Rodent Papillomaviruses
Abstract
:1. Introduction
“Our attention was recently called to a disease occurring in wild cottontail rabbits in northwestern Iowa. Rabbits shot there by hunters were said to have numerous horn-like protuberances on the skin over various parts of their bodies. The animals were referred to popularly as “horned” or “warty” rabbits”.Shope and Hurst [1].
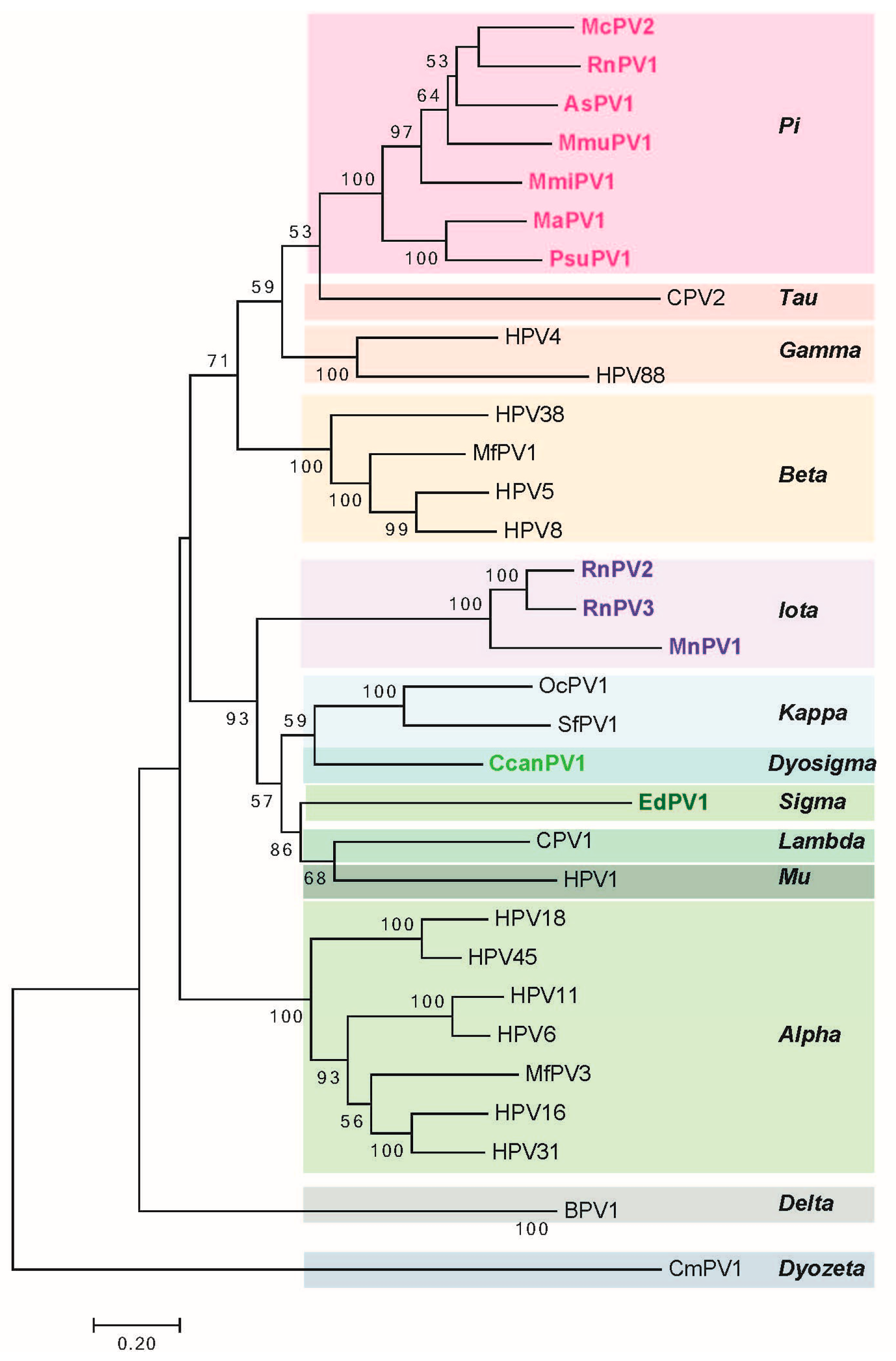
2. Genomic Analysis of Rodent Papillomaviruses
3. Pathogenesis by Rodent Papillomaviruses
3.1. Mastomys-Associated Papillomaviruses
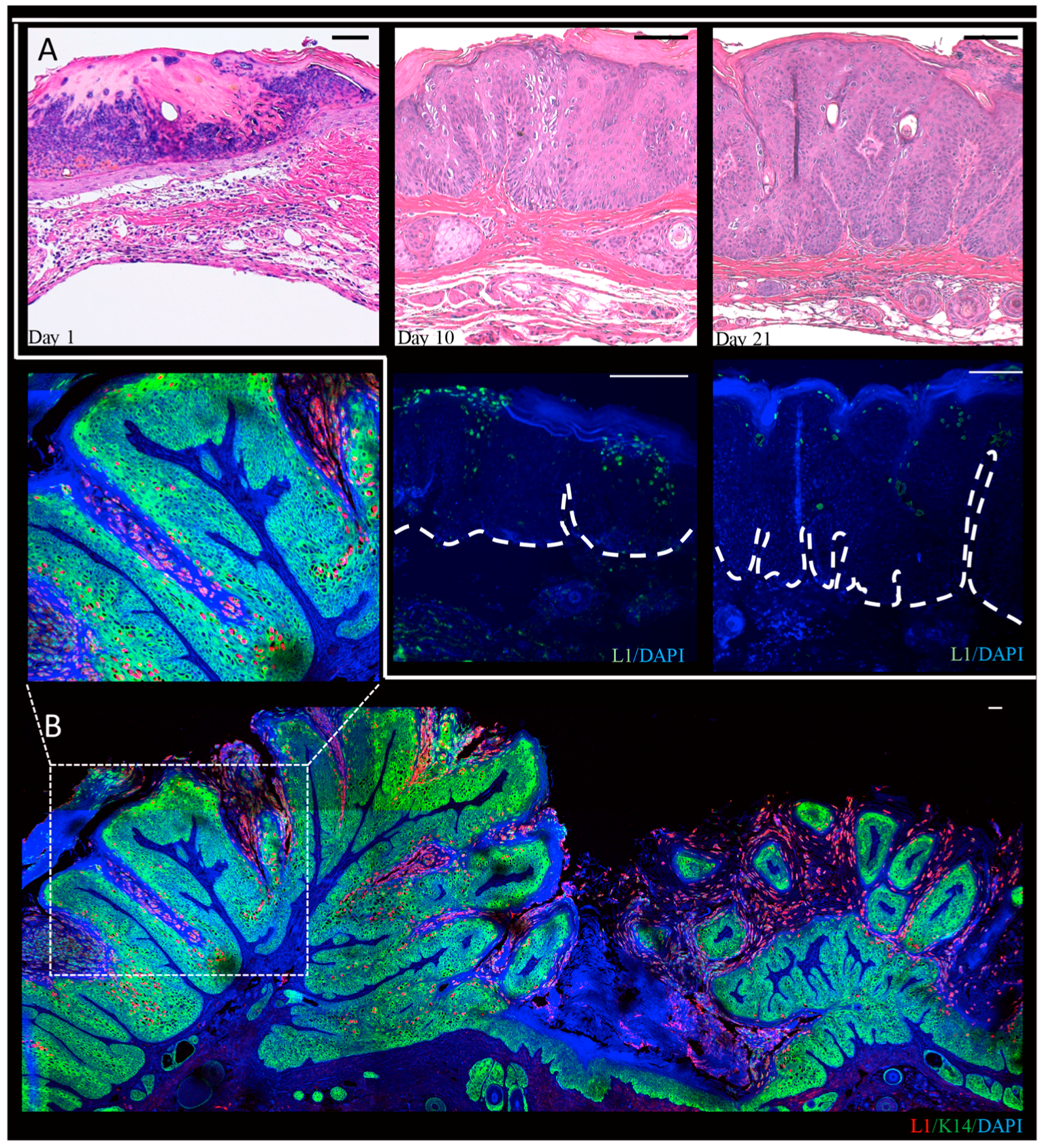
3.2. Mus Musculus-Associated Papillomaviruses
3.2.1. Discovery of the First Papillomavirus to Infect the Standard Laboratory Mouse Strain, Mus musculus
3.2.2. T-Cell Deficient Strains are Susceptible to MmuPV1 Infection
3.2.3. MmuPV1 Infection in Hairless Strains
3.2.4. Role of Ultraviolet Radiation (UVR) in MmuPV1-Associated Disease
3.3. Other Rodent Papillomaviruses
4. Insights into Vaccine Development
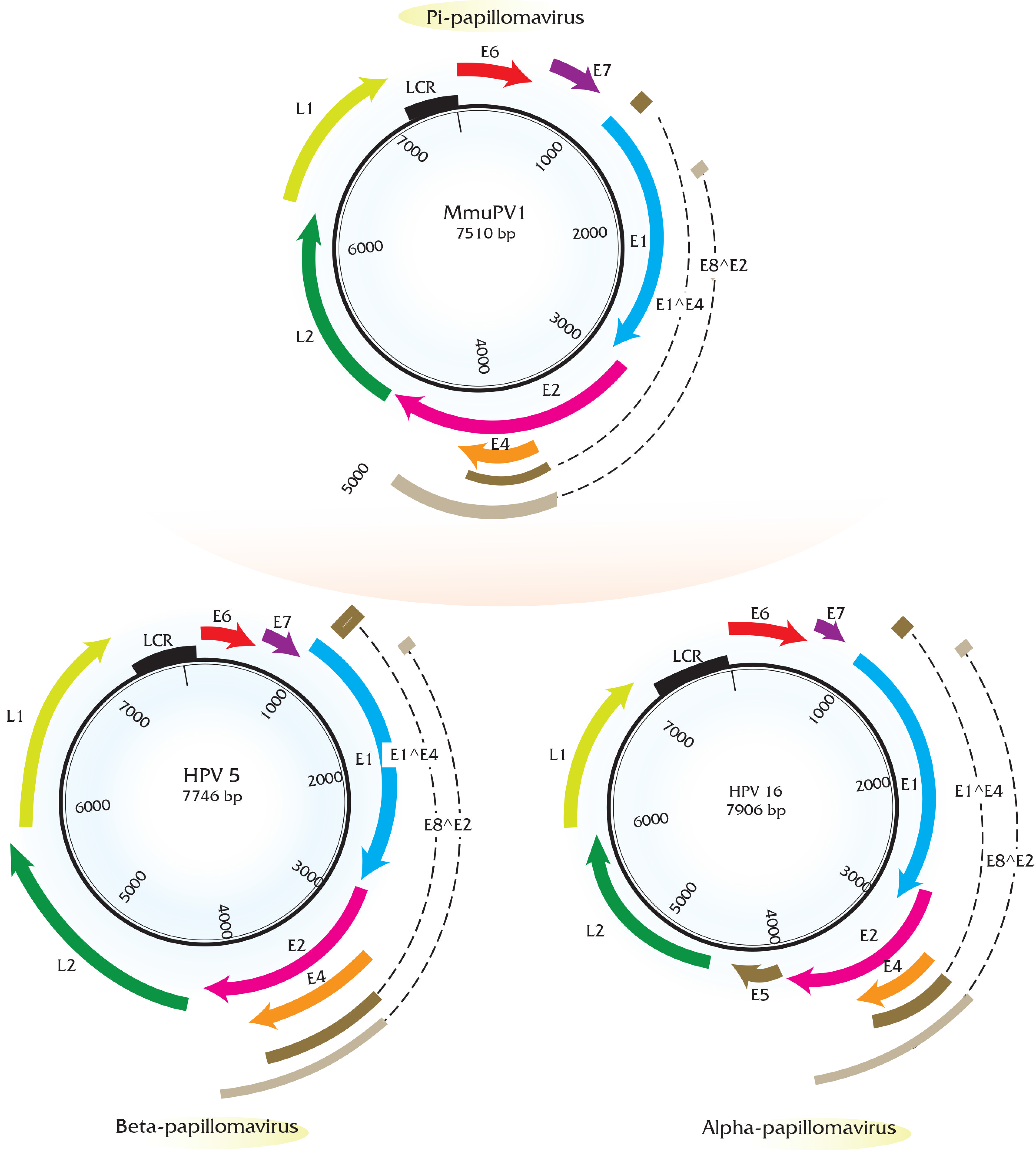
5. Transcript Maps of Rodent Papillomaviruses
6. Roles of Rodent PV’s E6 and E7 Proteins
7. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Shope, R.E.; Hurst, E.W. Infectious papillomatosis of rabbits: With a note on the histopathology. J. Exp. Med. 1933, 58, 607–624. [Google Scholar] [CrossRef] [PubMed]
- Boshart, M.; Gissmann, L.; Ikenberg, H.; Kleinheinz, A.; Scheurlen, W.; zur Hausen, H. A new type of papillomavirus DNA, its presence in genital cancer biopsies and in cell lines derived from cervical cancer. EMBO J. 1984, 3, 1151–1157. [Google Scholar] [PubMed]
- Durst, M.; Gissmann, L.; Ikenberg, H.; zur Hausen, H. A papillomavirus DNA from a cervical carcinoma and its prevalence in cancer biopsy samples from different geographic regions. Proc. Natl. Acad. Sci USA 1983, 80, 3812–3815. [Google Scholar] [CrossRef] [PubMed]
- DiMaio, D. Nuns, warts, viruses, and cancer. Yale J. Biol. Med. 2015, 88, 127–129. [Google Scholar] [PubMed]
- Van Doorslaer, K.; Li, Z.; Xirasagar, S.; Maes, P.; Kaminsky, D.; Liou, D.; Sun, Q.; Kaur, R.; Huyen, Y.; McBride, A.A. The papillomavirus episteme: A major update to the papillomavirus sequence database. Nucleic Acids Res. 2017, 45, D499–D506. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin-Drubin, M.E.; Meyers, J.; Munger, K. Cancer associated human papillomaviruses. Curr. Opin. Virol. 2012, 2, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Jablonska, S.; Majewski, S.; Obalek, S.; Orth, G. Cutaneous warts. Clin. Dermatol. 1997, 15, 309–319. [Google Scholar] [CrossRef]
- Jablonska, S.; Orth, G.; Obalek, S.; Croissant, O. Cutaneous warts Clinical, histologic, and virologic correlations. Clin. Dermatol. 1985, 3, 71–82. [Google Scholar] [CrossRef]
- Tschandl, P.; Rosendahl, C.; Kittler, H. Cutaneous human papillomavirus infection: Manifestations and diagnosis. Curr. Probl. Dermatol. 2014, 45, 92–97. [Google Scholar] [PubMed]
- Lebwohl, M.G.; Rosen, T.; Stockfleth, E. The role of human papillomavirus in common skin conditions: Current viewpoints and therapeutic options. Cutis 2010, 86. [Google Scholar]
- Mammas, I.N.; Spandidos, D.A.; Sourvinos, G. Genomic diversity of human papillomaviruses (HPV) and clinical implications: An overview in adulthood and childhood. Infect. Genet. Evol. 2014, 21, 220–226. [Google Scholar] [CrossRef] [PubMed]
- Van Haalen, F.M.; Bruggink, S.C.; Gussekloo, J.; Assendelft, W.J.; Eekhof, J.A. Warts in primary schoolchildren: Prevalence and relation with environmental factors. Br. J. Dermatol. 2009, 161, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Akgul, B.; Cooke, J.C.; Storey, A. HPV-associated skin disease. J. Pathol. 2006, 208, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Astori, G.; Lavergne, D.; Benton, C.; Hockmayr, B.; Egawa, K.; Garbe, C.; de Villiers, E.M. Human papillomaviruses are commonly found in normal skin of immunocompetent hosts. J. Investig. Dermatol. 1998, 110, 752–755. [Google Scholar] [CrossRef] [PubMed]
- Gassenmaier, A.; Fuchs, P.; Schell, H.; Pfister, H. Papillomavirus DNA in warts of immunosuppressed renal allograft recipients. Arch. Dermatol. Res. 1986, 278, 219–223. [Google Scholar] [CrossRef] [PubMed]
- Antonsson, A.; Forslund, O.; Ekberg, H.; Sterner, G.; Hansson, B.G. The ubiquity and impressive genomic diversity of human skin papillomaviruses suggest a commensalic nature of these viruses. J. Virol. 2000, 74, 11636–11641. [Google Scholar] [CrossRef] [PubMed]
- Howley, P.M.; Pfister, H.J. Beta genus papillomaviruses and skin cancer. Virology 2015, 479–480, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Connolly, K.; Manders, P.; Earls, P.; Epstein, R.J. Papillomavirus-associated squamous skin cancers following transplant immunosuppression: One notch closer to control. Cancer Treat. Rev. 2014, 40, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Meyers, C.; Laimins, L.A. In vitro systems for the study and propagation of human papillomaviruses. Curr. Top. Microbiol. Immunol. 1994, 186, 199–215. [Google Scholar] [PubMed]
- Christensen, N.D.; Budgeon, L.R.; Cladel, N.M.; Hu, J. Recent advances in preclinical model systems for papillomaviruses. Virus Res. 2017, 231, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Gil da Costa, R.M.; Peleteiro, M.C.; Pires, M.A.; DiMaio, D. An update on canine, feline and bovine papillomaviruses. Transbound. Emerg. Dis. 2017, 64, 1371–1379. [Google Scholar] [CrossRef] [PubMed]
- Doorbar, J. Model systems of human papillomavirus-associated disease. J. Pathol. 2016, 238, 166–179. [Google Scholar] [CrossRef] [PubMed]
- Rector, A.; van Ranst, M. Animal papillomaviruses. Virology 2013, 445, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Peh, W.L.; Middleton, K.; Christensen, N.; Nicholls, P.; Egawa, K.; Sotlar, K.; Brandsma, J.; Percival, A.; Lewis, J.; Liu, W.J.; et al. Life cycle heterogeneity in animal models of human papillomavirus-associated disease. J. Virol. 2002, 76, 10401–10416. [Google Scholar] [CrossRef] [PubMed]
- Campo, M.S. Animal models of papillomavirus pathogenesis. Virus Res. 2002, 89, 249–261. [Google Scholar] [CrossRef]
- Munday, J.S.; Kiupel, M. Papillomavirus-associated cutaneous neoplasia in mammals. Vet. Pathol. 2010, 47, 254–264. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.Y.; Howley, P.M.; Levinson, A.D.; Seeburg, P.H. The primary structure and genetic organization of the bovine papillomavirus type 1 genome. Nature 1982, 299, 529–534. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, W.D.; Olson, C.; Meinke, W. Quantitation of bovine papilloma viral DNA in viral-induced tumors. J. Virol. 1976, 17, 824–831. [Google Scholar] [PubMed]
- Olson, C. Cutaneous papillomatosis in cattle and other animals. Ann. N. Y. Acad. Sci. 1963, 108, 1042–1056. [Google Scholar] [CrossRef] [PubMed]
- Nasir, L.; Campo, M.S. Bovine papillomaviruses: Their role in the aetiology of cutaneous tumours of bovids and equids. Vet. Dermatol. 2008, 19, 243–254. [Google Scholar] [CrossRef] [PubMed]
- DiMaio, D.; Petti, L.M. The E5 proteins. Virology 2013, 445, 99–114. [Google Scholar] [CrossRef] [PubMed]
- Munday, J.S. Bovine and human papillomaviruses: A comparative review. Vet. Pathol. 2014, 51, 1063–1075. [Google Scholar] [CrossRef] [PubMed]
- Venuti, A.; Paolini, F.; Nasir, L.; Corteggio, A.; Roperto, S.; Campo, M.S.; Borzacchiello, G. Papillomavirus E5: The smallest oncoprotein with many functions. Mol. Cancer 2011, 10, 140. [Google Scholar] [CrossRef] [PubMed]
