Effective Detection of Porcine Cytomegalovirus Using Non-Invasively Taken Samples from Piglets
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Examined Samples
2.2. DNA Extraction
2.3. Conventional Polymerase Chain Reaction and Nested Polymerase Chain Reaction
2.4. Cloning and Sequence Analysis of the Amplicons
2.5. Real-Time Polymerase Chain Reaction
2.6. Software
3. Results
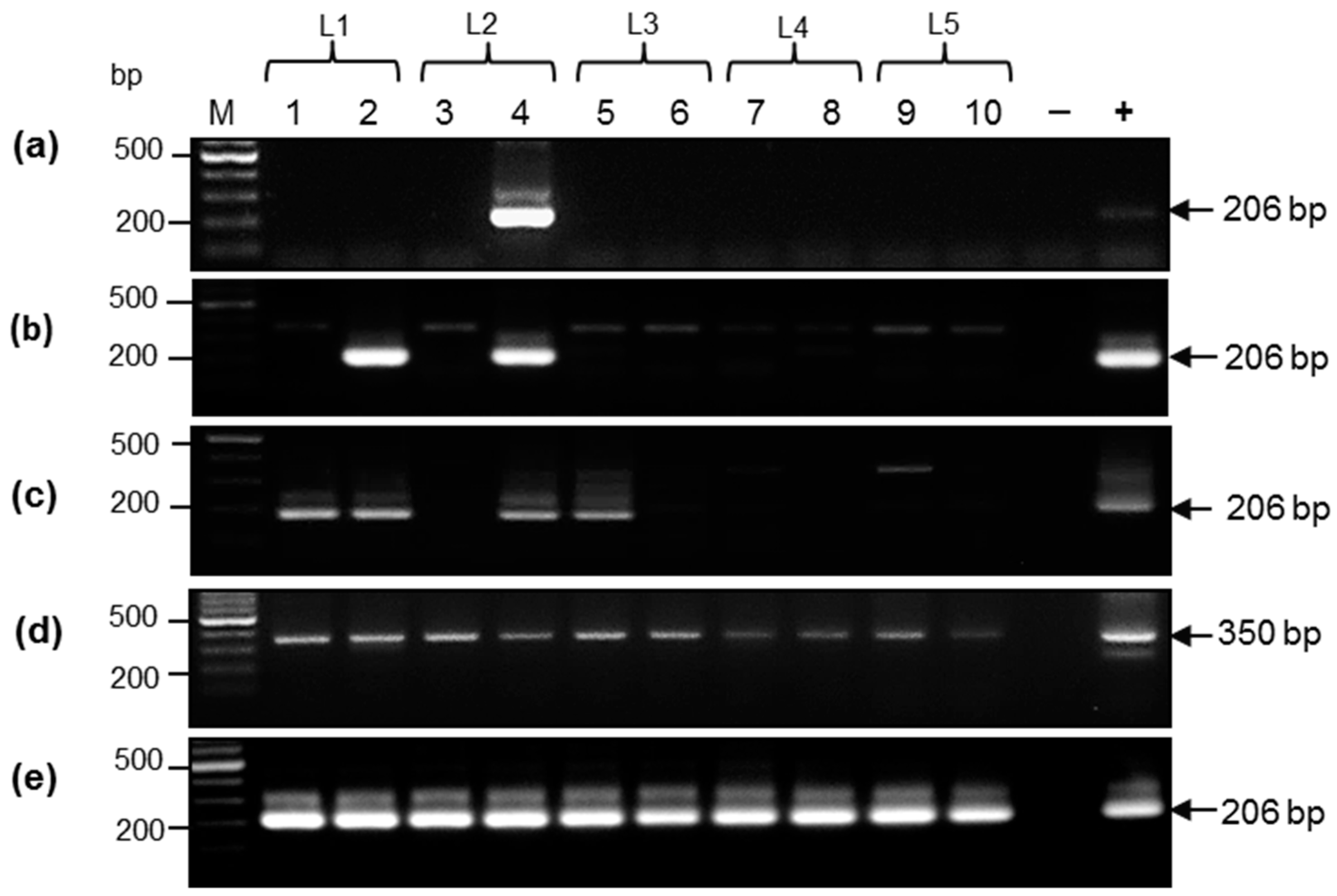
3.1. Characterization of DNA from Samples Collected by Non-Invasive and Low-Invasive Methods
3.2. Comparative Analysis of DNA Samples from Aachen Minipigs by Nested Polymerase Chain Reaction
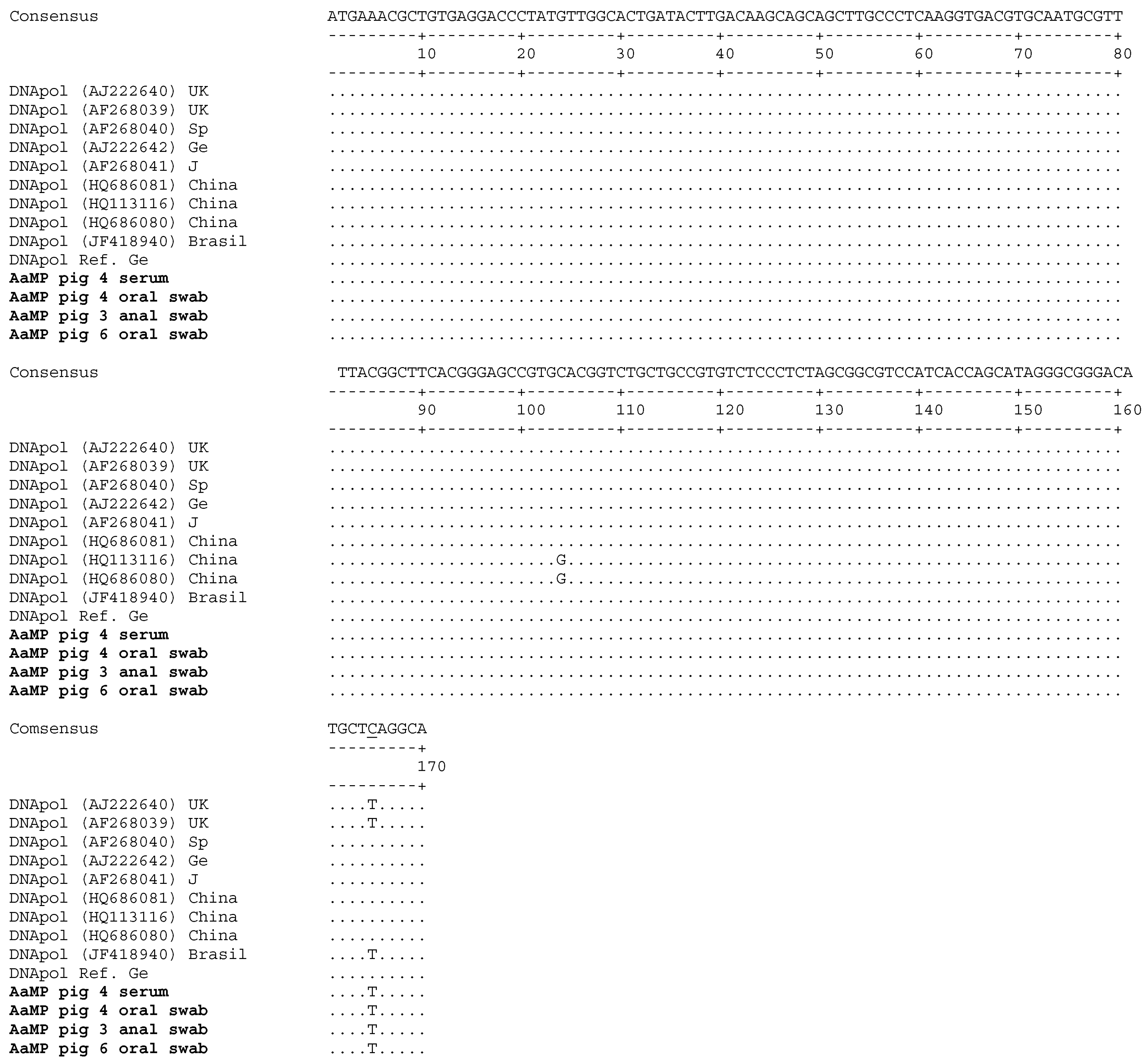
3.3. Porcine Cytomegalovirus Sequences Detected in Piglets
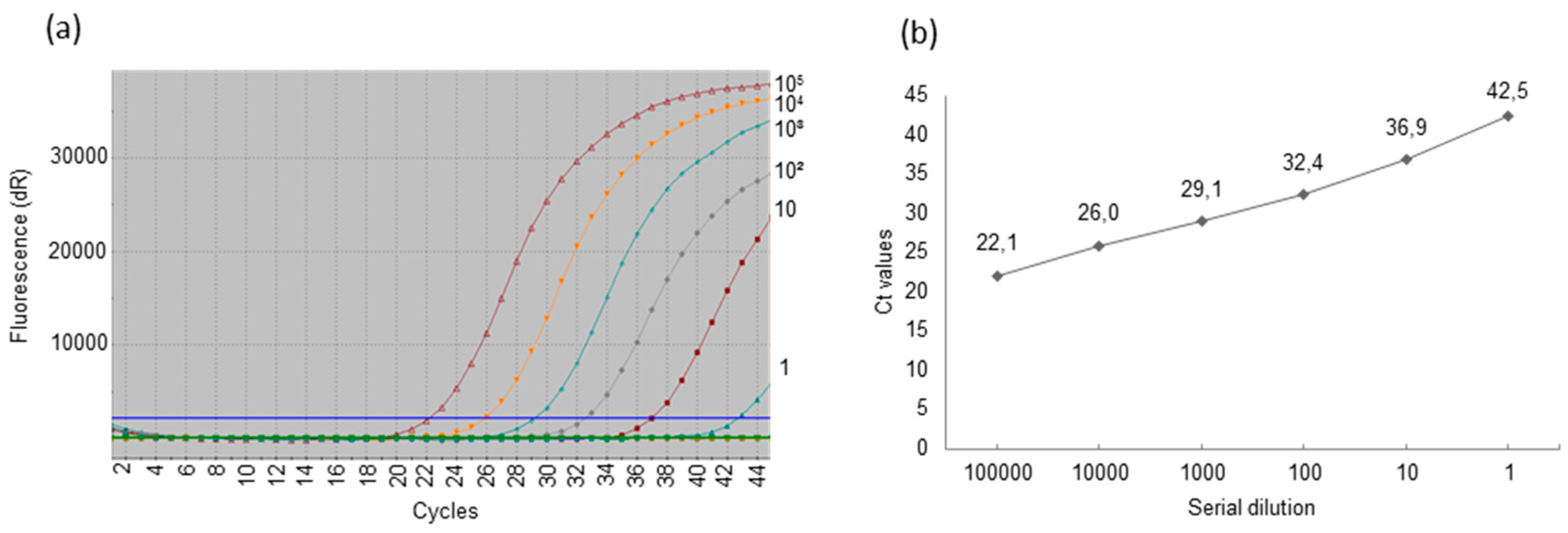
3.4. Detection of Porcine Cytomegalovirus by Real-Time Polymerase Chain Reaction and Comparison of the Sensitivity of Uniplex and Duplex Systems
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Denner, J. Recent Progress in Xenotransplantation, with Emphasis on Virological Safety. Ann. Transplant. 2016, 21, 717–727. [Google Scholar] [CrossRef]
- Denner, J.; Tönjes, R.R. Infection barriers to successful xenotransplantation focusing on porcine endogenous retroviruses. Clin. Microbiol. Rev. 2012, 25, 318–343. [Google Scholar] [CrossRef] [PubMed]
- Mueller, N.J.; Barth, R.N.; Yamamoto, S.; Kitamura, H.; Patience, C.; Yamada, K.; Cooper, D.K.; Sachs, D.H.; Kaur, A.; Fishman, J.A. Activation of cytomegalovirus in pig-to-primate organ xenotransplantation. J. Virol. 2002, 76, 4734–4764. [Google Scholar] [CrossRef] [PubMed]
- Mueller, N.J.; Livingston, C.; Knosalla, C.; Barth, R.N.; Yamamoto, S.; Gollackner, B.; Dor, F.J.; Buhler, L.; Sachs, D.H.; Yamada, K.; et al. Activation of porcine cytomegalovirus, but not porcine lymphotropic herpesvirus, in pig-to baboon xenotransplantation. J. Inf. Dis. 2004, 189, 1628–1633. [Google Scholar] [CrossRef] [PubMed]
