The Evolutionary History and Spatiotemporal Dynamics of the NC Lineage of Citrus Tristeza Virus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequence Dataset
2.2. Phylogenetic Analysis
2.3. Phylodynamic and Phylogeographic Approaches
3. Results
3.1. The Monophyly and Temporal Signal of the Data Set
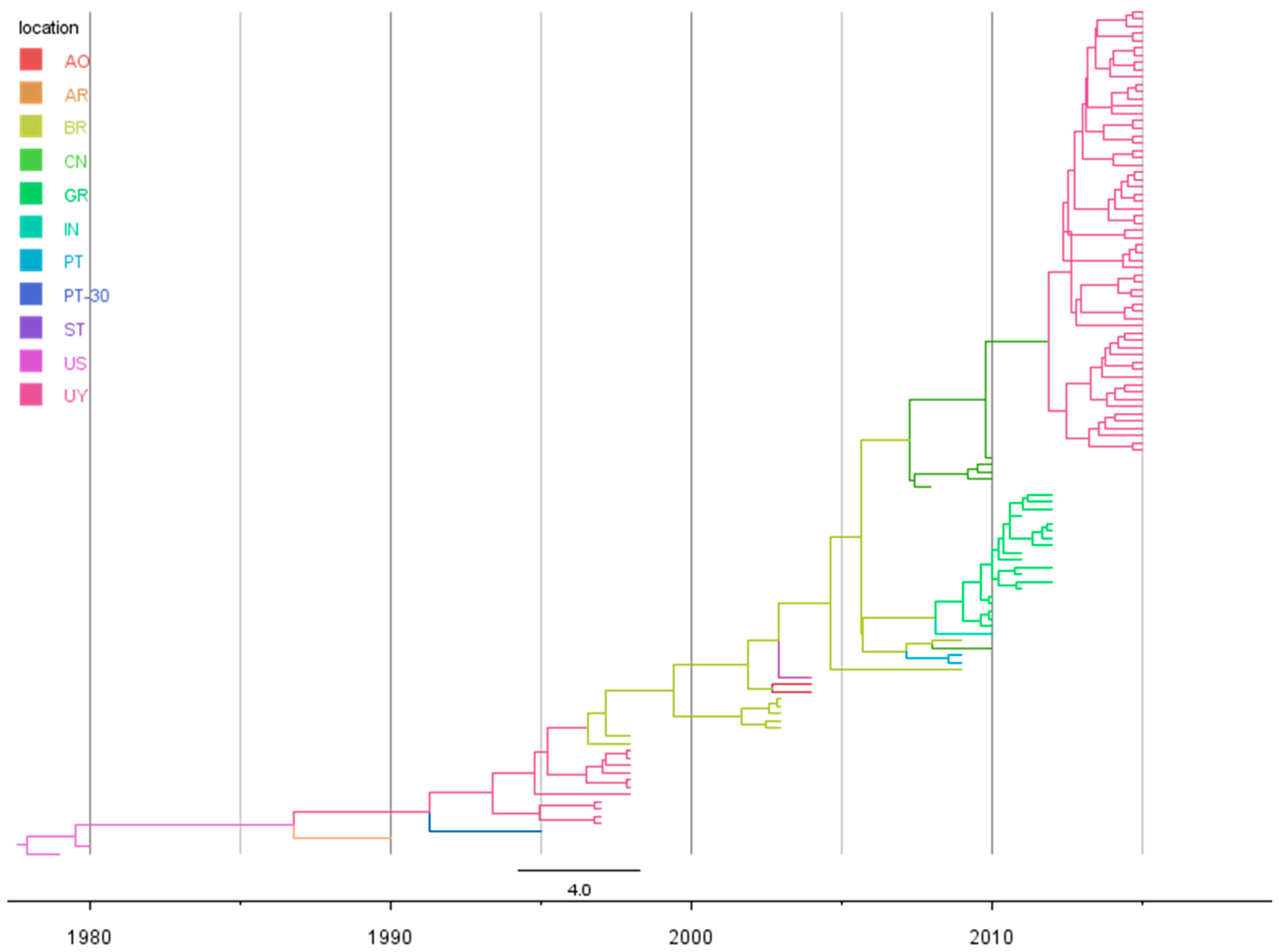
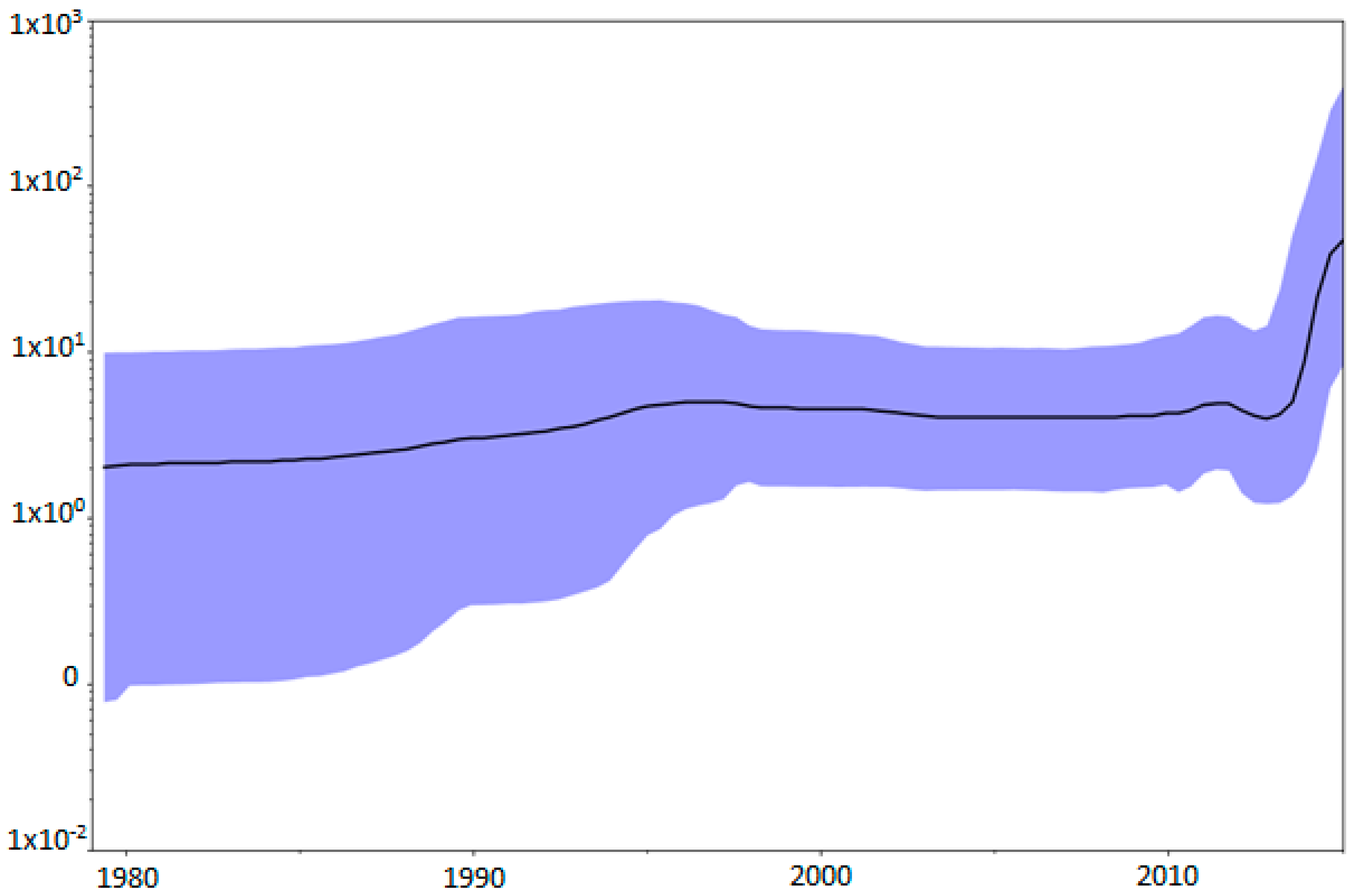
3.2. Rates and Dates of Evolution
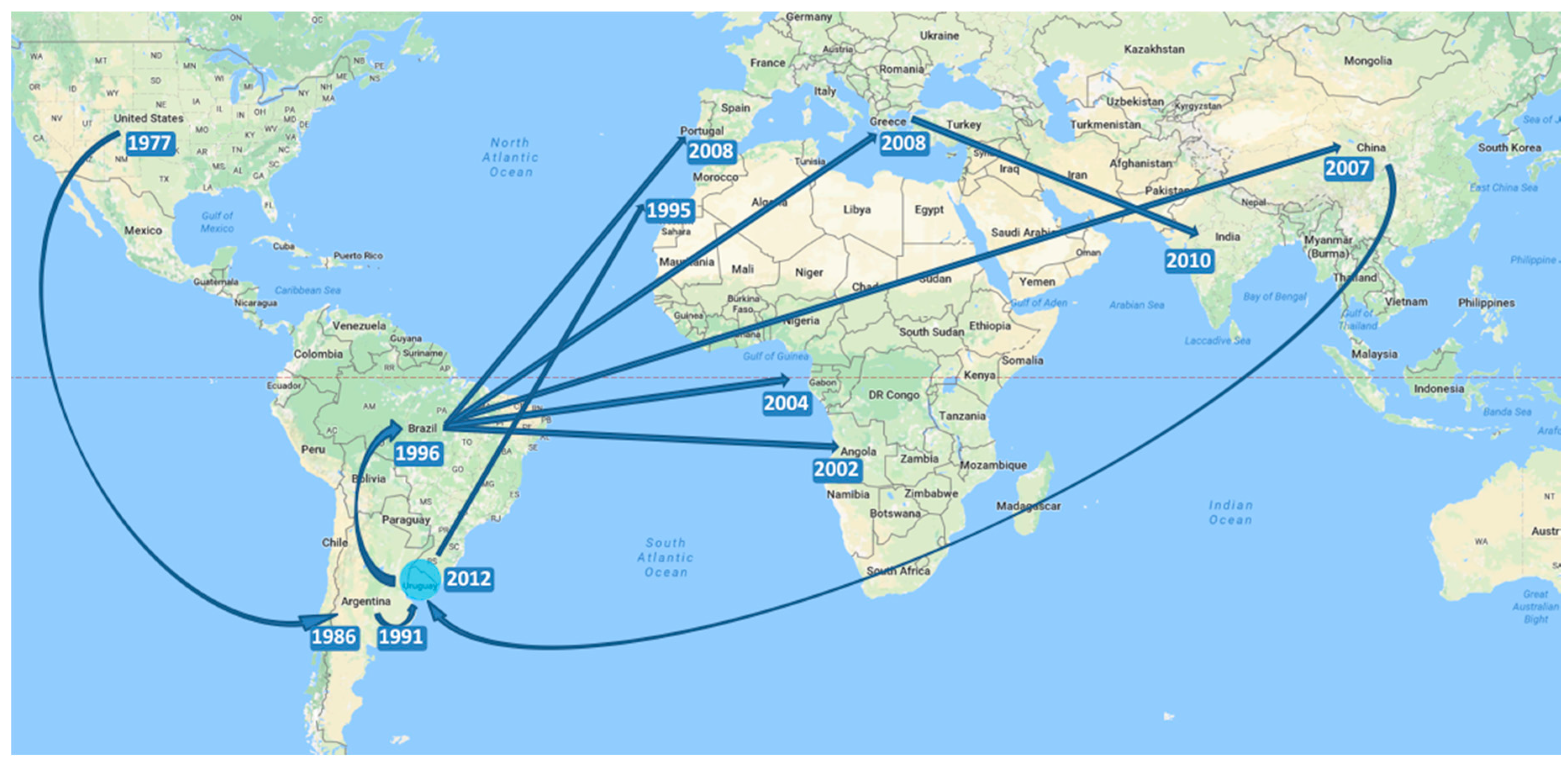
3.3. Evolutionary History of the Genotype
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Domingo, E.; Holland, J. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 1997, 51, 151–178. [Google Scholar] [CrossRef] [PubMed]
- Moreno, P.; Ambros, S.; Albiach-Marti, M.R.; Guerri, J.; Peña, L. Plant diseases that changed the world Citrus tristeza virus: A pathogen that changed the course of the citrus industry. Mol. Plant Pathol. 2008, 9, 251–268. [Google Scholar] [CrossRef] [PubMed]
- Karasev, A.V.; Boyko, V.P.; Gowda, S.; Nikolaeva, O.V.; Hilf, M.E.; Koonin, E.V.; Niblett, C.L.; Cline, K.; Gumpf, D.J.; Lee, R.F.; et al. Complete Sequence of the Citrus Tristeza Virus RNA Genome. Virology 1995, 208, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Satyanarayana, T.; Gowda, S.; Mawassi, M.; Albiach-Martí, M.R.; Ayllón, M.A.; Robertson, C.; Garnsey, S.M.; Dawson, W.O. Closterovirus encoded HSP70 homolog and p61 in addition to both coat proteins function in efficient virion assembly. Virology 2000, 278, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Ayllon, M.A.; Lopez, C.; Navas-Castillo, J.; Garnsey, S.M.; Guerri, J.; Flores, R.; Moreno, P. Polymorphism of the 5’ terminal region of Citrus tristeza virus (CTV) RNA: Incidence of three sequence types in isolates of different origin and pathogenicity. Arch. Virol. 2001, 146, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Folimonov, A.; Shintaku, M.; Wan-Xiang, L.; Falk, B.W.; Dawson, W.O.; Ding, S. Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proc. Natl. Acad. Sci. USA 2004, 101, 15742–15747. [Google Scholar] [CrossRef] [PubMed]