- Lange, C.E.; Favrot, C. Canine papillomaviruses. Vet. Clin. North Am. Small Anim. Pract. 2011, 41, 1183–1195. [Google Scholar] [CrossRef] [PubMed]
- Nicholls, P.K.; Stanley, M.A. Canine papillomavirus—A centenary review. J. Comp. Pathol. 1999, 120, 219–233. [Google Scholar] [CrossRef] [PubMed]
- Stanley, M.A. Progress in prophylactic and therapeutic vaccines for human papillomavirus infection. Expert Rev. Vaccines 2003, 2, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Frazer, I.H. The role of vaccines in the control of STDs: HPV vaccines. Genitourin. Med. 1996, 72, 398–403. [Google Scholar] [CrossRef] [PubMed]
- Nicholls, P.K.; Stanley, M.A. The immunology of animal papillomaviruses. Vet. Immunol. Immunopathol. 2000, 73, 101–127. [Google Scholar] [CrossRef]
- Frazer, I.H. Development and implementation of papillomavirus prophylactic vaccines. J. Immunol. 2014, 192, 4007–4011. [Google Scholar] [CrossRef] [PubMed]
- Rous, P.; Kidd, J.G. The carcinogenic effect of a papilloma virus on the tarred skin of rabbits: I. Description of the phenomenon. J. Exp. Med. 1938, 67, 399–428. [Google Scholar] [CrossRef] [PubMed]
- Kerr, P.J.; Donnelly, T.M. Viral infections of rabbits. Vet. Clin. North Am. Small Anim. Pract. 2013, 16, 437–468. [Google Scholar] [CrossRef] [PubMed]
- Christensen, N.D. Cottontail rabbit papillomavirus (CRPV) model system to test antiviral and immunotherapeutic strategies. Antivir. Chem. Chemother. 2005, 16, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Breitburd, F.; Salmon, J.; Orth, G. The rabbit viral skin papillomas and carcinomas: A model for the immunogenetics of HPV-associated carcinogenesis. Clin. Dermatol. 1997, 15, 237–247. [Google Scholar] [CrossRef]
- Peng, X.; Knouse, J.A.; Hernon, K.M. Rabbit models for studying human infectious diseases. Comp. Med. 2015, 65, 499–507. [Google Scholar] [PubMed]
- Doorbar, J. The papillomavirus life cycle. J. Clin. Virol. 2005, 32 (Suppl. 1), S7–S15. [Google Scholar] [CrossRef] [PubMed]
- Zur Hausen, H.; de Villiers, E.M. Cancer “causation” by infections—Individual contributions and synergistic networks. Semin. Oncol. 2014, 41, 860–875. [Google Scholar] [CrossRef] [PubMed]
- Schulz, E.; Gottschling, M.; Ulrich, R.G.; Richter, D.; Stockfleth, E.; Nindl, I. Isolation of three novel rat and mouse papillomaviruses and their genomic characterization. PLoS ONE 2012, 7, e47164. [Google Scholar] [CrossRef] [PubMed]
- Rogovskyy, A.S.; Chen, Z.; Burk, R.D.; Bankhead, T. Characterization of the north american beaver (castor canadensis) papillomavirus genome. Vet. Microbiol. 2014, 168, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Rector, A.; Tachezy, R.; van Doorslaer, K.; MacNamara, T.; Burk, R.D.; Sundberg, J.P.; van Ranst, M. Isolation and cloning of a papillomavirus from a north american porcupine by using multiply primed rolling-circle amplification: The erethizon dorsatum papillomavirus type 1. Virology 2005, 331, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, T.; Maeda, H.; Kameyama, Y.; Moriyama, M.; Kanai, S.; Kurata, T. Presence of a novel hamster oral papillomavirus in dysplastic lesions of hamster lingual mucosa induced by application of dimethylbenzanthracene and excisional wounding: Molecular cloning and complete nucleotide sequence. J. Gen. Virol. 1997, 78 Pt 5, 1087–1093. [Google Scholar] [CrossRef] [PubMed]
- Nafz, J.; Schafer, K.; Chen, S.F.; Bravo, I.G.; Ibberson, M.; Nindl, I.; Stockfleth, E.; Rosl, F. A novel rodent papillomavirus isolated from anogenital lesions in its natural host. Virology 2008, 374, 186–197. [Google Scholar] [CrossRef] [PubMed]
- Van Doorslaer, K.; Rector, A.; Jenson, A.B.; Sundberg, J.P.; van Ranst, M.; Ghim, S.J. Complete genomic characterization of a murine papillomavirus isolated from papillomatous lesions of a european harvest mouse (micromys minutus). J. Gen. Virol. 2007, 88, 1484–1488. [Google Scholar] [CrossRef] [PubMed]
- Sundberg, J.P.; O’Banion, M.K.; Shima, A.; Knupp, C.; Reichmann, M.E. Papillomas and carcinomas associated with a papillomavirus in european harvest mice (micromys minutus). Vet. Pathol. 1988, 25, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Ingle, A.; Ghim, S.; Joh, J.; Chepkoech, I.; Bennett Jenson, A.; Sundberg, J.P. Novel laboratory mouse papillomavirus (MusPV) infection. Vet. Pathol. 2011, 48, 500–505. [Google Scholar] [CrossRef] [PubMed]
- Joh, J.; Jenson, A.B.; Proctor, M.; Ingle, A.; Silva, K.A.; Potter, C.S.; Sundberg, J.P.; Ghim, S.J. Molecular diagnosis of a laboratory mouse papillomavirus (MusPV). Exp. Mol. Pathol. 2012, 93, 416–421. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.H.; Tachezy, R.; van Ranst, M.; Chan, S.Y.; Bernard, H.U.; Burk, R.D. The mastomys natalensis papillomavirus: Nucleotide sequence, genome organization, and phylogenetic relationship of a rodent papillomavirus involved in tumorigenesis of cutaneous epithelia. Virology 1994, 198, 534–541. [Google Scholar] [CrossRef] [PubMed]
- Amtmann, E.; Volm, M.; Wayss, K. Tumour induction in the rodent mastomys natalensis by activation of endogenous papilloma virus genomes. Nature 1984, 308, 291–292. [Google Scholar] [CrossRef] [PubMed]
- Kocjan, B.J.; Hosnjak, L.; Racnik, J.; Zadravec, M.; Poljak, M. Complete genome sequence of phodopus sungorus papillomavirus type 1 (PSPV1), a novel member of the pipapillomavirus genus, isolated from a siberian hamster. Genome Announc. 2014, 2, e00311–e00314. [Google Scholar] [CrossRef] [PubMed]
- Joh, J.; Jenson, A.B.; King, W.; Proctor, M.; Ingle, A.; Sundberg, J.P.; Ghim, S.J. Genomic analysis of the first laboratory-mouse papillomavirus. J. Gen. Virol. 2011, 92, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Stevens, H.; Rector, A.; van Ranst, M. Multiply primed rolling-circle amplification method for the amplification of circular DNA viruses. Cold Spring Harb. Protoc. 2010, 2010, pdb prot5415. [Google Scholar] [CrossRef] [PubMed]
- Rector, A.; Tachezy, R.; van Ranst, M. A sequence-independent strategy for detection and cloning of circular DNA virus genomes by using multiply primed rolling-circle amplification. J. Virol. 2004, 78, 4993–4998. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Cladel, N.M.; Budgeon, L.R.; Balogh, K.K.; Christensen, N.D. The mouse papillomavirus infection model. Viruses 2017, 9, 246. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.M.; Baker, C.C. Papillomavirus genome structure, expression, and post-transcriptional regulation. Front. Biosci. 2006, 11, 2286–2302. [Google Scholar] [CrossRef] [PubMed]
- Doorbar, J. Papillomavirus life cycle organization and biomarker selection. Dis. Markers 2007, 23, 297–313. [Google Scholar] [CrossRef] [PubMed]
- Doorbar, J.; Quint, W.; Banks, L.; Bravo, I.G.; Stoler, M.; Broker, T.R.; Stanley, M.A. The biology and life-cycle of human papillomaviruses. Vaccine 2012, 30 (Suppl. 5), F55–F70. [Google Scholar] [CrossRef] [PubMed]
- Howley, P.M. Warts, cancer and ubiquitylation: Lessons from the papillomaviruses. Trans. Am. Clin. Clim. Assoc. 2006, 117, 113–126; discussion 126–127. [Google Scholar]
- Bernard, H.U. Coevolution of papillomaviruses with human populations. Trends Microbiol. 1994, 2, 140–143. [Google Scholar] [CrossRef]
- Garcia-Vallve, S.; Alonso, A.; Bravo, I.G. Papillomaviruses: Different genes have different histories. Trends Microbiol. 2005, 13, 514–521. [Google Scholar] [CrossRef] [PubMed]
- Gottschling, M.; Goker, M.; Stamatakis, A.; Bininda-Emonds, O.R.; Nindl, I.; Bravo, I.G. Quantifying the phylodynamic forces driving papillomavirus evolution. Mol. Biol. Evol. 2011, 28, 2101–2113. [Google Scholar] [CrossRef] [PubMed]
