A Next-Generation Sequencing Approach Uncovers Viral Transcripts Incorporated in Poxvirus Virions
Abstract
:1. Introduction
2. Materials and Methods
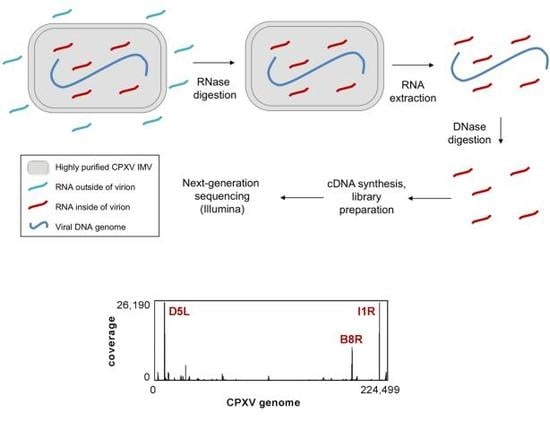
2.1. Virus Propagation and Purification
2.2. Sample Preparation for Transcript Analysis
2.3. Next-Generation Sequencing
2.4. NGS Data Analysis
3. Results
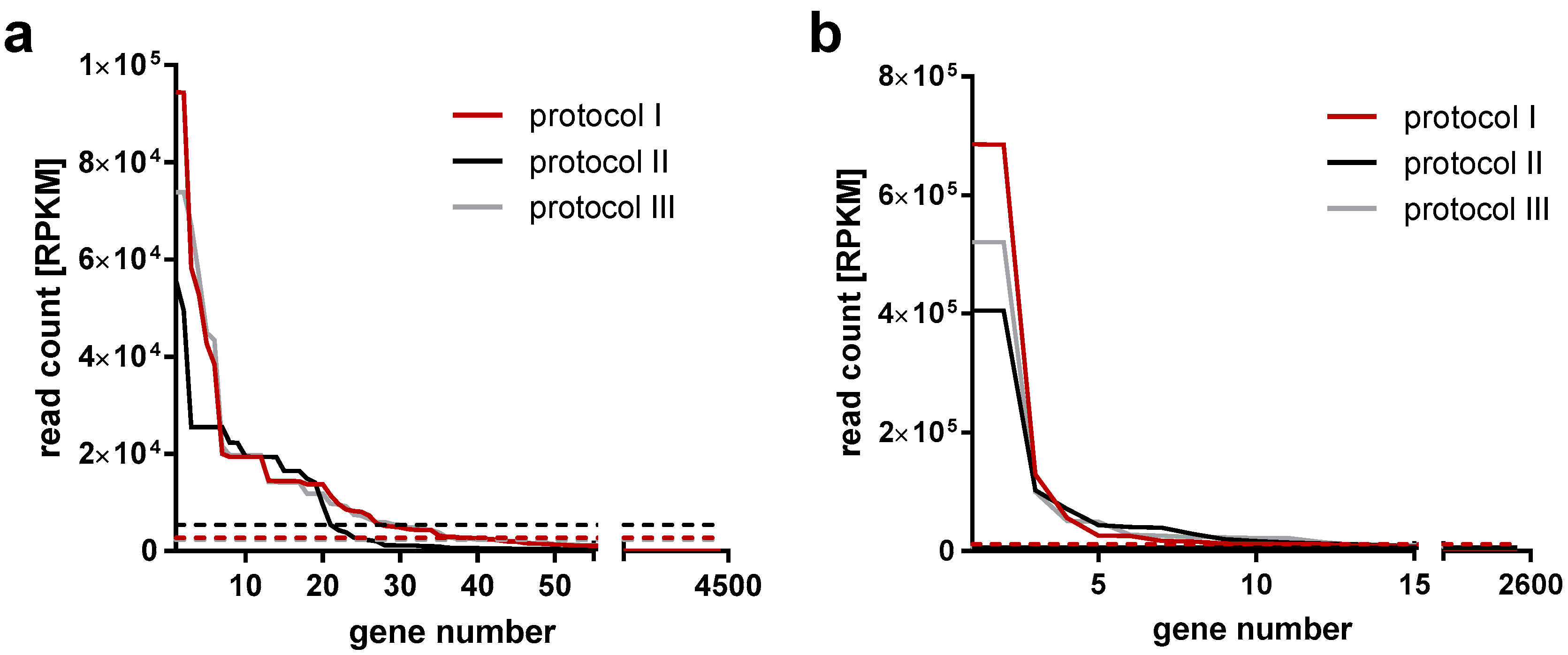
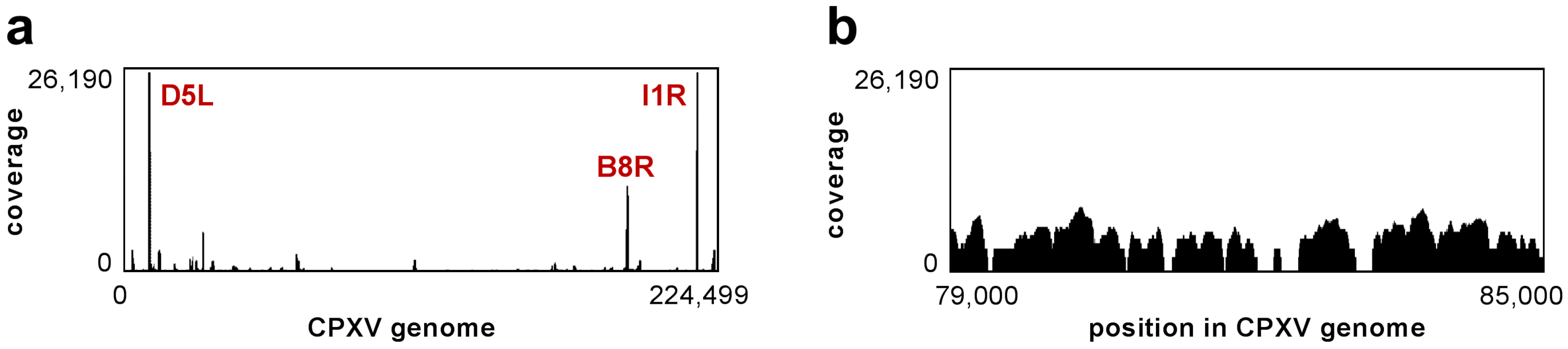
3.1. Data Overview
3.2. Incorporation of Viral Transcripts into Virus Particles
3.3. Incorporation of Host Transcripts
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Greijer, A.E.; Dekkers, C.A.; Middeldorp, J.M. Human cytomegalovirus virions differentially incorporate viral and host cell RNA during the assembly process. J. Virol. 2000, 74, 9078–9082. [Google Scholar] [CrossRef] [PubMed]
- Bresnahan, W.A.; Shenk, T. A subset of viral transcripts packaged within human cytomegalovirus particles. Science 2000, 288, 2373–2376. [Google Scholar] [CrossRef] [PubMed]
- Terhune, S.S.; Schroer, J.; Shenk, T. RNAs are packaged into human cytomegalovirus virions in proportion to their intracellular concentration. J. Virol. 2004, 78, 10390–10398. [Google Scholar] [CrossRef] [PubMed]
- Sciortino, M.T.; Suzuki, M.; Taddeo, B.; Roizman, B. RNAs extracted from herpes simplex virus 1 virions: Apparent selectivity of viral but not cellular RNAs packaged in virions. J. Virol. 2001, 75, 8105–8116. [Google Scholar] [CrossRef] [PubMed]
- Raoult, D.; Audic, S.; Robert, C.; Abergel, C.; Renesto, P.; Ogata, H.; La Scola, B.; Suzan, M.; Claverie, J.M. The 1.2-megabase genome sequence of mimivirus. Science 2004, 306, 1344–1350. [Google Scholar] [CrossRef] [PubMed]
- Suzan-Monti, M.; La Scola, B.; Raoult, D. Genomic and evolutionary aspects of mimivirus. Virus Res. 2006, 117, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Sciortino, M.T.; Taddeo, B.; Poon, A.P.; Mastino, A.; Roizman, B. Of the three tegument proteins that package mRNA in herpes simplex virions, one (vp22) transports the mRNA to uninfected cells for expression prior to viral infection. Proc. Natl. Acad. Sci. USA 2002, 99, 8318–8323. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Bresnahan, W.; Shenk, T. Human cytomegalovirus encodes a highly specific RANTES decoy receptor. Proc. Natl. Acad. Sci. USA 2004, 101, 16642–16647. [Google Scholar] [CrossRef] [PubMed]
- Schramm, B.; Krijnse Locker, J. Cytoplasmic organization of poxvirus DNA replication. Traffic 2005, 6, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Feore, S.M.; Bennett, M.; Chantrey, J.; Jones, T.; Baxby, D.; Begon, M. The effect of cowpox virus infection on fecundity in bank voles and wood mice. Proc. Biol. Sci. 1997, 264, 1457–1461. [Google Scholar] [CrossRef] [PubMed]
