The IMPORTance of the Nucleus during Flavivirus Replication
Abstract
:1. Introduction
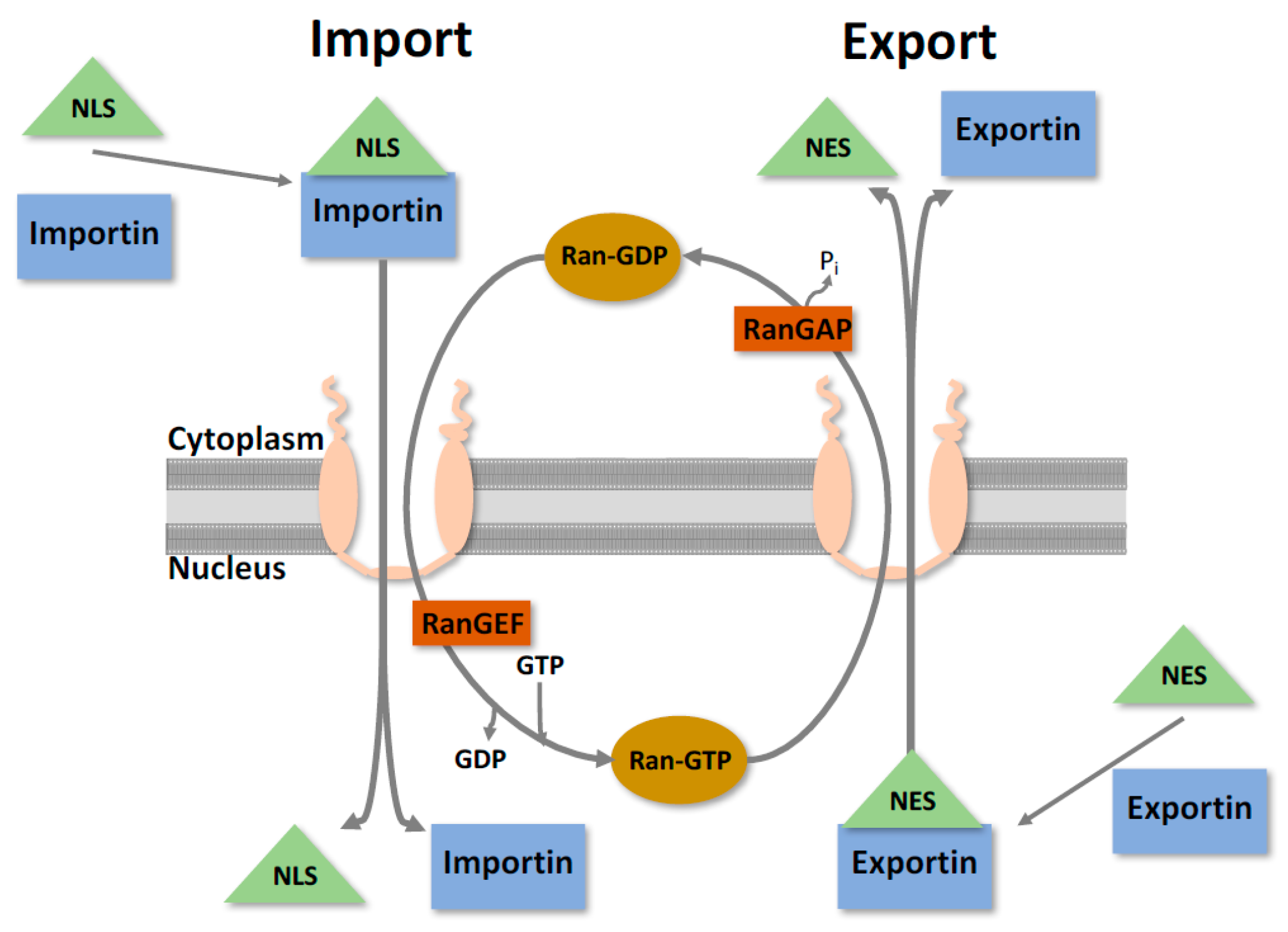
2. Nuclear Trafficking
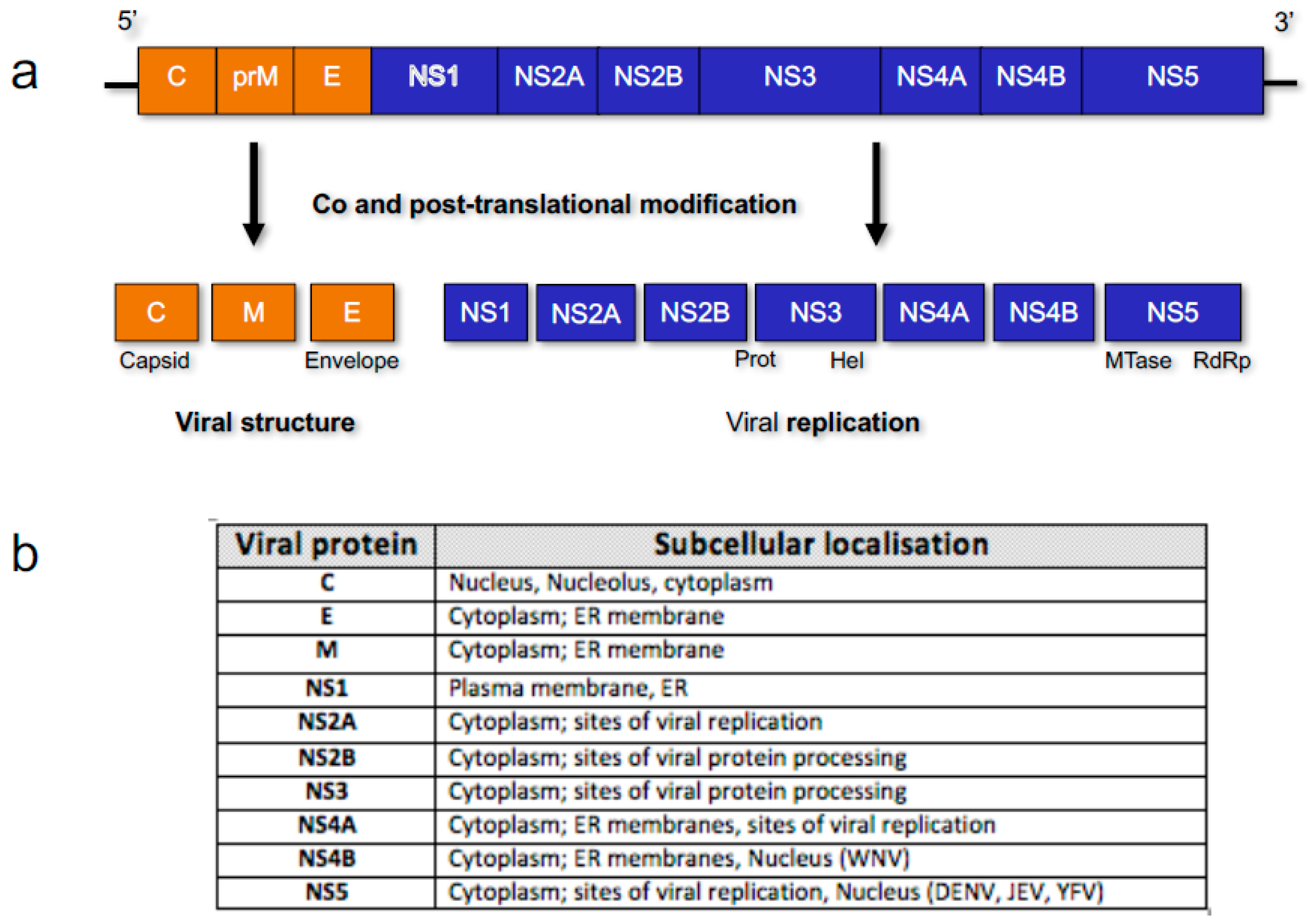
3. Flavivirus Protein Nuclear Localisation
3.1. Core/Capsid
3.2. Non-Structural (NS) Protein 5 (NS5)
3.3. NS Proteins NS3 and NS4B and RNA Synthesis
4. Sequestration of Host Nuclear Components
5. Future Considerations
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chambers, T.J.; Hahn, C.S.; Galler, R.; Rice, C.M. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 1990, 44, 649–688. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, J. Wrapping Things up about Virus RNA Replication. Traffic 2005, 6, 967–977. [Google Scholar] [CrossRef] [PubMed]
- Westaway, E.G.; Mackenzie, J.M.; Khromykh, A.A. Replication and gene function in Kunjin virus. Curr. Top. Microbiol. Immunol. 2002, 267, 323–351. [Google Scholar] [PubMed]
- Leary, K.; Blair, C.D. Sequential events in the morphogenesis of Japanese encephalitis virus. J. Ultrastruct. Res. 1980, 72, 123–129. [Google Scholar] [CrossRef]
- Gillespie, L.K.; Hoenen, A.; Morgan, G.; Mackenzie, J.M. The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. J. Virol. 2010, 84, 10438–10447. [Google Scholar] [CrossRef] [PubMed]
- Uchil, P.D.; Satchidanandam, V. Architecture of the flaviviral replication complex: Protease, nuclease, and detergents reveal encasement within double-layered membrane compartments. J. Biol. Chem. 2003, 278, 24388–24398. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, J.M.; Jones, M.K.; Westaway, E.G. Markers for trans-Golgi membranes and the intermediate compartment localize to induced membranes with distinct replication functions in flavivirus-infected cells. J. Virol. 1999, 73, 9555–9567. [Google Scholar] [PubMed]
- Welsch, S.; Miller, S.; Romero-Brey, I.; Merz, A.; Bleck, C.K.E.; Walther, P.; Fuller, S.D.; Antony, C.; Krijnse-Locker, J.; Bartenschlager, R. Composition and Three-Dimensional Architecture of the Dengue Virus Replication and Assembly Sites. Cell Host Microbe 2009, 5, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Wente, S.R.; Rout, M.P. The nuclear pore complex and nuclear transport. Cold Spring Harb. Perspect. Biol. 2010, 10, a000562. [Google Scholar] [CrossRef] [PubMed]
- Grossman, E.; Medalia, O.; Zwerger, M. Functional architecture of the nuclear pore complex. Annu. Rev. Biophys. 2012, 41, 557–584. [Google Scholar] [CrossRef] [PubMed]
- Hinshaw, J.E.; Carragher, B.O.; Milligan, R.A. Architecture and design of the nuclear pore complex. Cell 1992, 69, 1133–1141. [Google Scholar] [CrossRef]