- Rubio, L.; Ayllon, M.A.; Kong, P.; Fernandez, A.; Polek, M.L.; Guerri, J.; Moreno, P.; Falk, B.W. Genetic variation of Citrus tristeza virus isolates from California and Spain: Evidence for mixed infections and recombination. J. Virol. 2001, 75, 8054–8062. [Google Scholar] [CrossRef] [PubMed]
- Hilf, M.E.; Karasev, A.V.; Albiachi-Marti, M.R.; Dawson, W.O.; Garnsey, S.M. Two paths of sequence divergence in the Citrus tristeza virus complex. Phytopathology 1999, 89, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Hilf, M.E.; Mavrodieva, V.A.; Garnsey, S.M. Genetic marker analysis of a global collection of isolates of Citrus tristeza virus: Characterization and distribution of CTV genotypes and association with symptoms. Phytopathology 2005, 95, 909–917. [Google Scholar] [CrossRef] [PubMed]
- Harper, S.J.; Dawson, T.E.; Pearson, M.N. Isolates of Citrus tristeza virus that overcome Poncirus trifoliata resistance comprise a novel strain. Arch. Virol. 2010, 155, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Brlansky, R.H. Genome Analysis of an Orange Stem Pitting Citrus Tristeza Virus Isolate Reveals a Novel Recombinant Genotype. Virus Res. 2010, 151, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Harper, S.J. Citrus tristeza virus: Evolution of complex and varied genotypic groups. Front Microbiol. 2013, 4, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Benítez-Galeano, M.J.; Rubio, L.; Bertalmío, A.; Maeso, D.; Rivas, F.; Colina, R. Phylogenetic Studies of the Three RNA Silencing Suppressor Genes of South American CTV Isolates Reveal the Circulation of a Novel Genetic Lineage. Viruses 2015, 7, 4152–4168. [Google Scholar] [CrossRef] [PubMed]