- Morozov, V.A.; Abicht, J.M.; Reichart, B.; Mayr, T.; Guethoff, S.; Denner, J. Active replication of porcine cytomegalovirus (PCMV) following transplantation of a pig heart into a baboon despite undetected virus in the donor pig. Ann. Virol. Res. 2016, 2, 1018. [Google Scholar]
- Yamada, K.; Tasaki, M.; Sekijima, M.; Wilkinson, R.A.; Villani, V.; Moran, S.G.; Cormac, T.A.; Hanekamp, I.M.; Arn, J.S.; Fishman, J.A.; et al. Porcine cytomegalovirus infection is associated with early rejection of kidney grafts in a pig to baboon xenotransplantation model. Transplantation 2014, 98, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Sekijima, M.; Waki, S.; Sahara, H.; Tasaki, M.; Wilkinson, R.A.; Villani, V.; Shimatsu, Y.; Nakano, K.; Matsunari, H.; Nagashima, H.; et al. Results of life-supporting galactosyltransferase knockout kidneys in cynomolgus monkeys using two different sources of galactosyltransferase knockout swine. Transplantation 2014, 98, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Tucker, A.W.; Galbraith, D.; McEwan, P.; Onions, D. Evaluation of porcine cytomegalovirus as a potential zoonotic agent in Xenotransplantation. Transplant. Proc. 1999, 31, 915. [Google Scholar] [CrossRef]
- Fishman, J.A.; Patience, C. Xenotransplantation: Infectious risk revisited. Am. J. Transplant. 2004, 4, 1383–1390. [Google Scholar] [CrossRef] [PubMed]
- Fishman, J.A.; Scobie, L.; Takeuchi, Y. Xenotransplantation-associated infectious risk: A WHO consultation. Xenotransplantation 2012, 19, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Denner, J. Xenotransplantation and porcine cytomegalovirus. Xenotransplantation 2015, 22, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Zeng, N.; Zhou, L.; Ge, X.; Guo, X.; Yang, H. Genomic organization and molecular characterization of porcine cytomegalovirus. Virology 2014, 460–461, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Edington, N. Porcine cytomegalovirus. Dis. Swine 1986, 138, 330–336. [Google Scholar]