- Dingwall, C.; Robbins, J.; Dilworth, S.M.; Roberts, B.; Richardson, W.D. The nucleoplasmin nuclear location sequence is larger and more complex than that of SV-40 large T antigen. J. Cell Biol. 1988, 107, 841–849. [Google Scholar] [CrossRef] [PubMed]
- Goldfarb, D.S.; Gariepy, J.; Schoolnik, G.; Kornberg, R.D. Synthetic peptides as nuclear localization signals. Nature 1986, 322, 641–644. [Google Scholar] [CrossRef] [PubMed]
- Dingwall, C.; Laskey, R.A. Nuclear import: A tale of two sites. Curr. Biol. 1998, 8, R922–R924. [Google Scholar] [CrossRef]
- Pemberton, L.F.; Paschal, B.M. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic 2005, 6, 187–198. [Google Scholar]
- Yuh, M.C.; Blobel, G. Karyopherins and nuclear import. Curr. Opin. Struct. Biol. 2001, 11, 703–715. [Google Scholar]
- Macara, I.G. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 2001, 65, 570–594. [Google Scholar] [CrossRef] [PubMed]
- Westaway, E.G.; Khromykh, A.A.; Kenney, M.T.; Mackenzie, J.M.; Jones, M.K. Proteins C and NS4B of the flavivirus Kunjin translocate independently into the nucleus. Virology 1997, 234, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Bulich, R.; Aaskov, J.G. Nuclear localization of dengue 2 virus core protein detected with monoclonal antibodies. J. Gen. Virol. 1992, 73, 2999–3003. [Google Scholar] [CrossRef] [PubMed]
- Mori, Y.; Okabayashi, T.; Yamashita, T.; Zhao, Z.; Wakita, T.; Yasui, K.; Hasebe, F.; Tadano, M.; Konishi, E.; Moriishi, K.; et al. Nuclear Localization of Japanese Encephalitis Virus Core Protein Enhances Viral Replication. J. Virol. 2005, 79, 3448–3458. [Google Scholar] [CrossRef] [PubMed]
- Dokland, T.; Walsh, M.; Mackenzie, J.M.; Khromykh, A.A.; Ee, K.H.; Wang, S. West Nile Virus Core Protein. Structure 2004, 12, 1157–1163. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.T.; Ma, L.; Burgner, J.W.; Groesch, T.D.; Post, C.B.; Kuhn, R.J. Flavivirus Capsid Is a Dimeric Alpha-Helical Protein. J. Virol. 2003, 77, 7143–7149. [Google Scholar] [CrossRef] [PubMed]
- Schlick, P.; Kofler, R.M.; Schittl, B.; Taucher, C.; Nagy, E.; Meinke, A.; Mandl, C.W. Characterization of West Nile virus live vaccine candidates attenuated by capsid deletion mutations. Vaccine 2010, 28, 5903–5909. [Google Scholar] [CrossRef] [PubMed]
- Kofler, R.M.; Heinz, F.X.; Mandl, C.W. Capsid protein C of tick-borne encephalitis virus tolerates large internal deletions and is a favorable target for attenuation of virulence. J. Virol. 2002, 76, 3534–3543. [Google Scholar] [CrossRef] [PubMed]