- Schulz, E.; Gottschling, M.; Wibbelt, G.; Stockfleth, E.; Nindl, I. Isolation and genomic characterization of the first norway rat (rattus norvegicus) papillomavirus and its phylogenetic position within pipapillomavirus, primarily infecting rodents. J. Gen. Virol. 2009, 90, 2609–2614. [Google Scholar] [CrossRef] [PubMed]
- Egawa, N.; Egawa, K.; Griffin, H.; Doorbar, J. Human papillomaviruses; epithelial tropisms, and the development of neoplasia. Viruses 2015, 7, 3863–3890. [Google Scholar] [CrossRef] [PubMed]
- De Villiers, E.M.; Fauquet, C.; Broker, T.R.; Bernard, H.U.; zur Hausen, H. Classification of papillomaviruses. Virology 2004, 324, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- De Villiers, E.M. Cross-Roads in the classification of papillomaviruses. Virology 2013, 445, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Bergvall, M.; Melendy, T.; Archambault, J. The e1 proteins. Virology 2013, 445, 35–56. [Google Scholar] [CrossRef] [PubMed]
- McBride, A.A. The papillomavirus E2 proteins. Virology 2013, 445, 57–79. [Google Scholar] [CrossRef] [PubMed]
- Doorbar, J. The E4 protein; structure, function and patterns of expression. Virology 2013, 445, 80–98. [Google Scholar] [CrossRef] [PubMed]
- Orth, G. Host defenses against human papillomaviruses: Lessons from epidermodysplasia verruciformis. Curr. Top. Microbiol. Immunol. 2008, 321, 59–83. [Google Scholar] [PubMed]
- Roman, A.; Munger, K. The papillomavirus E7 proteins. Virology 2013, 445, 138–168. [Google Scholar] [CrossRef] [PubMed]
- Vande Pol, S.B.; Klingelhutz, A.J. Papillomavirus E6 oncoproteins. Virology 2013, 445, 115–137. [Google Scholar] [CrossRef] [PubMed]
- Rudolph, R.; Hundeiker, M. “Keratoacanthomas” in mastomys natalensis (author’s transl). Arch. Dermatol. Res. 1975, 254, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Rudolph, R.; Thiel, W. Pathological anatomy and histology of spontaneous, epithelial skin tumors in mastomys natalensis. Z. Fur Vet. Reihe A 1976, 23, 429–441. [Google Scholar]
- Muller, H.; Gissmann, L. Mastomys natalensis papilloma virus (MNPV), the causative agent of epithelial proliferations: Characterization of the virus particle. J. Gen. Virol. 1978, 41, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Reinacher, M.; Muller, H.; Thiel, W.; Rudolph, R.L. Localization of papillomavirus and virus-specific antigens in the skin of tumor-bearing mastomys natalensis (gra giessen). Med. Microbiol. Immunol. 1978, 165, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Rudolph, R.L. Neoplasms of the skin in a wild-colored inbred strain of mastomys natalensis (wsa giessen). Vet. Pathol. 1980, 17, 600–613. [Google Scholar] [CrossRef] [PubMed]
- Kruppa, T.F.; Iglauer, F.; Ihnen, E.; Miller, K.; Kunstyr, I. Mastomys natalensis or mastomys coucha. Correct species designation in animal experiments. Trop. Med. Parasitol. 1990, 41, 219–220. [Google Scholar] [PubMed]
- Schafer, K.; Neumann, J.; Waterboer, T.; Rosl, F. Serological markers for papillomavirus infection and skin tumour development in the rodent model mastomys coucha. J. Gen. Virol. 2011, 92, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Vinzon, S.E.; Braspenning-Wesch, I.; Muller, M.; Geissler, E.K.; Nindl, I.; Grone, H.J.; Schafer, K.; Rosl, F. Protective vaccination against papillomavirus-induced skin tumors under immunocompetent and immunosuppressive conditions: A preclinical study using a natural outbred animal model. PLoS Pathog. 2014, 10, e1003924. [Google Scholar] [CrossRef] [PubMed]
- Nafz, J.; Kohler, A.; Ohnesorge, M.; Nindl, I.; Stockfleth, E.; Rosl, F. Persistence of mastomys natalensis papillomavirus in multiple organs identifies novel targets for infection. J. Gen. Virol. 2007, 88, 2670–2678. [Google Scholar] [CrossRef] [PubMed]
- Blanpain, C.; Fuchs, E. Epidermal stem cells of the skin. Annu. Rev. Cell. Dev. Biol. 2006, 22, 339–373. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, A.; Rochat, A.; Zeltner, R.; Borenstein, L.; Barrandon, Y.; Wettstein, F.O.; Iftner, T. The primary target cells of the high-risk cottontail rabbit papillomavirus colocalize with hair follicle stem cells. J. Virol. 1996, 70, 1912–1922. [Google Scholar] [PubMed]
- Maglennon, G.A.; Doorbar, J. The biology of papillomavirus latency. Open Virol. J. 2012, 6, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Siegsmund, M.; Wayss, K.; Amtmann, E. Activation of latent papillomavirus genomes by chronic mechanical irritation. J. Gen. Virol. 1991, 72 Pt 11, 2787–2789. [Google Scholar] [CrossRef] [PubMed]
- Wayss, K.; Reyes-Mayes, D.; Volm, M. Chemical carcinogenesis by the two-stage protocol in the skin mastomys natalensis (muridae) using topical initiation with 7,12-dimethylbenz(a)anthracene and topical promotion with 12-0-tetradecanoylphorbol-13-acetate. Virchows Arch. B Cell. Pathol. Incl. Mol. Pathol. 1981, 38, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Helfrich, I.; Chen, M.; Schmidt, R.; Furstenberger, G.; Kopp-Schneider, A.; Trick, D.; Grone, H.J.; Zur Hausen, H.; Rosl, F. Increased incidence of squamous cell carcinomas in mastomys natalensis papillomavirus e6 transgenic mice during two-stage skin carcinogenesis. J. Virol. 2004, 78, 4797–4805. [Google Scholar] [CrossRef] [PubMed]
- Hasche, D.; Stephan, S.; Savelyeva, L.; Westermann, F.; Rosl, F.; Vinzon, S.E. Establishment of an immortalized skin keratinocyte cell line derived from the animal model mastomys coucha. PLoS ONE 2016, 11, e0161283. [Google Scholar] [CrossRef] [PubMed]
- Salvermoser, M.; Chotewutmontri, S.; Braspenning-Wesch, I.; Hasche, D.; Rosl, F.; Vinzon, S.E. Transcriptome analysis of mastomys natalensis papillomavirus in productive lesions after natural infection. J. Gen. Virol. 2016, 97, 1658–1669. [Google Scholar] [CrossRef] [PubMed]
- Handisurya, A.; Day, P.M.; Thompson, C.D.; Buck, C.B.; Pang, Y.Y.; Lowy, D.R.; Schiller, J.T. Characterization of mus musculus papillomavirus 1 infection in situ reveals an unusual pattern of late gene expression and capsid protein localization. J. Virol. 2013, 87, 13214–13225. [Google Scholar] [CrossRef] [PubMed]
- Sundberg, J.P.; Stearns, T.M.; Joh, J.; Proctor, M.; Ingle, A.; Silva, K.A.; Dadras, S.S.; Jenson, A.B.; Ghim, S.J. Immune status, strain background, and anatomic site of inoculation affect mouse papillomavirus (MmuPV1) induction of exophytic papillomas or endophytic trichoblastomas. PLoS ONE 2014, 9, e113582. [Google Scholar] [CrossRef] [PubMed]
- Florin, L.; Sapp, C.; Streeck, R.E.; Sapp, M. Assembly and translocation of papillomavirus capsid proteins. J. Virol. 2002, 76, 10009–10014. [Google Scholar] [CrossRef] [PubMed]
- Egawa, K.; Iftner, A.; Doorbar, J.; Honda, Y.; Iftner, T. Synthesis of viral DNA and late capsid protein L1 in parabasal spinous cell layers of naturally occurring benign warts infected with human papillomavirus type 1. Virology 2000, 268, 281–293. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.Y.; Majerciak, V.; Uberoi, A.; Kim, B.H.; Gotte, D.; Chen, X.; Cam, M.; Lambert, P.F.; Zheng, Z.M. The full transcription map of mouse papillomavirus type 1 (MmuPV1). PLoS Pathog. 2017. accepted. [Google Scholar]
- Cladel, N.M.; Budgeon, L.R.; Cooper, T.K.; Balogh, K.K.; Christensen, N.D.; Myers, R.; Majerciak, V.; Gotte, D.; Zheng, Z.M.; Hu, J. Mouse papillomavirus infections spread to cutaneous sites with progression to malignancy. J. Gen. Virol. 2017, 98, 2520–2529. [Google Scholar] [CrossRef] [PubMed]
