, 73, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Shojima, T.; Hoshino, S.; Abe, M.; Yasuda, J.; Shogen, H.; Kobayashi, T.; Miyazawa, T. Construction and characterization of an infectious molecular clone of koala retrovirus. J. Virol. 2013, 7, 5081–5088. [Google Scholar] [CrossRef] [PubMed]
- Fiebig, U.; Hartmann, M.G.; Bannert, N.; Kurth, R.; Denner, J. Transspecies transmission of the endogenous koala retrovirus. J. Virol. 2006, 80, 5651–5654. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Stadler, C.K.; Gorman, K.; Jensen, N.; Kim, D.; Zheng, H.; Tang, S.; Switzer, W.M.; Pye, G.W.; Eiden, M.V. An exogenous retrovirus isolated from koalas with malignant neoplasias in a US zoo. Proc. Natl. Acad. Sci. USA 2013, 110, 11547–11552. [Google Scholar] [CrossRef] [PubMed]
- Shojima, T.; Yoshikawa, R.; Hoshino, S.; Shimode, S.; Nakagawa, S.; Ohata, T.; Nakaoka, R.; Miyazawa, T. Identification of a novel subgroup of koala retrovirus from Koalas in Japanese zoos. J. Virol. 2013, 87, 9943–9948. [Google Scholar] [CrossRef] [PubMed]
- Fiebig, U.; Keller, M.; Denner, J. Detection of koala retrovirus subgroup B (KoRV-B) in animals housed at European zoos. Arch. Virol. 2016, 161, 3549–3553. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, M.; Pavasovic, A.; King, A.G.; Prentis, P.J.; Eldridge, M.D.B.; Chen, Z.; Colgan, D.J.; Polkinghorne, A.; Wilkins, M.R.; Flanagan, C. A transcriptome resource for the koala (Phascolarctos cinereus): Insights into koala retrovirus transcription and sequence diversity. BMC Genom. 2014, 15, 786. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Gorman, K.; Santiago, J.C.; Kluska, K.; Eiden, M.V. Genetic diversity of koala retroviral envelopes. Viruses 2015, 7, 1258–1270. [Google Scholar] [CrossRef] [PubMed]
- Denner, J. The transmembrane proteins contribute to immunodeficiencies induced by HIV-1 and other retroviruses. AIDS 2014, 28, 1081–1090. [Google Scholar] [CrossRef] [PubMed]
- Polkinghorne, A.; Hanger, J.; Timms, P. Recent advances in understanding the biology, epidemiology and control of chlamydial infections in koalas. Vet. Microbiol. 2013, 165, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Sparkes, A.H. Feline leukaemia virus and vaccination. J. Feline Med. Surg. 2003, 5, 97–100. [Google Scholar] [CrossRef]
- Fiebig, U.; Dieckhoff, B.; Wurzbacher, C.; Möller, A.; Kurth, R.; Denner, J. Induction of neutralizing antibodies specific for the envelope proteins of the koala retrovirus by immunization with recombinant proteins or with DNA. Virol. J. 2015, 12, 68. [Google Scholar] [CrossRef] [PubMed]
- Fiebig, U.; Keller, M.; Möller, A.; Timms, P.; Denner, J. Lack of antiviral antibody response in koalas infected with koala retroviruses (KoRV). Virus Res. 2015, 198, 30–34. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.D.; Bergholz, U.; Ziegler, M.; Stocking, C. Identification of the myelin protein plasmolipin as the cell entry receptor for Mus caroli endogenous retrovirus. J. Virol. 1998, 72, 7459–7466. [Google Scholar] [CrossRef] [PubMed]
- Callahan, R.; Meade, C.; Todaro, G.J. Isolation of an endogenous type C virus related to the infectious primate type C viruses from the Asian rodent Vandeleuria oleracea. J. Virol. 1979, 30, 124–131. [Google Scholar] [PubMed]
- Benveniste, R.E.; Callahan, R.; Sherr, C.J.; Chapman, V.; Todaro, G.J. Two distinct endogenous type C viruses isolated from the Asian rodent Mus cervicolor: Conservation of virogene sequences in related rodent species. J. Virol. 1977, 21, 849–862. [Google Scholar] [PubMed]
- Wolgamot, G.; Bonham, L.; Miller, A.D. Sequence analysis of Mus dunni endogenous virus reveals a hybrid VL30/gibbon ape leukemia virus-like structure and a distinct envelope. J. Virol. 1998, 72, 7459–7466. [Google Scholar] [PubMed]
- Bromham, L.; Clark, F.; McKee, J.J. Discovery of a novel murine type C retrovirus by data mining. J. Virol. 2001, 75, 3053–3057. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.; Lau, S.; Woo, P.; Yuen, K.Y. Bats as a continuing source of emerging infections in humans. Rev. Med. Virol. 2007, 17, 67–91. [Google Scholar] [CrossRef] [PubMed]
- Calisher, C.H.; Childs, J.E.; Field, H.E.; Holmes, K.V.; Schountz, T. Bats: Important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 2006, 19, 531–545. [Google Scholar] [CrossRef] [PubMed]