- Khromykh, A.A.; Westaway, E.G. Subgenomic replicons of the flavivirus Kunjin: Construction and applications. J. Virol. 1997, 71, 1497–1505. [Google Scholar] [PubMed]
- Kofler, R.M.; Leitner, A.; O’Riordain, G.; Heinz, F.X.; Mandl, C.W. Spontaneous mutations restore the viability of tick-borne encephalitis virus mutants with large deletions in protein C. J. Virol. 2003, 77, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Netsawang, J.; Noisakran, S.; Puttikhunt, C.; Kasinrerk, W.; Wongwiwat, W.; Malasit, P.; Yenchitsomanus, P.; Limjindaporn, T. Nuclear localization of dengue virus capsid protein is required for DAXX interaction and apoptosis. Virus Res. 2010, 147, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Limjindaporn, T.; Netsawang, J.; Noisakran, S.; Thiemmeca, S.; Wongwiwat, W.; Sudsaward, S.; Avirutnan, P.; Puttikhunt, C.; Kasinrerk, W.; Sriburi, R.; et al. Sensitization to Fas-mediated apoptosis by dengue virus capsid protein. Biochem. Biophys. Res. Commun. 2007, 362, 334–339. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, Y.; Mori, Y.; Abe, T.; Yamashita, T.; Okamoto, T.; Ichimura, T.; Moriishi, K.; Matsuura, Y. Nucleolar protein B23 interacts with Japanese encephalitis virus core protein and participates in viral replication. Microbiol. Immunol. 2006, 50, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Anderson, R.; Hobman, T.C. The capsid-binding nucleolar helicase DDX56 is important for infectivity of West Nile virus. J. Virol. 2011, 85, 5571–5580. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.R.; Lee, S.R.; Oh, W.; Lee, E.W.; Yeh, J.Y.; Nah, J.J.; Joo, Y.S.; Shin, J.; Lee, H.W.; Pyo, S.; et al. West Nile virus capsid protein induces p53-mediated apoptosis via the sequestration of HDM2 to the nucleolus. Cell. Microbiol. 2008, 10, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.H.; Syu, W.J.; Huang, K.J.; Lei, H.Y.; Yao, C.W.; King, C.C.; Hu, S.T. Intracellular localization and determination of a nuclear localization signal of the core protein of dengue virus. J. Gen. Virol. 2002, 83, 3093–3102. [Google Scholar] [CrossRef] [PubMed]
- Sangiambut, S.; Keelapang, P.; Aaskov, J.; Puttikhunt, C.; Kasinrerk, W.; Malasit, P.; Sittisombut, N. Multiple regions in dengue virus capsid protein contribute to nuclear localization during virus infection. J. Gen. Virol. 2008, 89, 1254–1264. [Google Scholar] [CrossRef] [PubMed]