- Nicholls, P.K.; Doorbar, J.; Moore, R.A.; Peh, W.; Anderson, D.M.; Stanley, M.A. Detection of viral DNA and E4 protein in basal keratinocytes of experimental canine oral papillomavirus lesions. Virology 2001, 284, 82–98. [Google Scholar] [CrossRef] [PubMed]
- Boxman, I.L.; Berkhout, R.J.; Mulder, L.H.; Wolkers, M.C.; Bouwes Bavinck, J.N.; Vermeer, B.J.; ter Schegget, J. Detection of human papillomavirus DNA in plucked hairs from renal transplant recipients and healthy volunteers. J. Investig. Dermatol. 1997, 108, 712–715. [Google Scholar] [CrossRef] [PubMed]
- Buck, C.B.; Day, P.M.; Trus, B.L. The papillomavirus major capsid protein L1. Virology 2013, 445, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.W.; Roden, R.B. L2, the minor capsid protein of papillomavirus. Virology 2013, 445, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Handisurya, A.; Day, P.M.; Thompson, C.D.; Buck, C.B.; Kwak, K.; Roden, R.B.; Lowy, D.R.; Schiller, J.T. Murine skin and vaginal mucosa are similarly susceptible to infection by pseudovirions of different papillomavirus classifications and species. Virology 2012, 433, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Day, P.M.; Thompson, C.D.; Lowy, D.R.; Schiller, J.T. The HPV16 and MusPV1 papillomaviruses initially interact with distinct host components on the basement membrane. Virology 2015, 481, 79–94. [Google Scholar] [CrossRef] [PubMed]
- Sapp, M.; Day, P.M. Structure, attachment and entry of polyoma- and papillomaviruses. Virology 2009, 384, 400–409. [Google Scholar] [CrossRef] [PubMed]
- Surviladze, Z.; Dziduszko, A.; Ozbun, M.A. Essential roles for soluble virion-associated heparan sulfonated proteoglycans and growth factors in human papillomavirus infections. PLoS Pathog. 2012, 8, e1002519. [Google Scholar] [CrossRef] [PubMed]
- Spoden, G.; Kuhling, L.; Cordes, N.; Frenzel, B.; Sapp, M.; Boller, K.; Florin, L.; Schelhaas, M. Human papillomavirus types 16, 18, and 31 share similar endocytic requirements for entry. J. Virol. 2013, 87, 7765–7773. [Google Scholar] [CrossRef] [PubMed]
- Flanagan, S.P. ‘Nude’, a new hairless gene with pleiotropic effects in the mouse. Genet. Res. 1966, 8, 295–309. [Google Scholar] [CrossRef] [PubMed]
- Pantelouris, E.M. Absence of thymus in a mouse mutant. Nature 1968, 217, 370–371. [Google Scholar] [CrossRef] [PubMed]
- Quigley, D.A.; Kandyba, E.; Huang, P.; Halliwill, K.D.; Sjolund, J.; Pelorosso, F.; Wong, C.E.; Hirst, G.L.; Wu, D.; Delrosario, R.; et al. Gene expression architecture of mouse dorsal and tail skin reveals functional differences in inflammation and cancer. Cell Rep. 2016, 16, 1153–1165. [Google Scholar] [CrossRef] [PubMed]
- Uberoi, A.; Yoshida, S.; Frazer, I.H.; Pitot, H.C.; Lambert, P.F. Role of ultraviolet radiation in papillomavirus-induced disease. PLoS Pathog. 2016, 12, e1005664. [Google Scholar] [CrossRef] [PubMed]
- Cladel, N.M.; Budgeon, L.R.; Balogh, K.K.; Cooper, T.K.; Hu, J.; Christensen, N.D. A novel pre-clinical murine model to study the life cycle and progression of cervical and anal papillomavirus infections. PLoS ONE 2015, 10, e0120128. [Google Scholar] [CrossRef] [PubMed]
- Cladel, N.M.; Budgeon, L.R.; Cooper, T.K.; Balogh, K.K.; Hu, J.; Christensen, N.D. Secondary infections, expanded tissue tropism, and evidence for malignant potential in immunocompromised mice infected with mus musculus papillomavirus 1 DNA and virus. J. Virol. 2013, 87, 9391–9395. [Google Scholar] [CrossRef] [PubMed]
- Cladel, N.M.; Budgeon, L.R.; Balogh, K.K.; Cooper, T.K.; Hu, J.; Christensen, N.D. Mouse papillomavirus MmuPV1 infects oral mucosa and preferentially targets the base of the tongue. Virology 2016, 488, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.W.; Jiang, R.; Peng, S.; Chang, Y.N.; Hung, C.F.; Roden, R.B. Immunologic control of mus musculus papillomavirus type 1. PLoS Pathog. 2015, 11, e1005243. [Google Scholar] [CrossRef] [PubMed]
- Bosma, M.; Schuler, W.; Bosma, G. The scid mouse mutant. Curr. Top. Microbiol. Immunol. 1988, 137, 197–202. [Google Scholar] [PubMed]
- Blunt, T.; Finnie, N.J.; Taccioli, G.E.; Smith, G.C.; Demengeot, J.; Gottlieb, T.M.; Mizuta, R.; Varghese, A.J.; Alt, F.W.; Jeggo, P.A.; et al. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell 1995, 80, 813–823. [Google Scholar] [CrossRef]
- Christianson, S.W.; Shultz, L.D.; Leiter, E.H. Adoptive transfer of diabetes into immunodeficient nod-scid/scid mice. Relative contributions of CD4+ and CD8+ T-cells from diabetic versus prediabetic NOD.NON-Thy-1a donors. Diabetes 1993, 42, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Nonoyama, S.; Smith, F.O.; Bernstein, I.D.; Ochs, H.D. Strain-dependent leakiness of mice with severe combined immune deficiency. J. Immunol. 1993, 150, 3817–3824. [Google Scholar] [PubMed]
- Shultz, L.D.; Schweitzer, P.A.; Christianson, S.W.; Gott, B.; Schweitzer, I.B.; Tennent, B.; McKenna, S.; Mobraaten, L.; Rajan, T.V.; Greiner, D.L.; et al. Multiple defects in innate and adaptive immunologic function in nod/ltsz-scid mice. J. Immunol. 1995, 154, 180–191. [Google Scholar] [PubMed]
- Handisurya, A.; Day, P.M.; Thompson, C.D.; Bonelli, M.; Lowy, D.R.; Schiller, J.T. Strain-specific properties and t cells regulate the susceptibility to papilloma induction by mus musculus papillomavirus 1. PLoS Pathog. 2014, 10, e1004314. [Google Scholar] [CrossRef] [PubMed]
- Nesnow, S.; Bergman, H.; Slaga, T.J. Comparison of the tumorigenic response of sencar and C57BL/6 mice to benzo(a)pyrene and the inter-experimental variability over a three-year period. Environ. Heal. Perspect. 1986, 68, 19–25. [Google Scholar] [CrossRef]
- Slaga, T.J. Sencar mouse skin tumorigenesis model versus other strains and stocks of mice. Environ. Heal. Perspect. 1986, 68, 27–32. [Google Scholar] [CrossRef]
- Joh, J.; Chilton, P.M.; Wilcher, S.A.; Zahin, M.; Park, J.; Proctor, M.L.; Ghim, S.J.; Jenson, A.B. T cell-mediated antitumor immune response eliminates skin tumors induced by mouse papillomavirus, MmuPV1. Exp. Mol. Pathol. 2017, 103, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.A.; Bevan, M.J. Effector and memory CTL differentiation. Annu. Rev. Immunol. 2007, 25, 171–192. [Google Scholar] [CrossRef] [PubMed]
- Wherry, E.J.; Ahmed, R. Memory CD8 t-cell differentiation during viral infection. J. Virol. 2004, 78, 5535–5545. [Google Scholar] [CrossRef] [PubMed]
- Whitmire, J.K. Induction and function of virus-specific CD4+ T cell responses. Virology 2011, 411, 216–228. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, Y.; Lu, B.; Gerard, C.; Iwasaki, A. CD8(+) T lymphocyte mobilization to virus-infected tissue requires CD4(+) T-cell help. Nature 2009, 462, 510–513. [Google Scholar] [CrossRef] [PubMed]
- Buller, R.M.; Holmes, K.L.; Hugin, A.; Frederickson, T.N.; Morse, H.C., 3rd. Induction of cytotoxic T-cell responses in vivo in the absence of CD4 helper cells. Nature 1987, 328, 77–79. [Google Scholar] [CrossRef] [PubMed]
- Ruedl, C.; Kopf, M.; Bachmann, M.F. CD8(+) T cells mediate CD40-independent maturation of dendritic cells in vivo. J. Exp. Med. 1999, 189, 1875–1884. [Google Scholar] [CrossRef] [PubMed]
- Yauch, L.E.; Prestwood, T.R.; May, M.M.; Morar, M.M.; Zellweger, R.M.; Peters, B.; Sette, A.; Shresta, S. CD4+ T cells are not required for the induction of dengue virus-specific CD8+ T cell or antibody responses but contribute to protection after vaccination. J. Immunol. 2010, 185, 5405–5416. [Google Scholar] [CrossRef] [PubMed]
- Wieland, U.; Kreuter, A.; Pfister, H. Human papillomavirus and immunosuppression. Curr. Probl. Dermatol. 2014, 45, 154–165. [Google Scholar] [PubMed]