- Kurth, A.; Wibbelt, G.; Gerber, H.P.; Petschaelis, A.; Pauli, G.; Nitsche, A. Rat-to-elephant-to-human transmission of cowpox virus. Emerg. Infect. Dis. 2008, 14, 670–671. [Google Scholar] [CrossRef] [PubMed]
- Vorou, R.M.; Papavassiliou, V.G.; Pierroutsakos, I.N. Cowpox virus infection: An emerging health threat. Curr. Opin. Infect. Dis. 2008, 21, 153–156. [Google Scholar] [CrossRef] [PubMed]
- Burgess, H.M.; Mohr, I. Cellular 5′-3′ mRNA exonuclease xrn1 controls double-stranded RNA accumulation and anti-viral responses. Cell Host Microbe 2015, 17, 332–344. [Google Scholar] [CrossRef] [PubMed]
- Doellinger, J.; Schaade, L.; Nitsche, A. Comparison of the cowpox virus and vaccinia virus mature virion proteome: Analysis of the species- and strain-specific proteome. PLoS ONE 2015, 10, e0141527. [Google Scholar] [CrossRef] [PubMed]
- Peters, N.E.; Ferguson, B.J.; Mazzon, M.; Fahy, A.S.; Krysztofinska, E.; Arribas-Bosacoma, R.; Pearl, L.H.; Ren, H.; Smith, G.L. A mechanism for the inhibition of DNA-pk-mediated DNA sensing by a virus. PLoS Pathog. 2013, 9, e1003649. [Google Scholar] [CrossRef] [PubMed]
- Earl, P.L.; Moss, B.; Wyatt, L.S.; Carroll, M.W. Generation of recombinant vaccinia viruses. Curr. Protoc. Mol. Biol. 2001. [Google Scholar] [CrossRef]
- Resch, W.; Hixson, K.K.; Moore, R.J.; Lipton, M.S.; Moss, B. Protein composition of the vaccinia virus mature virion. Virology 2007, 358, 233–247. [Google Scholar] [CrossRef] [PubMed]
- Wolfel, R.; Paweska, J.T.; Petersen, N.; Grobbelaar, A.A.; Leman, P.A.; Hewson, R.; Georges-Courbot, M.C.; Papa, A.; Gunther, S.; Drosten, C. Virus detection and monitoring of viral load in Crimean-Congo hemorrhagic fever virus patients. Emerg. Infect. Dis. 2007, 13, 1097–1100. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, K.; Nitsche, A. Multicolour, multiplex real-time PCR assay for the detection of human-pathogenic poxviruses. Mol. Cell. Probes 2010, 24, 110–113. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Reynolds, S.E.; Martens, C.A.; Bruno, D.P.; Porcella, S.F.; Moss, B. Expression profiling of the intermediate and late stages of poxvirus replication. J. Virol. 2011, 85, 9899–9908. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Bruno, D.P.; Martens, C.A.; Porcella, S.F.; Moss, B. Simultaneous high-resolution analysis of vaccinia virus and host cell transcriptomes by deep RNA sequencing. Proc. Natl. Acad. Sci. USA 2010, 107, 11513–11518. [Google Scholar] [CrossRef] [PubMed]