- Bhuvanakantham, R.; Chong, M.K.; Ng, M.L. Specific interaction of capsid protein and importin-a/b influences West Nile virus production. Biochem. Biophys. Res. Commun. 2009, 389, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Jans, D.A.; Hübner, S. Regulation of protein transport to the nucleus: Central role of phosphorylation. Physiol. Rev. 1996, 76, 651–685. [Google Scholar] [PubMed]
- Bhuvanakantham, R.; Cheong, Y.K.; Ng, M.L. West Nile virus capsid protein interaction with importin and HDM2 protein is regulated by protein kinase C-mediated phosphorylation. Microbes Infect. 2010, 12, 615–625. [Google Scholar] [CrossRef] [PubMed]
- Edward, Z.; Takegami, T. Localization and functions of Japanese encephalitis virus nonstructural proteins NS3 and NS5 for viral RNA synthesis in the infected cells. Microbiol. Immunol. 1993, 37, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Forwood, J.K.; Brooks, A.; Briggs, L.J.; Xiao, C.Y.; Jans, D.A.; Vasudevan, S.G. The 37-amino-acid interdomain of dengue virus NS5 protein contains a functional NLS and inhibitory CK2 site. Biochem. Biophys. Res. Commun. 1999, 257, 731–737. [Google Scholar] [CrossRef] [PubMed]
- Buckley, A.; Gaidamovich, S.; Turchinskaya, A.; Gould, E.A. Monoclonal antibodies identify the NS5 yellow fever virus non-structural protein in the nuclei of infected cells. J. Gen. Virol. 1992, 73, 1125–1130. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.; Brooks, A.J.; Jans, D.A.; Vasudevan, S.G. A small region of the dengue virus-encoded RNA-dependent RNA polymerase, NS5, confers interaction with both the nuclear transport receptor importin-β and the viral helicase, NS3. J. Gen. Virol. 2001, 82, 735–745. [Google Scholar] [CrossRef] [PubMed]
- Pryor, M.J.; Rawlinson, S.M.; Butcher, R.E.; Barton, C.L.; Waterhouse, T.A.; Vasudevan, S.G.; Bardin, P.G.; Wright, P.J.; Jans, D.A.; Davidson, A.D. Nuclear Localization of Dengue Virus Nonstructural Protein 5 Through Its Importin α/β-Recognized Nuclear Localization Sequences is Integral to Viral Infection. Traffic 2007, 8, 795–807. [Google Scholar] [CrossRef] [PubMed]
- Brooks, A.J.; Johansson, M.; John, A.V.; Xu, Y.; Jans, D.A.; Vasudevan, S.G. The interdomain region of dengue NS5 protein that binds to the viral helicase NS3 contains independently functional importin-β1 and importin α/β-recognized nuclear localization signals. J. Biol. Chem. 2002, 277, 36399–36407. [Google Scholar] [CrossRef] [PubMed]
- Rawlinson, S.M.; Pryor, M.J.; Wright, P.J.; Jans, D.A. CRM1-mediated nuclear export of dengue virus RNA polymerase NS5 modulates interleukin-8 induction and virus production. J. Biol. Chem. 2009, 284, 15589–15597. [Google Scholar] [CrossRef] [PubMed]