- Berman, A.; Winkelmann, R.K. Involuting common warts. Clinical and histopathologic findings. J. Am. Acad. Dermatol. 1980, 3, 356–362. [Google Scholar] [CrossRef]
- Iwatsuki, K.; Tagami, H.; Takigawa, M.; Yamada, M. Plane warts under spontaneous regression. Immunopathologic study on cellular constituents leading to the inflammatory reaction. Arch. Dermatol. 1986, 122, 655–659. [Google Scholar] [CrossRef] [PubMed]
- Thivolet, J.; Viac, J.; Staquet, M.J. Cell-mediated immunity in wart infection. Int. J. Dermatol 1982, 21, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Coleman, N.; Stanley, M.A. Analysis of HLA-DR expression on keratinocytes in cervical neoplasia. Int. J. Cancer 1994, 56, 314–319. [Google Scholar] [CrossRef] [PubMed]
- Steele, J.C.; Mann, C.H.; Rookes, S.; Rollason, T.; Murphy, D.; Freeth, M.G.; Gallimore, P.H.; Roberts, S. T-cell responses to human papillomavirus type 16 among women with different grades of cervical neoplasia. Br. J. Cancer 2005, 93, 248–259. [Google Scholar] [CrossRef] [PubMed]
- Stanley, M.A. Epithelial cell responses to infection with human papillomavirus. Clin. Microbiol. Rev. 2012, 25, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Amador-Molina, A.; Hernandez-Valencia, J.F.; Lamoyi, E.; Contreras-Paredes, A.; Lizano, M. Role of innate immunity against human papillomavirus (HPV) infections and effect of adjuvants in promoting specific immune response. Viruses 2013, 5, 2624–2642. [Google Scholar] [CrossRef] [PubMed]
- Yan, N.; Chen, Z.J. Intrinsic antiviral immunity. Nat. Immunol. 2012, 13, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Stepp, W.H.; Meyers, J.M.; McBride, A.A. Sp100 provides intrinsic immunity against human papillomavirus infection. MBio 2013, 4, e00845-13. [Google Scholar] [CrossRef] [PubMed]
- Lazarczyk, M.; Pons, C.; Mendoza, J.A.; Cassonnet, P.; Jacob, Y.; Favre, M. Regulation of cellular zinc balance as a potential mechanism of ever-mediated protection against pathogenesis by cutaneous oncogenic human papillomaviruses. J. Exp. Med. 2008, 205, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Gunasekharan, V.; Laimins, L.A. Human papillomaviruses modulate microrna 145 expression to directly control genome amplification. J. Virol. 2013, 87, 6037–6043. [Google Scholar] [CrossRef] [PubMed]
- Saikia, P.; Fensterl, V.; Sen, G.C. The inhibitory action of p56 on select functions of E1 mediates interferon’s effect on human papillomavirus DNA replication. J. Virol. 2010, 84, 13036–13039. [Google Scholar] [CrossRef] [PubMed]
- Lo Cigno, I.; de Andrea, M.; Borgogna, C.; Albertini, S.; Landini, M.M.; Peretti, A.; Johnson, K.E.; Chandran, B.; Landolfo, S.; Gariglio, M. The nuclear DNA sensor IFI16 acts as a restriction factor for human papillomavirus replication through epigenetic modifications of the viral promoters. J. Virol. 2015, 89, 7506–7520. [Google Scholar] [CrossRef] [PubMed]
- Porter, S.S.; Stepp, W.H.; Stamos, J.D.; McBride, A.A. Host cell restriction factors that limit transcription and replication of human papillomavirus. Virus Res. 2017, 231, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Warren, C.J.; Westrich, J.A.; Doorslaer, K.V.; Pyeon, D. Roles of APOBEC3A and APOBEC3B in human papillomavirus infection and disease progression. Viruses 2017, 9, 233. [Google Scholar] [CrossRef] [PubMed]
- Vartanian, J.P.; Guetard, D.; Henry, M.; Wain-Hobson, S. Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions. Science 2008, 320, 230–233. [Google Scholar] [CrossRef] [PubMed]
- Warren, C.J.; Xu, T.; Guo, K.; Griffin, L.M.; Westrich, J.A.; Lee, D.; Lambert, P.F.; Santiago, M.L.; Pyeon, D. APOBEC3A functions as a restriction factor of human papillomavirus. J. Virol. 2015, 89, 688–702. [Google Scholar] [CrossRef] [PubMed]
- Mirabello, L.; Yeager, M.; Yu, K.; Clifford, G.M.; Xiao, Y.; Zhu, B.; Cullen, M.; Boland, J.F.; Wentzensen, N.; Nelson, C.W.; et al. HPV16 E7 genetic conservation is critical to carcinogenesis. Cell 2017, 170, 1164–1174. [Google Scholar] [CrossRef] [PubMed]
- Randelia, H.P.; Sanghvi, L. ‘Bare’, a new hairless mutant in the mouse—genetics and histology. Genet. Res. 1961, 2, 283–289. [Google Scholar] [CrossRef]
- Bhisey, R.A.; Veturkar, P.L.; Borges, A. Comparison of sensitivity of swiss albino and hairless swiss bare mice to two stage skin tumorigenesis. Indian J.Exp. Biol. 1987, 25, 90–96. [Google Scholar] [PubMed]
- Bhisey, R.A.; Veturkar, P.L.; Jeyapaul, J.; Borges, A. Effects of TPA dose variation and mezerein on skin tumorigenesis by initiation promotion protocol in S/RV cri mice. Indian J.Exp. Biol. 1988, 26, 764–767. [Google Scholar] [PubMed]
- Bhisey, R.A.; Veturkar, P.L. S/RV Cri-ba, a hairless mouse strain sensitive to skin tumorigenesis by suboptimal doses of 7,12-dimethylbenz[a]anthracene, initiation-promotion and two stage promotion protocols. Cancer Lett. 1990, 52, 63–69. [Google Scholar] [CrossRef]
- Benavides, F.; Oberyszyn, T.M.; VanBuskirk, A.M.; Reeve, V.E.; Kusewitt, D.F. The hairless mouse in skin research. J. Dermatol. Sci. 2009, 53, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.C.; McMillan, N.A.; Antonsson, A. Human papillomavirus type spectrum in normal skin of individuals with or without a history of frequent sun exposure. J. Gen. Virol. 2008, 89, 2891–2897. [Google Scholar] [CrossRef] [PubMed]
- Hampras, S.S.; Giuliano, A.R.; Lin, H.Y.; Fisher, K.J.; Abrahamsen, M.E.; Sirak, B.A.; Iannacone, M.R.; Gheit, T.; Tommasino, M.; Rollison, D.E. Natural history of cutaneous human papillomavirus (HPV) infection in men: The him study. PLoS ONE 2014, 9, e104843. [Google Scholar] [CrossRef] [PubMed]
- Iannacone, M.R.; Michael, K.M.; Giuliano, A.R.; Waterboer, T.; Pawlita, M.; Rollison, D.E. Risk factors for cutaneous human papillomavirus seroreactivity among patients undergoing skin cancer screening in florida. J. Infect. Dis. 2010, 201, 760–769. [Google Scholar] [CrossRef] [PubMed]
- Iannacone, M.R.; Wang, W.; Stockwell, H.G.; O’Rourke, K.; Giuliano, A.R.; Sondak, V.K.; Messina, J.L.; Roetzheim, R.G.; Cherpelis, B.S.; Fenske, N.A.; et al. Sunlight exposure and cutaneous human papillomavirus seroreactivity in basal cell and squamous cell carcinomas of the skin. J. Infect. Dis. 2012, 206, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Snell, G.D. Inheritance in the house mouse, the linkage relations of short-ear, hairless, and naked. Genetics 1931, 16, 42–74. [Google Scholar] [PubMed]
- Tilbrook, P.A.; Greenoak, G.E.; Reeve, V.E.; Canfield, P.J.; Gissmann, L.; Gallagher, C.H.; Kulski, J.K. Identification of papillomaviral DNA sequences in hairless mouse tumours induced by ultraviolet irradiation. J. Gen. Virol. 1989, 70 Pt 4, 1005–1009. [Google Scholar] [CrossRef] [PubMed]
- Reeve, V.E.; Greenoak, G.E.; Canfield, P.J.; Boehm-Wilcox, C.; Tilbrook, P.A.; Kulski, J.K.; Gallagher, C.H. Enhancement of u.v.-induced skin carcinogenesis in the hairless mouse by inoculation with cell-free extracts of skin tumours. Immunol. Cell Biol. 1989, 67 Pt 6, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Nouri, M.; Brandsma, J.L.; Iftner, T.; Steinberg, B.M. Induction of E6/E7 expression in cottontail rabbit papillomavirus latency following UV activation. Virology 1999, 263, 388–394. [Google Scholar] [CrossRef] [PubMed]
- Marcuzzi, G.P.; Hufbauer, M.; Kasper, H.U.; Weissenborn, S.J.; Smola, S.; Pfister, H. Spontaneous tumour development in human papillomavirus type 8 E6 transgenic mice and rapid induction by UV-light exposure and wounding. J. Gen. Virol. 2009, 90, 2855–2864. [Google Scholar] [CrossRef] [PubMed]