- Goltz, M.; Widen, F.; Banks, M.; Belak, S.; Ehlers, B. Characterization of the DNA polymerase loci of porcine cytomegalovirus from diverse geographical origins. Virus Genes 2000, 21, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Clark, D.A.; Fryer, J.F.L.; Tucker, A.W.; McArdle, P.D.; Hughes, A.E.; Emery, V.C.; Griffiths, P.D. Porcine cytomegalovirus in pigs being bred for xenograft organs; progress towards control. Xenotransplantation 2003, 10, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liao, S.; Zhu, L.; Xu, Z.; Zhou, Y. Molecular epidemiology of porcine cytomegalovirus (PCMV) in Sichuan province, China: 2010–2013. PLoS ONE 2013, 8, e64648. [Google Scholar]
- Edington, N.; Watt, R.G.; Plowright, W. Experimental transplacental transmission of porcine cytomegalovirus. J. Hyg. 1977, 78, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Edington, N.; Broad, S.; Wrathall, A.E.; Done, J.T. Superinfection with porcine cytomegalovirus initiate infection. Vet. Microbiol. 1988, 16, 189–193. [Google Scholar] [CrossRef]
- Mueller, N.J.; Kuwaki, K.; Knosalla, C.; Dor, F.J.; Gollackner, B.; Wilkinson, R.A.; Arn, S.; Sachs, D.H.; Cooper, D.K.; Fishman, J.A. Early weaning of piglets fails to exclude porcine lymphotropic herpesvirus. Xenotransplantation 2005, 12, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Whitteker, J.L.; Dudani, A.K.; Tackaberry, E.S. Human fibroblasts are permissive for porcine cytomegalovirus in vitro. Transplantation 2008, 86, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Fryer, J.F.L.; Griffiths, P.D.; Fishman, J.A.; Emery, V.C.; Clark, D.A. Quantitation of porcine cytomegalovirus in pig tissues by PCR. J. Clin. Microbiol. 2001, 39, 1155–1156. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Xu, Z.; Zhu, L.; Liao, S.; Guo, W. Transcriptome analysis of porcine thymus following porcine cytomegalovirus infection. PLos ONE 2014, 9, e113921. [Google Scholar] [CrossRef] [PubMed]