- Medin, C.L.; Fitzgerald, K.A.; Alan, L.; Rothman, A.L. Dengue Virus Nonstructural Protein NS5 Induces Interleukin-8 Transcription and Secretion. J. Virol. 2005, 79, 11053–11061. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Bühler, S.; Selisko, B.; Davidson, A.; Mulder, K.; Canard, B.; Miller, S.; Bartenschlager, R. Nuclear localization of dengue virus nonstructural protein 5 does not strictly correlate with efficient viral RNA replication and inhibition of type I interferon signaling. J. Virol. 2013, 87, 4545–4557. [Google Scholar] [CrossRef] [PubMed]
- Tay, M.Y.F.; Smith, K.; Ng, I.H.W.; Chan, K.W.K.; Zhao, Y.; Ooi, E.E.; Lescar, J.; Luo, D.; Jans, D.A.; Forwood, J.K.; et al. The C-terminal 18 Amino Acid Region of Dengue Virus NS5 Regulates its Subcellular Localization and Contains a Conserved Arginine Residue Essential for Infectious Virus Production. PLoS Pathog. 2016, 12, e1005886. [Google Scholar] [CrossRef] [PubMed]
- Fraser, J.E.; Rawlinson, S.M.; Heaton, S.M.; Jans, D.A. Dynamic nucleolar targeting of dengue virus polymerase NS5 in response to extracellular pH. J. Virol. 2016, 90, 5797–5807. [Google Scholar] [CrossRef] [PubMed]
- Tay, M.Y.F.; Fraser, J.E.; Chan, W.K.K.; Moreland, N.J.; Rathore, A.P.; Wang, C.; Vasudevan, S.G.; Jans, D.A. Nuclear localization of dengue virus (DENV) 1–4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin. Antivir. Res. 2013, 99, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Hannemann, H.; Sung, P.Y.; Chiu, H.C.; Yousuf, A.; Bird, J.; Lim, S.P.; Davidson, A.D. Serotype-specific differences in dengue virus non-structural protein 5 nuclear localization. J. Biol. Chem. 2013, 288, 22621–22635. [Google Scholar] [CrossRef] [PubMed]
- Wagstaff, K.M.; Rawlinson, S.M.; Hearps, A.C.; Jans, D.A. An AlphaScreen®-based assay for high-throughput screening for specific inhibitors of nuclear import. J. Biomol. Screen. 2011, 16, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Wagstaff, K.M.; Sivakumaran, H.; Heaton, S.M.; Harrich, D.; Jans, D.A. Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem. J. 2012, 443, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Fraser, J.E.; Watanabe, S.; Wang, C.; Chan, W.K.K.; Maher, B.; Lopez-Denman, A.; Hick, C.; Wagstaff, K.M.; Mackenzie, J.M.; Sexton, P.M.; et al. A nuclear transport inhibitor that modulates the unfolded protein response and provides in vivo protection against lethal dengue virus infection. J. Infect. Dis. 2014, 210, 1780–1791. [Google Scholar] [CrossRef] [PubMed]
- Faustino, R.S.; Nelson, T.J.; Terzic, A.; Perez-Terzic, C. Nuclear transport: Target for therapy. Clin. Pharmacol. Ther. 2007, 81, 880–886. [Google Scholar] [CrossRef] [PubMed]