- Michel, A.; Kopp-Schneider, A.; Zentgraf, H.; Gruber, A.D.; de Villiers, E.M. E6/E7 expression of human papillomavirus type 20 (HPV-20) and HPV-27 influences proliferation and differentiation of the skin in UV-irradiated SKH-hr1 transgenic mice. J. Virol. 2006, 80, 11153–11164. [Google Scholar] [CrossRef] [PubMed]
- Viarisio, D.; Mueller-Decker, K.; Kloz, U.; Aengeneyndt, B.; Kopp-Schneider, A.; Grone, H.J.; Gheit, T.; Flechtenmacher, C.; Gissmann, L.; Tommasino, M. E6 and E7 from beta HPV38 cooperate with ultraviolet light in the development of actinic keratosis-like lesions and squamous cell carcinoma in mice. PLoS Pathog. 2011, 7, e1002125. [Google Scholar] [CrossRef] [PubMed]
- Norval, M. The effect of ultraviolet radiation on human viral infections. Photochem. Photobiol. 2006, 82, 1495–1504. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, S.E.; Byrne, S.N. The immunologic revolution: Photoimmunology. J. Investig. Dermatol. 2012, 132, 896–905. [Google Scholar] [CrossRef] [PubMed]
- Hart, P.H.; Gorman, S.; Finlay-Jones, J.J. Modulation of the immune system by UV radiation: More than just the effects of vitamin D? Nat. Rev. Immunol. 2011, 11, 584–596. [Google Scholar] [CrossRef] [PubMed]
- Norval, M.; Halliday, G.M. The consequences of UV-induced immunosuppression for human health. Photochem. Photobiol. 2011, 87, 965–977. [Google Scholar] [CrossRef] [PubMed]
- Schade, N.; Esser, C.; Krutmann, J. Ultraviolet B radiation-induced immunosuppression: Molecular mechanisms and cellular alterations. Photochem. Photobiol. Sci. 2005, 4, 699–708. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, T. The dark and the sunny sides of UVR-induced immunosuppression: Photoimmunology revisited. J. Investig. Dermatol. 2010, 130, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, T.; Beissert, S. Milestones in photoimmunology. J. Investig. Dermatol. 2013, 133, E7–E10. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, T.; Schwarz, A. Molecular mechanisms of ultraviolet radiation-induced immunosuppression. Eur. J. Cell Biol. 2011, 90, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, N.K.; Norval, M. Photoimmunosuppression: A brief overview. Photodermatol. Photoimmunol. Photomed. 2013, 29, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Poon, T.S.; Barnetson, R.S.; Halliday, G.M. Sunlight-induced immunosuppression in humans is initially because of UVB, then UVA, followed by interactive effects. J. Investig. Dermatol. 2005, 125, 840–846. [Google Scholar] [CrossRef] [PubMed]
- Byrne, S.N.; Spinks, N.; Halliday, G.M. Ultraviolet a irradiation of C57BL/6 mice suppresses systemic contact hypersensitivity or enhances secondary immunity depending on dose. J. Investig. Dermatol. 2002, 119, 858–864. [Google Scholar] [CrossRef] [PubMed]
- Kripke, M.L. Immunological unresponsiveness induced by ultraviolet radiation. Immunol. Rev. 1984, 80, 87–102. [Google Scholar] [CrossRef] [PubMed]
- Mottram, P.L.; Mirisklavos, A.; Clunie, G.J.; Noonan, F.P. A single dose of UV radiation suppresses delayed type hypersensitivity responses to alloantigens and prolongs heart allograft survival in mice. Immunol. Cell Biol. 1988, 66 Pt 5–6, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Noonan, F.P.; de Fabo, E.C.; Kripke, M.L. Suppression of contact hypersensitivity by uv radiation and its relationship to UV-induced suppression of tumor immunity. Photochem. Photobiol. 1981, 34, 683–689. [Google Scholar] [CrossRef] [PubMed]
- De Fabo, E.C.; Noonan, F.P. Mechanism of immune suppression by ultraviolet irradiation in vivo. I. Evidence for the existence of a unique photoreceptor in skin and its role in photoimmunology. J. Exp. Med. 1983, 158, 84–98. [Google Scholar] [CrossRef] [PubMed]
- Walterscheid, J.P.; Nghiem, D.X.; Kazimi, N.; Nutt, L.K.; McConkey, D.J.; Norval, M.; Ullrich, S.E. Cis-urocanic acid, a sunlight-induced immunosuppressive factor, activates immune suppression via the 5-HT2A receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 17420–17425. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Wu, S.B.; Hong, C.H.; Yu, H.S.; Wei, Y.H. Molecular mechanisms of uv-induced apoptosis and its effects on skin residential cells: The implication in uv-based phototherapy. Int. J. Mol. Sci. 2013, 14, 6414–6435. [Google Scholar] [CrossRef] [PubMed]
- Rana, S.; Byrne, S.N.; MacDonald, L.J.; Chan, C.Y.; Halliday, G.M. Ultraviolet b suppresses immunity by inhibiting effector and memory T cells. Am. J. Pathol. 2008, 172, 993–1004. [Google Scholar] [CrossRef] [PubMed]
- Elmets, C.A.; Bergstresser, P.R.; Tigelaar, R.E.; Wood, P.J.; Streilein, J.W. Analysis of the mechanism of unresponsiveness produced by haptens painted on skin exposed to low dose ultraviolet radiation. J. Exp. Med. 1983, 158, 781–794. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, T. 25 years of uv-induced immunosuppression mediated by T cells-from disregarded T suppressor cells to highly respected regulatory t cells. Photochem. Photobiol. 2008, 84, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Clydesdale, G.J.; Dandie, G.W.; Muller, H.K. Ultraviolet light induced injury: Immunological and inflammatory effects. Immunol. Cell Biol. 2001, 79, 547–568. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, S.E. Does exposure to UV radiation induce a shift to a TH-2-like immune reaction? Photochem. Photobiol. 1996, 64, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Chapman, R.S.; Cooper, K.D.; de Fabo, E.C.; Frederick, J.E.; Gelatt, K.N.; Hammond, S.P.; Hersey, P.; Koren, H.S.; Ley, R.D.; Noonan, F.; et al. Solar ultraviolet radiation and the risk of infectious disease: Summary of a workshop. Photochem. Photobiol. 1995, 61, 223–247. [Google Scholar] [PubMed]
- Patra, V.; Byrne, S.N.; Wolf, P. The skin microbiome: Is it affected by uv-induced immune suppression? Front. Microbiol. 2016, 7, 1235. [Google Scholar] [CrossRef] [PubMed]
- Tseng, C.C.; Li, C.S. Inactivation of viruses on surfaces by ultraviolet germicidal irradiation. J. Occup. Environ. Hyg. 2007, 4, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Lytle, C.D.; Knott, D.C. Enhanced mutagenesis parallels enhanced reactivation of herpes virus in a human cell line. EMBO J. 1982, 1, 701–703. [Google Scholar] [PubMed]
- Cornelis, J.J.; Su, Z.Z.; Rommelaere, J. Direct and indirect effects of ultraviolet light on the mutagenesis of parvovirus H-1 in human cells. EMBO J. 1982, 1, 693–699. [Google Scholar] [PubMed]
- Chodosh, L.A. UV crosslinking of proteins to nucleic acids. Curr. Protoc. Mol. Biol. 2001, Chapter 12, Unit 12.5. [Google Scholar] [CrossRef]
- Rainbow, A.J.; Castillo, J.E. Homologous recombination of adenovirus DNA in mammalian cells: Enhanced recombination following UV-irradiation of the virus. Mutat. Res. 1992, 274, 201–210. [Google Scholar] [CrossRef]
- Dasgupta, U.B.; Summers, W.C. Genetic recombination of herpes simplex virus, the role of the host cell and UV-irradiation of the virus. Mol. Gen. Genet. 1980, 178, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Stein, B.; Kramer, M.; Rahmsdorf, H.J.; Ponta, H.; Herrlich, P. UV-induced transcription from the human immunodeficiency virus type 1 (HIV-1) long terminal repeat and UV-induced secretion of an extracellular factor that induces HIV-1 transcription in nonirradiated cells. J. Virol. 1989, 63, 4540–4544. [Google Scholar] [PubMed]