- Silva, G.; Marques, N.; Nolasco, G. The evolutionary rate of citrus tristeza virus ranks among the rates of the slowest RNA viruses. J. Gen. Virol. 2012, 93, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Davino, S.; Willemsen, A.; Panno, S.; Davino, M.; Catara, A.; Elena, S.F.; Rubio, L. Emergence and Phylodynamics of Citrus Tristeza Virus in Sicily, Italy. PLoS ONE 2013, 8, e66700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Rybicki, E. RDP: Detection of Recombination Amongst Aligned Sequences. Bioinformatics 2000, 16, 562–563. [Google Scholar] [CrossRef] [PubMed]
- Padidam, M.; Sawyer, S.; Fauquet, C.M. Possible Emergence of New Geminiviruses by Frequent Recombination. Virology 1999, 265, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Maynard Smith, J. Analyzing the Mosaic Structure of Genes. J. Mol. Evol. 1992, 34, 126–129. [Google Scholar]
- Posada, D.; Crandall, K.A. Evaluation of Methods for Detecting Recombination from DNA Sequences: Computer Simulations. Proc. Natl. Acad. Sci. USA 2001, 98, 13757–13762. [Google Scholar] [CrossRef] [PubMed]
- Boni, M.F.; Posada, D.; Feldman, M.W. An Exact Nonparametric Method for Inferring Mosaic Structure in Sequence Triplets. Genetics 2007, 176, 1035–1047. [Google Scholar] [CrossRef] [PubMed]
- Salminen, M.O.; Carr, J.K.; Burke, D.S.; McCutchan, F.E. Identification of Breakpoints in Intergenotypic Recombinants of HIV Type 1 by BOOTSCANning. AIDS Res. Hum. Retrovir. 1995, 11, 1423–14215. [Google Scholar] [CrossRef] [PubMed]
- Holmes, E.C.; Worobey, M.; Rambaut, A. Phylogenetic Evidence for Recombination in Dengue Virus. Mol. Biol. Evol. 1999, 16, 405. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, M.J.; Armstrong, J.S.; Gibbs, A.J. Sister-Scanning: A Monte Carlo Procedure for Assessing Signals in Recombinant Sequences. Bioinformatics 2000, 16, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef] [PubMed]