- Plotzki, E.; Heinrichs, G.; Kubicková, B.; Ulrich, R.G.; Denner, J. Microbiological characterization of a newly established pig breed, Aachen minipigs. Xenotransplantation 2016, 23, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Morozov, V.A.; Morozov, A.V.; Denner, J. New nested and real-time PCR systems for porcine cytomegalovirus (PCMV) detection and quantification. Arch. Virol. 2016, 161, 1159–1168. [Google Scholar] [CrossRef] [PubMed]
- Duvigneau, J.C.; Hartl, R.T.; Groiss, S.; Gemeiner, M. Quantitative simultaneous multiplex real-time PCR for the detection of porcine cytokines. J. Immunol. Methods 2005, 30, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Basic Local Alignment Search Tool (BLAST). Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 11 January 2017).
- Abraham, J.E.; Maranian, M.J.; Spiteri, I.; Russell, R.; Ingle, S.; Luccarini, C.; Earl, H.M.; Pharoah, P.P.; Dunning, A.M.; Caldas, C. Saliva samples are a viable alternative to blood samples as a source of DNA for high throughput genotyping. BMC Med. Genom. 2012, 5, 19. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.S.; Moon, H.J.; Yang, J.S.; Park, S.J.; Song, D.S.; Kang, B.K.; Park, B.K. Multiplex PCR for the simultaneous detection of pseudorabies virus, porcine cytomegalovirus, and porcine circovirus in pigs. J. Virol. Methods 2007, 139, 39–43. [Google Scholar] [CrossRef] [PubMed]
- Cibulski, S.P.; Pasqualim, G.; Teixera, T.; Varela, A.P.M.; Dezen, D.; Holz, C.L.; Franco, A.C.; Roehe, P.M. Porcine cytomegalovirus infection is not associated to the occurrence of post-weaning multisystemic wasting syndrome. Vet. Med. Sci. 2016, 1, 23–29. [Google Scholar] [CrossRef]