- Loiacono, C.M.; Taus, N.S.; Mitchell, W.J. The herpes simplex virus type 1 ICP0 promoter is activated by viral reactivation stimuli in trigeminal ganglia neurons of transgenic mice. J. Neurovirol. 2003, 9, 336–345. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.; Harwood, C.; Thomas, M.; Banks, L.; Storey, A. Role of bak in UV-induced apoptosis in skin cancer and abrogation by HPV E6 proteins. Genes Dev. 2000, 14, 3065–3073. [Google Scholar] [CrossRef] [PubMed]
- Underbrink, M.P.; Howie, H.L.; Bedard, K.M.; Koop, J.I.; Galloway, D.A. E6 proteins from multiple human betapapillomavirus types degrade BAK and protect keratinocytes from apoptosis after UVB irradiation. J. Virol. 2008, 82, 10408–10417. [Google Scholar] [CrossRef] [PubMed]
- Giampieri, S.; Garcia-Escudero, R.; Green, J.; Storey, A. Human papillomavirus type 77 E6 protein selectively inhibits p53-dependent transcription of proapoptotic genes following UV-B irradiation. Oncogene 2004, 23, 5864–5870. [Google Scholar] [CrossRef] [PubMed]
- Maglennon, G.A.; McIntosh, P.B.; Doorbar, J. Immunosuppression facilitates the reactivation of latent papillomavirus infections. J. Virol. 2014, 88, 710–716. [Google Scholar] [CrossRef] [PubMed]
- O’Banion, M.K.; Reichmann, M.E.; Sundberg, J.P. Cloning and characterization of a papillomavirus associated with papillomas and carcinomas in the european harvest mouse (micromys minutus). J. Virol. 1988, 62, 226–233. [Google Scholar] [PubMed]
- Nafz, J.; Ohnesorge, M.; Stockfleth, E.; Rosl, F.; Nindl, I. Imiquimod treatment of papilloma virus and DMBA /TPA-induced cutaneous skin cancer in mastomys coucha: An unique animal model system useful for preclinical studies. Br. J. Dermatol. 2007, 157 (Suppl. 2), 14–17. [Google Scholar] [CrossRef] [PubMed]
- Garland, S.M.; Brotherton, J.M.L.; Moscicki, A.B.; Kaufmann, A.M.; Stanley, M.; Bhatla, N.; Sankaranarayanan, R.; de Sanjosé, S.; Palefsky, J.M. HPV vaccination of immunocompromised hosts. Papillomavirus Res. 2017, 4, 35–38. [Google Scholar] [CrossRef]
- Jiang, R.T.; Wang, J.W.; Peng, S.; Huang, T.C.; Wang, C.; Cannella, F.; Chang, Y.N.; Viscidi, R.P.; Best, S.R.A.; Hung, C.F.; et al. Spontaneous and vaccine-induced clearance of mus musculus papillomavirus 1 infection. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Meyers, C.; Wang, H.K.; Chow, L.T.; Zheng, Z.M. Construction of a full transcription map of human papillomavirus type 18 during productive viral infection. J. Virol. 2011, 85, 8080–8092. [Google Scholar] [CrossRef] [PubMed]
- Ozbun, M.A.; Meyers, C. Characterization of late gene transcripts expressed during vegetative replication of human papillomavirus type 31B. J. Virol. 1997, 71, 5161–5172. [Google Scholar] [PubMed]
- Grassmann, K.; Rapp, B.; Maschek, H.; Petry, K.U.; Iftner, T. Identification of a differentiation-inducible promoter in the E7 open reading frame of human papillomavirus type 16 (HPV-16) in raft cultures of a new cell line containing high copy numbers of episomal HPV-16 DNA. J. Virol. 1996, 70, 2339–2349. [Google Scholar] [PubMed]
- Sankovski, E.; Mannik, A.; Geimanen, J.; Ustav, E.; Ustav, M. Mapping of betapapillomavirus human papillomavirus 5 transcription and characterization of viral-genome replication function. J. Virol. 2014, 88, 961–973. [Google Scholar] [CrossRef] [PubMed]
- Isok-Paas, H.; Mannik, A.; Ustav, E.; Ustav, M. The transcription map of HPV11 in U2OS cells adequately reflects the initial and stable replication phases of the viral genome. Virol. J. 2015, 12, 59. [Google Scholar] [CrossRef] [PubMed]
- Jia, R.; Zheng, Z.M. Regulation of bovine papillomavirus type 1 gene expression by rna processing. Front. Biosci. (Landmark Ed.) 2009, 14, 1270–1282. [Google Scholar] [CrossRef] [PubMed]
- Probst-Hunczek, S.; Jager, G.; Schneider, M.; Notz, E.; Stubenrauch, F.; Iftner, T. Rna sequencing analysis identifies novel spliced transcripts but does not indicate quantitative or qualitative changes of viral transcripts during progression of cottontail rabbit papillomavirus-induced tumours. J. Gen. Virol. 2015, 96, 3083–3089. [Google Scholar] [CrossRef] [PubMed]
- Palermo-Dilts, D.A.; Broker, T.R.; Chow, L.T. Human papillomavirus type 1 produces redundant as well as polycistronic mrnas in plantar warts. J. Virol. 1990, 64, 3144–3149. [Google Scholar] [PubMed]
- Lambert, P.F.; Spalholz, B.A.; Howley, P.M. A transcriptional repressor encoded by BPV-1 shares a common carboxy-terminal domain with the E2 transactivator. Cell 1987, 50, 69–78. [Google Scholar] [CrossRef]
- Lambert, P.F.; Monk, B.C.; Howley, P.M. Phenotypic analysis of bovine papillomavirus type 1 E2 repressor mutants. J. Virol. 1990, 64, 950–956. [Google Scholar] [PubMed]
- Jeckel, S.; Loetzsch, E.; Huber, E.; Stubenrauch, F.; Iftner, T. Identification of the E9/E2C cDNA and functional characterization of the gene product reveal a new repressor of transcription and replication in cottontail rabbit papillomavirus. J. Virol. 2003, 77, 8736–8744. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.M.; Broker, T.R.; Chow, L.T. An E1M–E2C fusion protein encoded by human papillomavirus type 11 is asequence-specific transcription repressor. J. Virol. 1991, 65, 3317–3329. [Google Scholar] [PubMed]
- Lace, M.J.; Anson, J.R.; Thomas, G.S.; Turek, L.P.; Haugen, T.H. The E8–E2 gene product of human papillomavirus type 16 represses early transcription and replication but is dispensable for viral plasmid persistence in keratinocytes. J. Virol. 2008, 82, 10841–10853. [Google Scholar] [CrossRef] [PubMed]
- Stubenrauch, F.; Hummel, M.; Iftner, T.; Laimins, L.A. The E8E2C protein, a negative regulator of viral transcription and replication, is required for extrachromosomal maintenance of human papillomavirus type 31 in keratinocytes. J. Virol. 2000, 74, 1178–1186. [Google Scholar] [CrossRef] [PubMed]
- Straub, E.; Dreer, M.; Fertey, J.; Iftner, T.; Stubenrauch, F. The viral E8^E2C repressor limits productive replication of human papillomavirus 16. J. Virol. 2014, 88, 937–947. [Google Scholar] [CrossRef] [PubMed]
- Meyers, J.M.; Uberoi, A.; Grace, M.; Lambert, P.F.; Munger, K. Cutaneous HPV8 and MmuPV1 E6 proteins target the notch and tgf-beta tumor suppressors to inhibit differentiation and sustain keratinocyte proliferation. PLoS Pathog. 2017, 13, e1006171. [Google Scholar] [CrossRef] [PubMed]
- Zanier, K.; Charbonnier, S.; Sidi, A.O.; McEwen, A.G.; Ferrario, M.G.; Poussin-Courmontagne, P.; Cura, V.; Brimer, N.; Babah, K.O.; Ansari, T.; et al. Structural basis for hijacking of cellular LXXLL motifs by papillomavirus E6 oncoproteins. Science 2013, 339, 694–698. [Google Scholar] [CrossRef] [PubMed]
- Brimer, N.; Lyons, C.; Vande Pol, S.B. Association of E6AP (UBE3A) with human papillomavirus type 11 E6 protein. Virology 2007, 358, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Huibregtse, J.M.; Scheffner, M.; Howley, P.M. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991, 10, 4129–4135. [Google Scholar] [PubMed]
- White, E.A.; Kramer, R.E.; Tan, M.J.; Hayes, S.D.; Harper, J.W.; Howley, P.M. Comprehensive analysis of host cellular interactions with human papillomavirus E6 proteins identifies new E6 binding partners and reflects viral diversity. J. Virol. 2012, 86, 13174–13186. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.; Howley, P.M. The bovine papillomavirus E6 oncoprotein interacts with paxillin and disrupts the actin cytoskeleton. Proc. Natl. Acad. Sci. USA 1997, 94, 4412–4417. [Google Scholar] [CrossRef] [PubMed]