- Hayward, J.A.; Tachedjian, M.; Cui, J.; Field, H.; Holmes, E.C.; Wang, L.F.; Tachedjian, G. Identification of diverse full-length endogenous betaretroviruses in megabats and microbats. Retrovirology 2013, 10, 35. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Tachedjian, G.; Wang, L.F. Bats and Rodents Shape Mammalian Retroviral Phylogeny. Sci. Rep. 2015, 5, 16561. [Google Scholar] [CrossRef] [PubMed]
- Alfano, N.; Kolokotronis, S.O.; Tsangaras, K.; Roca, A.L.; Xu, W.; Eiden, M.V.; Greenwood, A.D. Episodic Diversifying Selection Shaped the Genomes of Gibbon Ape Leukemia Virus and Related Gammaretroviruses. J. Virol. 2015, 90, 1757–1772. [Google Scholar] [CrossRef] [PubMed]
- Ávila-Arcos, M.C.; Ho, S.Y.; Ishida, Y.; Nikolaidis, N.; Tsangaras, K.; Hönig, K.; Medina, R.; Rasmussen, M.; Fordyce, S.L.; Calvignac-Spencer, S.; et al. One hundred twenty years of koala retrovirus evolution determined from museum skins. Mol. Biol. Evol. 2013, 30, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.K.; Martin, R. The Koala: A Natural History; New South Wales University Press: Kensington, Australia, 1988. [Google Scholar]
- Ekser, B.; Cooper, D.K.; Tector, A.J. The need for xenotransplantation as a source of organs and cells for clinical transplantation. Int. J. Surg. 2015, 23, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Denner, J.; Tönjes, R.R. Infection barriers to successful xenotransplantation focusing on porcine endogenous retroviruses. Clin. Microbiol. Rev. 2012, 25, 318–343. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, N.M.; Farrell, K.B.; Eiden, M.V. In vitro characterization of a koala retrovirus. J. Virol. 2006, 80, 3104–3107. [Google Scholar] [CrossRef] [PubMed]
- Sarma, P.S.; Huebner, R.J.; Basker, J.F.; Vernon, L.; Gilden, R.V. Feline leukemia and sarcoma viruses: Susceptibility of human cells to infection. Science 1970, 168, 1098–1100. [Google Scholar] [CrossRef] [PubMed]
- Butera, S.T.; Brown, J.; Callahan, M.E.; Owen, S.M.; Matthews, A.L.; Weigner, D.D.; Chapman, L.E.; Sandstrom, P.A. Survey of veterinary conference attendees for evidence of zoonotic infection by feline retroviruses. J. Am. Vet. Med. Assoc. 2000, 217, 1475–1479. [Google Scholar] [CrossRef] [PubMed]
- Wynyard, S.; Nathu, D.; Garkavenko, O.; Denner, J.; Elliott, R. Microbiological safety of the first clinical pig islet xenotransplantation trial in New Zealand. Xenotransplantation 2014, 21, 309–323. [Google Scholar] [CrossRef] [PubMed]
- Morozov, V.A.; Wynyard, S.; Matsumoto, S.; Abalovich, A.; Denner, J.; Elliott, R. No PERV transmission during a clinical trial of pig islet cell transplantation. Virus Res. 2016, 227, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Semaan, M.; Ivanusic, D.; Denner, J. Cytotoxic Effects during Knock Out of Multiple Porcine Endogenous Retrovirus (PERV) Sequences in the Pig Genome by Zinc Finger Nucleases (ZFN). PLoS ONE 2015, 10, e0122059. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Güell, M.; Niu, D.; George, H.; Lesha, E.; Grishin, D.; Aach, J.; Shrock, E.; Xu, W.; Poci, J.; et al. Genome-wide inactivation of porcine endogenous retroviruses (PERVs). Science 2015, 350, 1101–1104. [Google Scholar] [CrossRef] [PubMed]
- Denner, J. Elimination of porcine endogenous retroviruses from pig cells. Xenotransplantation 2015, 22, 411–412. [Google Scholar] [CrossRef] [PubMed]
| Viruses | Receptor Usage | Distribution | Reference |
|---|---|---|---|
| KoRV-A | Pit-1 | Australia | [34,35] |
| Japanese Zoos | [37,38] | ||
| German Zoos | [39] | ||
| San Diego Zoos | [40] | ||
| KoRV-B | THTR1 | Los Angeles Zoos | [40] |
| Japanese Zoos | [41] | ||
| European Zoos | [42] | ||
| Australian wild-living koalas | [43] | ||
| GaLV | Pit-1 | Gibbons; various locations, cell culture contaminations | [16,17,18,19,20,21,22,23,24,25,26,27,28,29] |
© 2016 by the author; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Denner, J. Transspecies Transmission of Gammaretroviruses and the Origin of the Gibbon Ape Leukaemia Virus (GaLV) and the Koala Retrovirus (KoRV). Viruses 2016, 8, 336. https://doi.org/10.3390/v8120336
Denner J. Transspecies Transmission of Gammaretroviruses and the Origin of the Gibbon Ape Leukaemia Virus (GaLV) and the Koala Retrovirus (KoRV). Viruses. 2016; 8(12):336. https://doi.org/10.3390/v8120336
Chicago/Turabian StyleDenner, Joachim. 2016. "Transspecies Transmission of Gammaretroviruses and the Origin of the Gibbon Ape Leukaemia Virus (GaLV) and the Koala Retrovirus (KoRV)" Viruses 8, no. 12: 336. https://doi.org/10.3390/v8120336






