Bat Astroviruses: Towards Understanding the Transmission Dynamics of a Neglected Virus Family
Abstract
:1. Introduction and Background
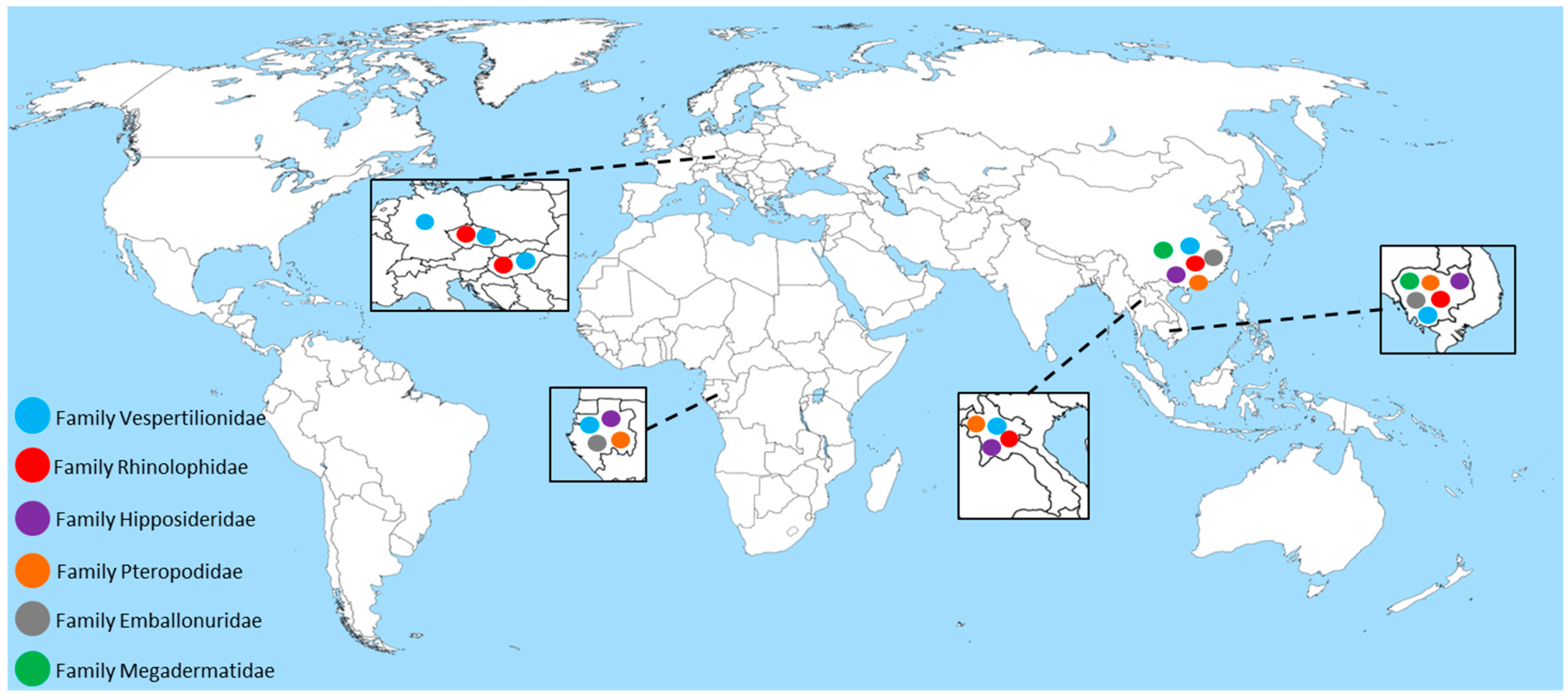
2. Astroviruses Detected in Bats across the Globe
3. Astroviruses Detected in Livestock
4. Human Astroviruses (HAstV)
5. Phylogenetic Analyses and Future Directions
6. Zoonotic Potential and Risk Assessment
7. Conclusions
Author Contributions
Conflicts of Interest
References
- Appleton, H.; Higgins, P.G. Letter: Viruses and gastroenteritis in infants. Lancet 1975, 1, 1297. [Google Scholar] [PubMed]
- Madeley, C.R.; Cosgrove, B.P. Letter: Viruses in infantile gastroenteritis. Lancet 1975, 2, 124. [Google Scholar] [CrossRef]
- Tse, H.; Chan, W.M.; Tsoi, H.W.; Fan, R.Y.; Lau, C.C.; Lau, S.K.; Woo, P.C.; Yuen, K.Y. Rediscovery and genomic characterization of bovine astroviruses. J. Gen. Virol. 2011, 92, 1888–1898. [Google Scholar] [CrossRef] [PubMed]
- Méndez, E.; Arias, C. Astroviruses. In Fields Virology; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007; pp. 981–1000. [Google Scholar]
- Finkbeiner, S.R.; Kirkwood, C.D.; Wang, D. Complete genome sequence of a highly divergent astrovirus isolated from a child with acute diarrhea. Virol. J. 2008, 5, 117. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Pan, M.; Wang, X.; Xu, Y.; Xie, X.; Knowles, N.J.; Yang, H.; Zhang, D. Complete sequence of a duck astrovirus associated with fatal hepatitis in ducklings. J. Gen. Virol. 2009, 90, 1104–1108. [Google Scholar] [CrossRef] [PubMed]
- Strain, E.; Kelley, L.A.; Schultz-Cherry, S.; Muse, S.V.; Koci, M.D. Genomic analysis of closely related astroviruses. J. Virol. 2008, 82, 5099–5103. [Google Scholar] [CrossRef] [PubMed]
- Finkbeiner, S.R.; Le, B.M.; Holtz, L.R.; Storch, G.A.; Wang, D. Detection of newly described astrovirus MLB1 in stool samples from children. Emerg. Infect. Dis. 2009, 15, 441–444. [Google Scholar] [CrossRef] [PubMed]
- Finkbeiner, S.R.; Li, Y.; Ruone, S.; Conrardy, C.; Gregoricus, N.; Toney, D.; Virgin, H.W.; Anderson, L.J.; Vinje, J.; Wang, D.; et al. Identification of a novel astrovirus (astrovirus VA1) associated with an outbreak of acute gastroenteritis. J. Virol. 2009, 83, 10836–10839. [Google Scholar] [CrossRef] [PubMed]
- Schlottau, K.; Schulze, C.; Bilk, S.; Hanke, D.; Hoper, D.; Beer, M.; Hoffmann, B. Detection of a Novel Bovine Astrovirus in a Cow with Encephalitis. Transbound. Emerg. Dis. 2016, 63, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.K.W.; Poon, L.L.M.; Guan, Y.; Peiris, J.S.M. Novel astroviruses in insectivorous bats. J. Virol. 2008, 82, 9107–9114. [Google Scholar] [CrossRef] [PubMed]