- Ronco, L.V.; Karpova, A.Y.; Vidal, M.; Howley, P.M. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 1998, 12, 2061–2072. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.J.; White, E.A.; Sowa, M.E.; Harper, J.W.; Aster, J.C.; Howley, P.M. Cutaneous beta-human papillomavirus E6 proteins bind mastermind-like coactivators and repress notch signaling. Proc. Natl. Acad. Sci. USA 2012, 109, E1473–E1480. [Google Scholar] [CrossRef] [PubMed]
- Meyers, J.M.; Spangle, J.M.; Munger, K. The human papillomavirus type 8 E6 protein interferes with notch activation during keratinocyte differentiation. J. Virol. 2013, 87, 4762–4767. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Zapien, D.; Ruiz, F.X.; Poirson, J.; Mitschler, A.; Ramirez, J.; Forster, A.; Cousido-Siah, A.; Masson, M.; Vande Pol, S.; Podjarny, A.; et al. Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53. Nature 2016, 529, 541–545. [Google Scholar] [CrossRef] [PubMed]
- Brimer, N.; Lyons, C.; Wallberg, A.E.; Vande Pol, S.B. Cutaneous papillomavirus E6 oncoproteins associate with MAML1 to repress transactivation and notch signaling. Oncogene 2012, 31, 4639–4646. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Aster, J.C.; Blacklow, S.C.; Lake, R.; Artavanis-Tsakonas, S.; Griffin, J.D. MAML1, a human homologue of drosophila mastermind, is a transcriptional co-activator for notch receptors. Nat. Genet. 2000, 26, 484–489. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhou, D.; Prabhu, A.; Schlegel, R.; Yuan, H. The canine papillomavirus and gamma HPV E7 proteins use an alternative domain to bind and destabilize the retinoblastoma protein. PLoS Pathog. 2010, 6, e1001089. [Google Scholar] [CrossRef] [PubMed]
- Pim, D.; Bergant, M.; Boon, S.S.; Ganti, K.; Kranjec, C.; Massimi, P.; Subbaiah, V.K.; Thomas, M.; Tomaic, V.; Banks, L. Human papillomaviruses and the specificity of PDZ domain targeting. FEBS J. 2012, 279, 3530–3537. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin-Drubin, M.E.; Munger, K. The human papillomavirus E7 oncoprotein. Virology 2009, 384, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Halbert, C.L.; Demers, G.W.; Galloway, D.A. The E6 and E7 genes of human papillomavirus type 6 have weak immortalizing activity in human epithelial cells. J. Virol. 1992, 66, 2125–2134. [Google Scholar] [PubMed]
- Chan, H.M.; Smith, L.; La Thangue, N.B. Role of lxcxe motif-dependent interactions in the activity of the retinoblastoma protein. Oncogene 2001, 20, 6152–6163. [Google Scholar] [CrossRef] [PubMed]
- Narechania, A.; Terai, M.; Chen, Z.; DeSalle, R.; Burk, R.D. Lack of the canonical pRB-binding domain in the E7 ORF of artiodactyl papillomaviruses is associated with the development of fibropapillomas. J. Gen. Virol. 2004, 85, 1243–1250. [Google Scholar] [CrossRef] [PubMed]
- Van Bressem, M.F.; Cassonnet, P.; Rector, A.; Desaintes, C.; van Waerebeek, K.; Alfaro-Shigueto, J.; van Ranst, M.; Orth, G. Genital warts in burmeister’s porpoises: Characterization of phocoena spinipinnis papillomavirus type 1 (PSPV-1) and evidence for a second, distantly related PSPV. J. Gen. Virol. 2007, 88, 1928–1933. [Google Scholar] [CrossRef] [PubMed]
- Stevens, H.; Rector, A.; van der Kroght, K.; van Ranst, M. Isolation and cloning of two variant papillomaviruses from domestic pigs: Sus scrofa papillomaviruses type 1 variants a and b. J. Gen. Virol. 2008, 89, 2475–2481. [Google Scholar] [CrossRef] [PubMed]
- Stevens, H.; Rector, A.; Bertelsen, M.F.; Leifsson, P.S.; van Ranst, M. Novel papillomavirus isolated from the oral mucosa of a polar bear does not cluster with other papillomaviruses of carnivores. Vet. Microbiol. 2008, 129, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Munger, K.; Basile, J.R.; Duensing, S.; Eichten, A.; Gonzalez, S.L.; Grace, M.; Zacny, V.L. Biological activities and molecular targets of the human papillomavirus E7 oncoprotein. Oncogene 2001, 20, 7888–7898. [Google Scholar] [CrossRef] [PubMed]
- White, E.A.; Munger, K.; Howley, P.M. High-risk human papillomavirus E7 proteins target PTPN14 for degradation. MBio 2016, 7, e01530-16. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
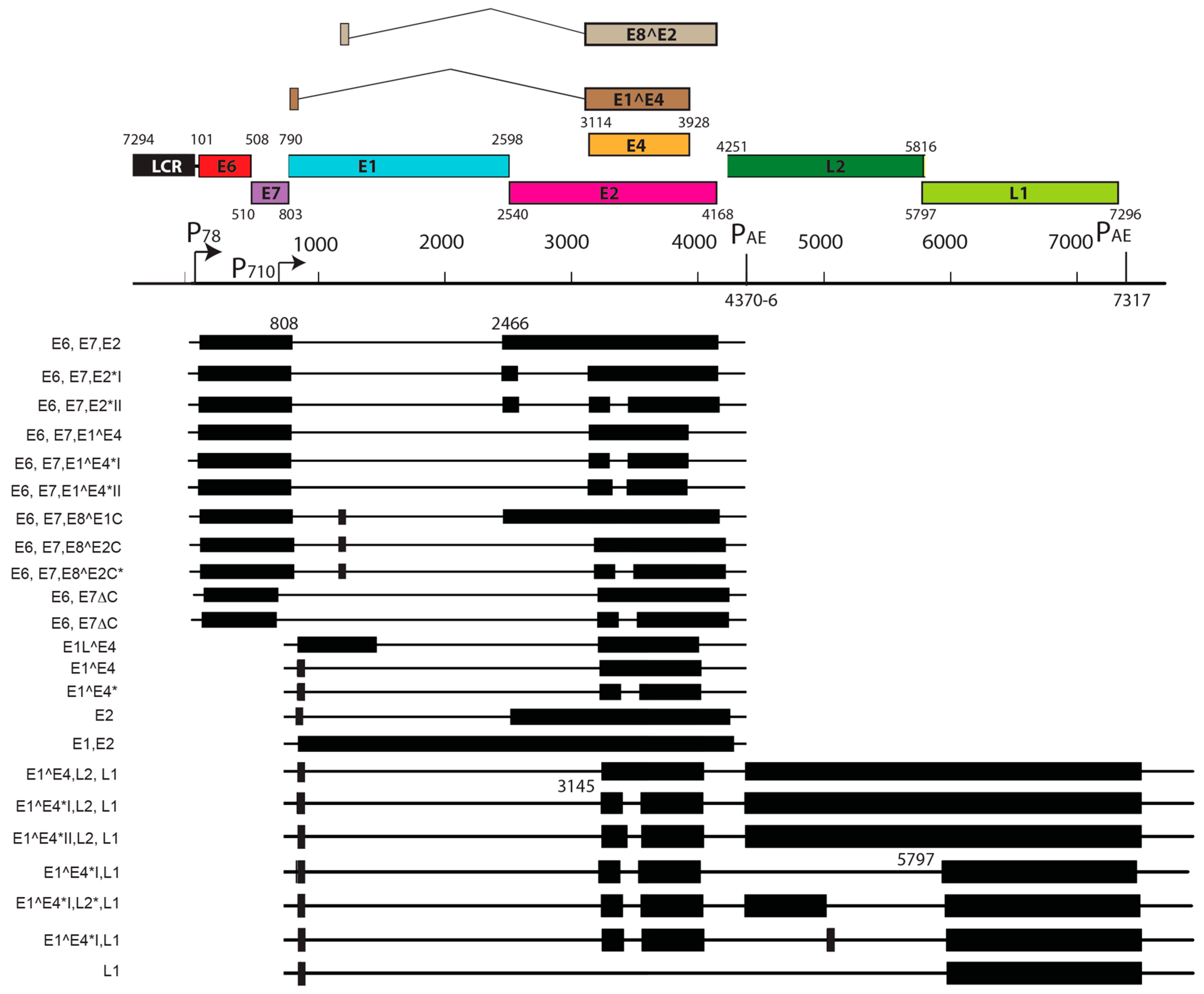
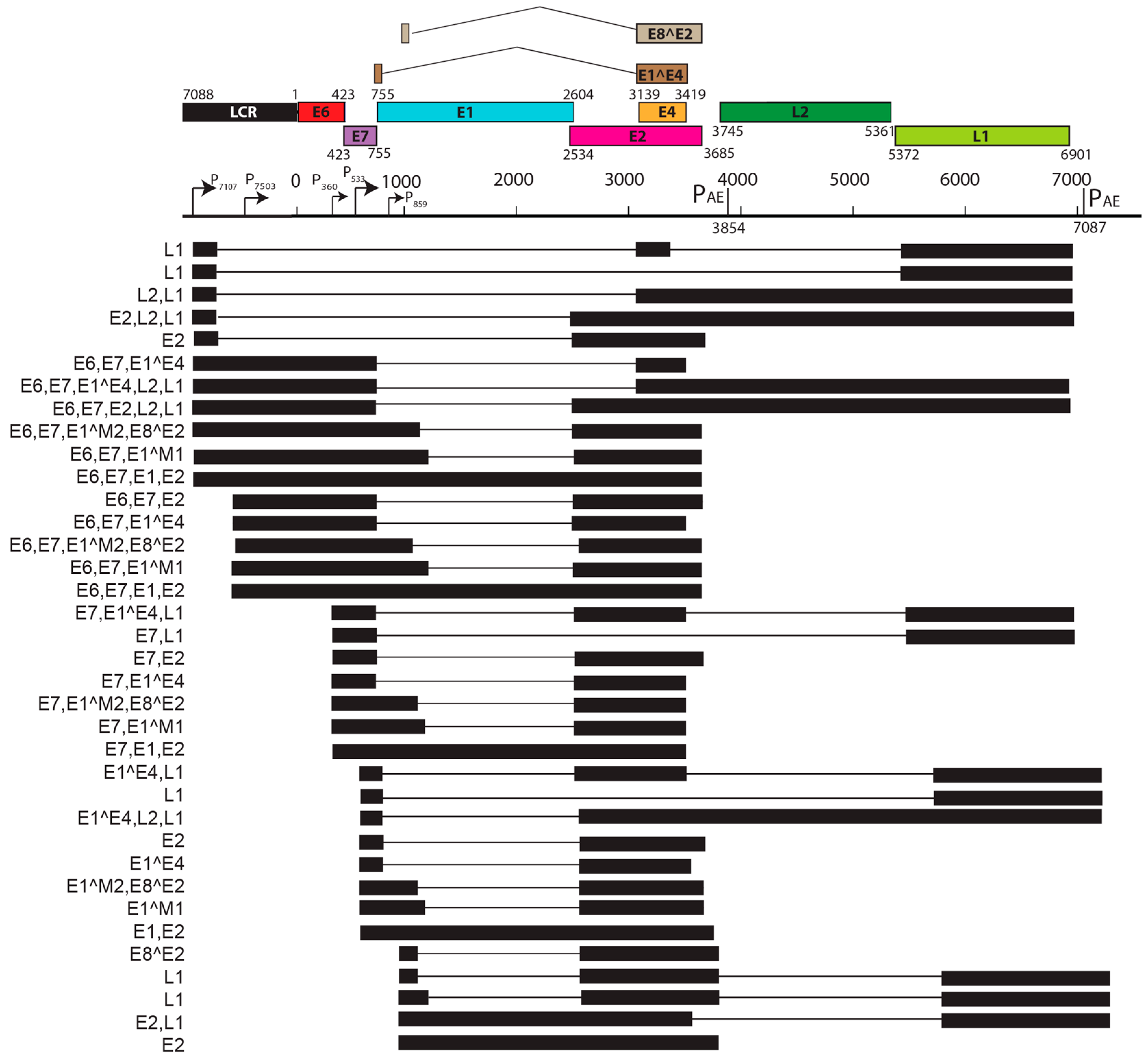
| S. No. | PV | Genome Size bp | PAVE 1 Name | Host Common Name | Genus | Reference |
|---|---|---|---|---|---|---|
| 1 | AsPV1 | 7589 | Apodemus sylvaticus Papillomavirus 1 | Long-tailed field mouse | π | [47] |
| 2 | CcanPV1 | 7435 | Castor canadensis Papillomavirus 1 | North-American beaver | Dyosigma | [48] |
| 3 | EdPV1 | 7428 | Erethizon dorsatum Papillomavirus 1 | North-American porcupine | σ | [49] |
| 4 | MaPV1 | 7647 | Mesocricetus auratus Papillomavirus 1 | Syrian golden hamster | π | [50] |
| 5 | McPV2 | 7522 | Mastomys coucha Papillomavirus 2 | Southern multimammate rat | π | [51] |
| 6 | MmiPV1 | 7393 | Micromys minutus Papillomavirus 1 | European harvest mouse | π | [52,53] |
| 7 | MmuPV1 2 | 7501 | Mus musculus Papillomavirus 1 | House mouse | π | [54,55] |
| 8 | MnPV1 | 7687 | Mastomys natalensis Papillomavirus 1 | Southern multimammate rat | ι | [56,57] |
| 9 | PsuPV1 | 7630 | Phodopus sungorus Papillomavirus 1 | Siberian hamster | π | [58] |
| 10 | RnPV1 | 7378 | Rattus norvegicus Papillomavirus 1 | Norwegian rat | π | [47] |
| 11 | RnPV2 | 7724 | Rattus norvegicus Papillomavirus 2 | Norwegian rat | ι | |
| 12 | RnPV3 | 7707 | Rattus norvegicus Papillomavirus 3 | Norwegian rat | ι |
| S. No. | Deficiency | Strain-Mutation | Reference |
|---|---|---|---|
| 1 | Lack T cells | B6.Cg-Foxn1nu/J | [55,99] |
| 2 | BALB/c-Foxn1nu | [116] | |
| 3 | Hsd:NU-Foxn1nu | [119] | |
| 4 | NCr nu/nu | [98] | |
| 5 | NMRI-Foxn1nu/Foxn1nu | [54] | |
| 6 | NU-Foxn1nu/Foxn1nu | [103] | |
| 7 | Lack T & B cells | B6.129S7-Rag1tm1Mom/J | [99,120] |
| 8 | B6.CB17-Prkdcscid/SzJ | [99] | |
| 9 | NCI SCID/Ncr | [120] | |
| 10 | SHO-PrkdcscidHrhr | [118] |
| S. No. | Deficiency | Strain/Mutation | Reference |
|---|---|---|---|
| 1 | Lack T & B cells | NOD.CB17-Prkdcscid/SzJ | [99] |
| 2 | Lack B cells | B6.129S2-Ighmtm1Cgn/J | [99] |
| 3 | Lack CD4+ T cells | B6.129S2-Cd4tm1Mak/J | [99] |
| 4 | Lack CD8+ T cells | B6.129S2-Cd8tm1Mak/J | [99] |
| 5 | Lack T helper cells/CD40 ligand K | B6.129S2-Cd40lgtm1Imx/J | [120] |
| 6 | Lack NK (CD1d) cells | B6.129S6-Del (3Cd1d2-Cd1d1)1Sbp/J | [126] |
| 7 | Decreased Anti-viral innate immunity, lack Type I Interferon | IFNAR KO | [120] |
| S. No. | Strain | Intervention | Reference |
|---|---|---|---|
| 1 | Cr:ORL SENCAR | Cyclosporine | [126] |
| 2 | FVB/NCr | ||
| 3 | BALB/cAnNCr | ||
| 4 | A/JCr | ||
| 5 | Cr:ORL SENCAR | Anti-CD3 | [126] |
| 6 | Anti-CD4 | ||
| 7 | Anti-CD8 | ||
| 8 | C57BL/6 | Anti-CD3 | [120,126] |
| 9 | Anti-CD4+Anti-CD8 * | ||
| 10 | BALB/c | Anti-CD3 * | [120] |
| Feature | MnPV1 | MmuPV1 |
|---|---|---|
| Source | Skin papillomas from M. Natalensis colonized with MnPV1 | Skin papillomas from immunodeficient mice infected with MmuPV1 |
| Early Promoters | Single promoter (P78) for E6 and E7 | Three promoters: P7503 for E6, P360 for E7, P7503 for E2 and/or E8^E2 |
| Late Promoters | Single late promoter (within E7 ORF) | Two late promoters (within LCR and E7 ORF) |
| Notable splice isoforms | E8^E2, E2*I, E2*II | E1^M1, E1^M2, E8^E2 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uberoi, A.; Lambert, P.F. Rodent Papillomaviruses. Viruses 2017, 9, 362. https://doi.org/10.3390/v9120362
Uberoi A, Lambert PF. Rodent Papillomaviruses. Viruses. 2017; 9(12):362. https://doi.org/10.3390/v9120362
Chicago/Turabian StyleUberoi, Aayushi, and Paul F. Lambert. 2017. "Rodent Papillomaviruses" Viruses 9, no. 12: 362. https://doi.org/10.3390/v9120362