- Elnifro, E.M.; Ashshi, A.M.; Cooper, R.J.; Klapper, P.E. Multiplex PCR: Optimization and application in diagnostic virology. Clin. Microbiol. Rev. 2000, 13, 559–570. [Google Scholar] [CrossRef] [PubMed]
| Primers (nt) | Sequence (5′-3′) | Tm (°C) | Oligonucleotide Length | Homo-Dimers. ΔG below −7 kcal/mol | Tm Hairpin (°C) |
|---|---|---|---|---|---|
| Conventional Polymerase Chain Reaction (PCR) | |||||
| F1 (63–81) * | ACGGGGATCGACGAGAAAG | 66.4 | 19 | none | 49 |
| R1 (412–390) * | CTAGACGAGAGGACATTGTTGAT | 61.1 | 23 | none | 29 |
| F2 (182–201)* | GAAGAGAAAGGAAGTGAAGG | 57.1 | 20 | none | None |
| R2 (386–368) * | GTCACTCGTCTGCCTAAGC | 60.2 | 19 | none | 30 |
| Real-time PCR | |||||
| Fr-t (279–299) * | AATGCGTTTTACAACTTCACG | 61.5 | 21 | none | 29 |
| Rr-t (373–354) * | CTGAGCATGTCCCGCCCTAT | 67.4 | 20 | none | 19 |
| Probe (331–350) * | 6FAM-CTCTAGCGGCGTCCATCACC-BHQ | 69.2 | 20 | none | 17 |
| F cyclophilin A (174–196) ** | TGCTTTCACAGAATAATTCCAGGATTTA | 59.1 | 28 | none | 23 |
| R cyclophilin A (250–230) ** | GACTTGCCACCAGTGCCATTA | 61.7 | 21 | none | 22 |
| Probe cyclophilin A (205–228) ** | Cy5-TGCCAGGGTGGTGACTTCACACGCC-BHQ | 67.1 | 25 | none | 44 |
| Piglet #/Material | Sera | Anal Swabs | Ears Biopsies | Oral Swabs |
|---|---|---|---|---|
| 1 | 29.7 | 22.2 | 22.3 | 26.1 |
| 2 | 29.6 | 23.4 | 24.6 | 24.3 |
| 3 | 27.8 | 21.5 | 23.0 | 25.5 |
| 4 | 26.8 | 21.8 | 22.0 | 25.5 |
| 5 | 27.7 | 21.6 | 20.5 | 28.5 |
| 6 | 27.8 | 21.7 | 20.7 | 26.4 |
| 7 | 28.3 | 21.9 | 21.3 | 28.5 |
| 8 | 28.3 | 22.3 | 23.5 | 28.0 |
| 9 | 28.2 | 25.6 | 21.6 | 26.7 |
| 10 | 27.1 | 23.6 | 21.0 | 25.9 |
| Mean values: | 28.13 | 22.56 | 22.05 | 26.54 |
| Copies/reaction: | 6 × 103 | 5 × 106 | 5 × 106 | 2 × 104 |
| Copy/µg: | 1.2 × 105 | 2.5 × 107 | 2.5 × 107 | 1 × 105 |
| Piglet # | Sera Uniplex Duplex | Oral Swabs Uniplex Duplex | Ears Biopsies Uniplex Duplex | Anal Swabs Uniplex Duplex | ||||
|---|---|---|---|---|---|---|---|---|