- Uchil, P.D.; Kumar, A.V.A.; Satchidanandam, V. Nuclear Localization of Flavivirus RNA Synthesis in Infected Cells. J. Virol. 2006, 80, 5451–5464. [Google Scholar] [CrossRef] [PubMed]
- Westaway, E.G.; Mackenzie, J.M.; Kenney, M.T.; Jones, M.K.; Khromykh, A.A. Ultrastructure of Kunjin virus-infected cells: Colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 1997, 71, 6650–6661. [Google Scholar] [PubMed]
- Westaway, E.G.; Khromykh, A.A.; Mackenzie, J.M. Nascent flavivirus RNA colocalized in situ with double-stranded RNA in stable replication complexes. Virology 1999, 258, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, J.M.; Jones, M.K.; Young, P.R. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology 1996, 220, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Kopek, B.G.; Perkins, G.; Miller, D.J.; Ellisman, M.H.; Ahlquist, P. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 2007, 5, e220. [Google Scholar] [CrossRef] [PubMed]
- Overby, A.K.; Popov, V.L.; Niedrig, M.; Weber, F. Tick-borne encephalitis virus delays interferon induction and hides its double-stranded RNA in intracellular membrane vesicles. J. Virol. 2010, 84, 8470–8483. [Google Scholar] [CrossRef] [PubMed]
- Barbier, V.; Lang, D.; Valois, S.; Rothman, A.L.; Medin, C.L. Dengue virus induces mitochondrial elongation through impairment of Drp1-triggered mitochondrial fission. Virology 2016, 500, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Chatel-Chaix, L.; Cortese, M.; Romero-Brey, I.; Bender, S.; Neufeldt, C.J.; Fischl, W.; Scaturro, P.; Schieber, N.; Schwab, Y.; Fischer, B.; et al. Dengue Virus Perturbs Mitochondrial Morphodynamics to Dampen Innate Immune Responses. Cell Host Microbe 2016, 20, 342–356. [Google Scholar] [CrossRef] [PubMed]
- Randall, R.E.; Goodbourn, S. Interferons and viruses: An interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 2008, 89, 1–47. [Google Scholar] [CrossRef] [PubMed]
- Levy, D.E.; García-Sastre, A. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 2001, 12, 143–156. [Google Scholar] [CrossRef]
- Fensterl, V.; Sen, G.C. Interferons and viral infections. BioFactors 2009, 35, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Laurent-Rolle, M.; Boer, E.F.; Lubick, K.J.; Wolfinbarger, J.B.; Carmody, A.B.; Rockx, B.; Liu, W.; Ashour, J.; Shupert, W.L.; Holbrook, M.R.; et al. The NS5 protein of the virulent West Nile virus NY99 strain is a potent antagonist of type I interferon-mediated JAK-STAT signaling. J. Virol. 