- Posada, D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef] [PubMed]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Autom. Control 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Anisimova, M.; Gascuel, O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 2006, 55, 539–552. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, S.Y.; Phillips, M.J.; Drummond, A.J.; Cooper, A. Accuracy of rate estimation using relaxed-clock models with a critical focus on the early metazoan radiation. Mol. Biol. Evol. 2005, 22, 1355–1363. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Ho, S.Y.W.; Phillips, M.J.; Rambaut, A. Relaxed Phylogenetics and Dating with Confidence. PLoS Biol. 2006, 4, e88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drummond, A.J.; Rambaut, A.; Shapiro, B.; Pybus, O.G. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 2005, 22, 1185–1192. [Google Scholar] [CrossRef] [PubMed]
- Bielejec, F.; Rambaut, A.; Suchard, M.A.; Lemey, P. SPREAD: Spatial phylogenetic reconstruction of evolutionary dynamics. Bioinformatics 2011, 27, 2910–2912. [Google Scholar] [CrossRef] [PubMed]
- Duffy, S.; Holmes, E.C. Validation of high rates of nucleotide substitution in geminiviruses: Phylogenetic evidence from East African cassava mosaic viruses. J. Gen. Virol. 2009, 90, 1539–1547. [Google Scholar] [CrossRef] [PubMed]
- Ramsden, C.; Holmes, E.C.; Charleston, M.A. Hantavirus evolution in relation to its rodent and insectivore hosts: No evidence for co-divergence. Mol. Biol. Evol. 2009, 26, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Lam, T.T.; de Carvalho, L.M.; Pybus, O.G. Exploring the temporal structure of heterochronous sequences using TempEst. Virus Evol. 2016, 2, vew007. [Google Scholar] [CrossRef] [PubMed]
- Duffy, S.; Shackelton, L.A.; Holmes, E.C. Rates of evolutionary change in viruses: Patterns and determinants. Nat. Rev. 2008, 9, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Meyer, A.G.; Spielman, S.J.; Bedford, T.; Wilke, C.O. Time dependence of evolutionary metrics during the 2009 pandemic influenza virus outbreak. Virus Evol. 2015, 1. [Google Scholar] [CrossRef] [PubMed]