| 1 | Neg. | Neg. | 30.0 | 30.0 | 35.8 | Neg. | Neg. | Neg. |
| 2 | Neg. | Neg. | 27.5 | 28.2 | 30.9 | 31.8 | 39.7 | Neg. |
| 3 | 38.6 | Neg. | 29.2 | 30.9 | Neg. | Neg. | Neg. | Neg. |
| 4 | 36.6 | Neg. | 28.4 | 30.8 | 39.4 | Neg. | 37.0 | Neg. |
| 5 | Neg. | Neg. | 27.3 | 29.1 | 34.0 | 41.3 | 36.2 | Neg. |
| 6 | 37.1 | Neg. | 28.1 | 28.6 | Neg. | Neg. | 41.3 | Neg. |
| 7 | Neg. | Neg. | 30.1 | 31.1 | Neg. | Neg. | Neg. | Neg. |
| 8 | 40.1 | Neg. | 29.4 | 31.4 | 32.3 | 39.7 | 39.5 | Neg. |
| 9 | 35.2 | Neg. | 26.1 | 26.9 | 35.2 | Neg. | 42.0 | Neg. |
| 10 | 37.2 | Neg. | 30.6 | Neg. | 43.0 | Neg. | 40.4 | Neg. |
| Piglet # | Sample | DNA Load (ng) | PCR | Nested PCR | Uniplex Real-Time PCR (Ct Values) | G.e./Reaction |
|---|---|---|---|---|---|---|
| 1 | Serum | 50 | Neg. | Neg. | Neg. | Neg. |
| Anal swab | 200 | Neg. | Neg. | Neg. | Neg. | |
| Oral swab | 200 | + | + | 30.0 | ~103 | |
| Ear biopsies | 200 | Neg. | + | 35.8 | ~40 | |
| 2 | Serum | 50 | Neg. | Neg. | Neg. | Neg. |
| Anal swab | 200 | Neg. | + | 39.7 | ~2–5 | |
| Oral swab | 200 | + | + | 27.5 | 104 | |
| Ear biopsies | 200 | Neg. | + | 30.9 | ~103 | |
| 3 | Serum | 50 | Neg. | Neg. | 38.6 | ~10 |
| Anal swab | 200 | Neg. | Neg. | Neg. | Neg. | |
| Oral swab | 200 | + | + | 29.2 | ~2 × 103 | |
| Ear biopsies | 200 | Neg. | Neg. | Neg. | Neg. | |
| 4 | Serum | 50 | + | + | 36.6 | ~20 |
| Anal swab | 200 | Neg. | + | 36.9 | ~20 | |
| Oral swab | 200 | + | + | 28.4 | ~7 × 103 | |
| Ear biopsies | 200 | Neg. | + | 39.4 | ~2–5 | |
| 5 | Serum | 50 | Neg. | Neg. | Neg. | Neg. |
| Anal swab | 200 | Neg. | Neg. | 36.2 | ~20 | |
| Oral swab | 200 | + | + | 27.3 | ~104 | |
| Ear biopsies | 200 | Neg. | + | 34.0 | 10e2 | |
| 6 | Serum | 50 | Neg. | Neg. | 37.1 | 10 |
| Anal swab | 200 | Neg. | Neg. | 41.3 | ~1–2 | |
| Oral swab | 200 | + | + | 28.1 | ~7 × 103 | |
| Ear biopsies | 200 | Neg. | Neg. | Neg. | Neg. | |