2010, 84, 3503–3515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashour, J.; Laurent-Rolle, M.; Shi, P.Y.; García-Sastre, A. NS5 of dengue virus mediates STAT2 binding and degradation. J. Virol. 2009, 83, 5408–5418. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.J.; Chang, B.L.; Yu, H.P.; Liao, C.L.; Lin, Y.L. Blocking of interferon-induced Jak-Stat signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. J. Virol. 2006, 80, 5908–5918. [Google Scholar] [CrossRef] [PubMed]
- Mazzon, M.; Jones, M.; Davidson, A.; Chain, B.; Jacobs, M. Dengue virus NS5 inhibits interferon-alpha signaling by blocking signal transducer and activator of transcription 2 phosphorylation. J. Infect. Dis. 2009, 200, 1261–1270. [Google Scholar] [CrossRef] [PubMed]
- Reid, C.R.; Hobman, T.C. The nucleolar helicase DDX56 redistributes to West Nile virus assembly sites. Virology 2017, 500, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Ye, W.; Yang, J.; Han, P.; Wang, Y.; Ye, C.; Weng, D.; Zhang, F.; Xu, Z.; Lei, Y. DDX21 translocates from nucleus to cytoplasm and stimulates the innate immune response due to dengue virus infection. Biochem. Biophys. Res. Commun. 2016, 473, 648–653. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.W.; Cheng, C.W.; Yang, T.C.; Li, S.W.; Cheng, M.H.; Wan, L.; Lin, Y.J.; Lai, C.H.; Lin, W.Y.; Kao, M.C. Interferon antagonist function of Japanese encephalitis virus NS4A and its interaction with DEAD-box RNA helicase DDX42. Virus Res. 2008, 137, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Ge, L.L.; Li, P.P.; Wang, Y.; Dai, J.J.; Sun, M.X.; Huang, L.; Shen, Z.Q.; Hu, X.C.; Ishag, H.; et al. Cellular DDX3 regulates Japanese encephalitis virus replication by interacting with viral un-translated regions. Virology 2014, 449, 70–81. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Hobman, T.C. The helicase activity of DDX56 is required for its role in assembly of infectious West Nile virus particles. Virology 2012, 433, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Kim, T.; Bao, M.; Facchinetti, V.; Jung, S.Y.; Ghaffari, A.A.; Qin, J.; Cheng, G.; Liu, Y.J. DDX1, DDX21, and DHX36 Helicases Form a Complex with the Adaptor Molecule TRIF to Sense dsRNA in Dendritic Cells. Immunity 2011, 34, 866–878. [Google Scholar] [CrossRef] [PubMed]
- De Nova-Ocampo, M.; Villegas-Sepúlveda, N.; del Angel, R.M. Translation elongation factor-1alpha, La, and PTB interact with the 3′ untranslated region of dengue 4 virus RNA. Virology 2002, 295, 337–347. [Google Scholar] [CrossRef] [PubMed]