- Yuen, L.; Moss, B. Oligonucleotide sequence signaling transcriptional termination of vaccinia virus early genes. Proc. Natl. Acad. Sci. USA 1987, 84, 6417–6421. [Google Scholar] [CrossRef] [PubMed]
- Hansen, K.D.; Brenner, S.E.; Dudoit, S. Biases in Illumina transcriptome sequencing caused by random hexamer priming. Nucleic Acids Res. 2010, 38, e131. [Google Scholar] [CrossRef] [PubMed]
- Costa, V.; Angelini, C.; de Feis, I.; Ciccodicola, A. Uncovering the complexity of transcriptomes with RNA-Seq. J. Biomed. Biotechnol. 2010, 2010, 853916. [Google Scholar] [CrossRef] [PubMed]
- Tolonen, N.; Doglio, L.; Schleich, S.; Krijnse Locker, J. Vaccinia virus DNA replication occurs in endoplasmic reticulum-enclosed cytoplasmic mini-nuclei. Mol. Biol. Cell 2001, 12, 2031–2046. [Google Scholar] [CrossRef] [PubMed]
- Jackson, R.; Standart, N. The awesome power of ribosome profiling. RNA 2015, 21, 652–654. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Ranganathan, P.N.; Feitelson, M.A. Direct synthesis of full length p53 cDNA from frozen liver. Biochem. Mol. Biol. Int. 1997, 41, 861–867. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.W.; Katsafanas, G.C.; Liu, R.K.; Wyatt, L.S.; Moss, B. Poxvirus decapping enzymes enhance virulence by preventing the accumulation of dsRNA and the induction of innate antiviral responses. Cell Host Microbe 2015, 17, 320–331. [Google Scholar] [CrossRef] [PubMed]
- Rubins, K.H.; Hensley, L.E.; Bell, G.W.; Wang, C.; Lefkowitz, E.J.; Brown, P.O.; Relman, D.A. Comparative analysis of viral gene expression programs during poxvirus infection: A transcriptional map of the vaccinia and monkeypox genomes. PLoS ONE 2008, 3, e2628. [Google Scholar] [CrossRef] [PubMed]
- Assarsson, E.; Greenbaum, J.A.; Sundstrom, M.; Schaffer, L.; Hammond, J.A.; Pasquetto, V.; Oseroff, C.; Hendrickson, R.C.; Lefkowitz, E.J.; Tscharke, D.C.; et al. Kinetic analysis of a complete poxvirus transcriptome reveals an immediate-early class of genes. Proc. Natl. Acad. Sci. USA 2008, 105, 2140–2145. [Google Scholar] [CrossRef] [PubMed]
- Cochran, M.A.; Puckett, C.; Moss, B. In vitro mutagenesis of the promoter region for a vaccinia virus gene: Evidence for tandem early and late regulatory signals. J. Virol. 1985, 54, 30–37. [Google Scholar] [PubMed]
- Garces, J.; Masternak, K.; Kunz, B.; Wittek, R. Reactivation of transcription from a vaccinia virus early promoter late in infection. J. Virol. 1993, 67, 5394–5401. [Google Scholar] [PubMed]
- Masternak, K.; Wittek, R. Cis- and trans-acting elements involved in reactivation of vaccinia virus early transcription. J. Virol. 1996, 70, 8737–8746. [Google Scholar] [PubMed]