| 7 | Serum | 50 | Neg. | Neg. | Neg. | Neg. |
| Anal swab | 200 | Neg. | Neg. | Neg. | Neg. | |
| Oral swab | 200 | + | + | 30.1 | 103 | |
| Ear biopsies | 200 | Neg. | Neg. | Neg. | Neg. | |
| 8 | Serum | 50 | Neg. | Neg. | 40.1 | ~2 |
| Anal swab | 200 | Neg. | +(weak) | 39.5 | ~2 | |
| Oral swab | 200 | + | + | 30.4 | 103 | |
| Ear biopsies | 200 | Neg. | Neg. | 40.1 | ~2 | |
| 9 | Serum | 50 | Neg. | Neg. | 35.2 | ~30 |
| Anal swab | 200 | Neg. | Neg. | 41.9 | ~1–2 | |
| Oral swab | 200 | + | + | 26.1 | 2 × 104 | |
| Ear biopsies | 200 | Neg. | Neg. | 35.2 | ~40 | |
| 10 | Serum | 50 | Neg. | Neg. | 37.2 | ~10 |
| Anal swab | 200 | Neg. | Neg. | 40.4 | ~2 | |
| Oral swab | 200 | + | + | 30.6 | 103 | |
| Ear biopsies | 200 | Neg. | Neg. | 38.1 | ~5 |
| Samples/Methods | PCR (Positive/Total) | Nested PCR (Positive/Total) | Uniplex Real-Time PCR (Positive/Total) | Duplex Real-Time PCR (Positive/Total) | G.e., Detected |
|---|---|---|---|---|---|
| Sera | 1/10 | 1/10 | 6/10 | 0/10 | 10–150/mL |
| Anal swabs | 0/10 | 3/10 | 7/10 | 0/10 | 5–100/μg |
| Oral swabs | 10/10 | 10/10 | 10/10 | 9/10 | 5 × 103–1 × 105/μg |
| Ear biopsies | 0/10 | 1/10 | 7/10 | 3/10 | 25–5 × 103/μg |
| Total | 11/40 | 15/40 | 30/40 | 12/40 |
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morozov, V.A.; Heinrichs, G.; Denner, J. Effective Detection of Porcine Cytomegalovirus Using Non-Invasively Taken Samples from Piglets. Viruses 2017, 9, 9. https://doi.org/10.3390/v9010009
Morozov VA, Heinrichs G, Denner J. Effective Detection of Porcine Cytomegalovirus Using Non-Invasively Taken Samples from Piglets. Viruses. 2017; 9(1):9. https://doi.org/10.3390/v9010009
Chicago/Turabian StyleMorozov, Vladimir A., Gerd Heinrichs, and Joachim Denner. 2017. "Effective Detection of Porcine Cytomegalovirus Using Non-Invasively Taken Samples from Piglets" Viruses 9, no. 1: 9. https://doi.org/10.3390/v9010009