- Park, D.J.; Dudas, G.; Wohl, S.; Goba, A.; Whitmer, S.L.; Andersen, K.G.; Sealfon, R.S.; Ladner, J.T.; Kugelman, J.R.; Matranga, C.B.; et al. Ebola Virus Epidemiology, Transmission, and Evolution during Seven Months in Sierra Leone. Cell 2015, 161, 1516–1526. [Google Scholar] [CrossRef] [PubMed]
- Silva, G.; Fonseca, F.; Santos, C.; Nolasco, G. Presence of Citrus Tristeza Virus in Angola and São Tomé e Príncipe: Characterization of isolates based on coat protein gene analysis. J. Plant Pathol. 2007, 89, 149–152. [Google Scholar]
- Owen, C.; Mathioudakis, M.; Gazivoda, A.; Gal, P.; Nol, N.; Kalliampakou, K.; Figas, A.; Bellan, A.; Iparaguirre, A.; Rubio, L.; et al. Evolution and molecular epidemiology of Citrus Tristeza virus on Crete: Recent introduction of a severe strain. J. Phytopathol. 2014, 162, 839–843. [Google Scholar] [CrossRef]
- Cowell, S.J.; Harper, S.J.; Dawson, W.O. Some like it hot: Citrus tristeza virus strains react differently to elevated temperature. Arch. Virol. 2016, 161, 3567–3570. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benítez-Galeano, M.J.; Castells, M.; Colina, R. The Evolutionary History and Spatiotemporal Dynamics of the NC Lineage of Citrus Tristeza Virus. Viruses 2017, 9, 272. https://doi.org/10.3390/v9100272
Benítez-Galeano MJ, Castells M, Colina R. The Evolutionary History and Spatiotemporal Dynamics of the NC Lineage of Citrus Tristeza Virus. Viruses. 2017; 9(10):272. https://doi.org/10.3390/v9100272
Chicago/Turabian StyleBenítez-Galeano, María José, Matías Castells, and Rodney Colina. 2017. "The Evolutionary History and Spatiotemporal Dynamics of the NC Lineage of Citrus Tristeza Virus" Viruses 9, no. 10: 272. https://doi.org/10.3390/v9100272