- García-Montalvo, B.M.; Medina, F.; Del Angel, R.M. La protein binds to NS5 and NS3 and to the 5′ and 3′ ends of Dengue 4 virus RNA. Virus Res. 2004, 102, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Paranjape, S.M.; Harris, E. Y box-binding protein-1 binds to the dengue virus 3′-untranslated region and mediates antiviral effects. J. Biol. Chem. 2007, 282, 30497–30508. [Google Scholar] [CrossRef] [PubMed]
- Agis-Juárez, R.A.; Galván, I.; Medina, F.; Daikoku, T.; Padmanabhan, R.; Ludert, J.E.; del Angel, R.M. Polypyrimidine tract-binding protein is relocated to the cytoplasm and is required during dengue virus infection in Vero cells. J. Gen. Virol. 2009, 90, 2893–2901. [Google Scholar] [CrossRef] [PubMed]
- Van Venrooij, W.J.; Pruijn, G.J. Ribonucleoprotein complexes as autoantigens. Curr. Opin. Immunol. 1995, 7, 819–824. [Google Scholar] [CrossRef]
- Li, W.; Kedersha, N.; Anderson, P.; Emara, M.; Swiderek, K.M.; Moreno, G.T.; Brinton, M.A. Cell proteins TIA-1 and TIAR interact with the 3′stem-loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. J. Virol. 2002, 76, 11989. [Google Scholar] [CrossRef] [PubMed]
- Emara, M.M.; Liu, H.; Davis, W.G.; Brinton, M.A. Mutation of mapped TIA-1/TIAR binding sites in the 3′ terminal stem-loop of West Nile virus minus-strand RNA in an infectious clone negatively affects genomic RNA amplification. J. Virol. 2008, 82, 10657–10670. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, R.E. Nuclear proteins hijacked by mammalian cytoplasmic plus strand RNA viruses. Virology 2015, 479, 457–474. [Google Scholar] [CrossRef] [PubMed]
| Virus | Host Factor | Role in Sequestration |
|---|---|---|
| Flavi | La | IRES conformation, translation-replication switching, genome circularization |
| Flavi | PTB | IRES conformation, eIF4G1 recruitment to IRES, genome circularization |
| DENV | RNA Helicase A | Genome circularization protein bridge, promote RNA replication |
| Flavi/WNV | Tia1, TIAR | Promotes + strand RNA synthesis |
| Flavi/DENV | Tudor-DN/p-100 | Promotes RNA synthesis |
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lopez-Denman, A.J.; Mackenzie, J.M. The IMPORTance of the Nucleus during Flavivirus Replication. Viruses 2017, 9, 14. https://doi.org/10.3390/v9010014
Lopez-Denman AJ, Mackenzie JM. The IMPORTance of the Nucleus during Flavivirus Replication. Viruses. 2017; 9(1):14. https://doi.org/10.3390/v9010014
Chicago/Turabian StyleLopez-Denman, Adam J., and Jason M. Mackenzie. 2017. "The IMPORTance of the Nucleus during Flavivirus Replication" Viruses 9, no. 1: 14. https://doi.org/10.3390/v9010014