- Rice, A.P.; Roberts, B.E. Vaccinia virus induces cellular mRNA degradation. J. Virol. 1983, 47, 529–539. [Google Scholar] [PubMed]

| Gene CPXV GRI-90 | Gene CPXV BR | Gene VACV Cop | Description a | Function | Host b | Expression c |
|---|---|---|---|---|---|---|
| none | 002/228 | no ORF | Uncharacterized protein | unknown | H/R | unknown |
| D5L/I1R | 009/222 | C16L/B22R | Inhibitor host DNA-PK d | immune evasion | H/R | unknown |
| D6L | 010 | no ORF | Uncharacterized protein | unknown | H | unknown |
| D10L | 012 | no ORF | Uncharacterized protein | unknown | H/R | unknown |
| C6L | 022 | C10L | Uncharacterized protein | unknown | H | early |
| C8L | 024 | no ORF | Interleukin 18-binding | immune evasion | H/R | early |
| C10L | 025 | no ORF | Uncharacterized protein | unknown | H/R | early |
| C14L | 030 | C6L | Uncharacterized protein | unknown | H/R | early |
| P2L | 040 | M2L | Uncharacterized protein | unknown | H | early |
| M1L | 041 | K1L | Interferon antagonist, hr | immune evasion | R | early |
| F3L | 069 | E3L | dsRNA-binding protein, hr | immune evasion | H/R | early |
| F4L | 070 | E4L | Viral RNA pol 30 kDa | transcription | H | early |
| J5R | 114 | H5R | Late transcription factor 4 | transcription | H/R | early |
| A34 | 168 | A33R | EEV glycoprotein | entry | H | early |
| A36R | 171 | A35R | Inhibitor MHC class II antigen presentation | immune evasion | H/R | early |
| A47R | 182 | A44R | 3β-Hydroxysteroid-dehydrogenase/Δ5-4-isomerase | immune evasion | H | early |
| B8R | 203 | B9R | Uncharacterized protein | unknown | H/R | intermediate |
| B13R | 208 | no ORF | Inhibitor of host NF-κB | immune evasion | H/R | early |
| D1L/I5R | vCCI | B29R/C23L | Chemokine-binding protein | immune evasion | H/R | early |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grossegesse, M.; Doellinger, J.; Haldemann, B.; Schaade, L.; Nitsche, A. A Next-Generation Sequencing Approach Uncovers Viral Transcripts Incorporated in Poxvirus Virions. Viruses 2017, 9, 296. https://doi.org/10.3390/v9100296
Grossegesse M, Doellinger J, Haldemann B, Schaade L, Nitsche A. A Next-Generation Sequencing Approach Uncovers Viral Transcripts Incorporated in Poxvirus Virions. Viruses. 2017; 9(10):296. https://doi.org/10.3390/v9100296
Chicago/Turabian StyleGrossegesse, Marica, Joerg Doellinger, Berit Haldemann, Lars Schaade, and Andreas Nitsche. 2017. "A Next-Generation Sequencing Approach Uncovers Viral Transcripts Incorporated in Poxvirus Virions" Viruses 9, no. 10: 296. https://doi.org/10.3390/v9100296