- Reuter, G.; Pankovics, P.; Boros, A. Identification of a novel astrovirus in a domestic pig in Hungary. Arch. Virol. 2011, 156, 125–128. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Roi, S.; Dastor, M.; Gallice, E.; Laurin, M.A.; L’Homme, Y. Multiple novel and prevalent astroviruses in pigs. Vet. Microbiol. 2011, 149, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, E.A.; Small, C.T.; Freiden, P.; Feeroz, M.M.; Matsen, F.A.T.; San, S.; Hasan, M.K.; Wang, D.; Jones-Engel, L.; Schultz-Cherry, S. Non-Human Primates Harbor Diverse Mammalian and Avian Astroviruses Including Those Associated with Human Infections. PLoS Pathog. 2015, 11, e1005225. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, A.; Duong, V.; Hul, V.; San, S.; Davun, H.; Omaliss, K.; Chea, S.; Hassanin, A.; Theppangna, W.; Silithammavong, S.; et al. Diversity of bat astroviruses in Lao PDR and Cambodia. Infect. Genet. Evol. 2016, 47, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Lekana-Douki, S.E.; Kombila-Koumavor, C.; Nkoghe, D.; Drosten, C.; Drexler, J.F.; Leroy, E.M. Molecular epidemiology of enteric viruses and genotyping of rotavirus A, adenovirus and astrovirus among children under 5 years old in Gabon. Int. J. Infect. Dis. 2015, 34, 90–95. [Google Scholar] [CrossRef] [PubMed]
- De Benedictis, P.; Schultz-Cherry, S.; Burnham, A.; Cattoli, G. Astrovirus infections in humans and animals—Molecular biology, genetic diversity, and interspecies transmissions. Infect. Genet. Evol. 2011, 11, 1529–1544. [Google Scholar] [CrossRef] [PubMed]
- Quan, P.L.; Wagner, T.A.; Briese, T.; Torgerson, T.R.; Hornig, M.; Tashmukhamedova, A.; Firth, C.; Palacios, G.; Baisre-De-Leon, A.; Paddock, C.D.; et al. Astrovirus encephalitis in boy with X-linked agammaglobulinemia. Emerg. Infect. Dis. 2010, 16, 918–925. [Google Scholar] [CrossRef] [PubMed]
- Fischer, K.; Zeus, V.; Kwasnitschka, L.; Kerth, G.; Haase, M.; Groschup, M.H.; Balkema-Buschmann, A. Insectivorous bats carry host specific astroviruses and coronaviruses across different regions in Germany. Infect. Genet. Evol. 2016, 37, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Kemenesi, G.; Dallos, B.; Gorfol, T.; Boldogh, S.; Estok, P.; Kurucz, K.; Kutas, A.; Foldes, F.; Oldal, M.; Nemeth, V.; et al. Molecular survey of RNA viruses in Hungarian bats: Discovering novel astroviruses, coronaviruses, and caliciviruses. Vector Borne Zoonotic Dis 2014, 14, 846–855. [Google Scholar] [CrossRef] [PubMed]
- Kemenesi, G.; Dallos, B.; Gorfol, T.; Boldogh, S.; Estok, P.; Kurucz, K.; Oldal, M.; Nemeth, V.; Madai, M.; Banyai, K.; et al. Novel European lineages of bat astroviruses identified in Hungary. Acta Virol. 2014, 58, 95–98. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.C.; Chu, D.K.W.; Liu, W.; Dong, B.O.; Zhang, S.Y.; Zhang, J.X.; Li, L.F.; Vijaykrishna, D.; Smith, G.J.D.; Chen, H.L.; et al. Detection of diverse astroviruses from bats in China. J. Gen. Virol. 2009, 90, 883–887. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Li, J.; Hu, G.; Chen, Z.; Wu, Y.; Chen, Y.; Chen, Z.; Liao, Y.; Zhou, J.; Ke, X.; et al. Isolation and phylogenetic characterization of bat astroviruses in southern China. Arch. Virol. 2011, 156, 1415–1423. [Google Scholar] [CrossRef] [PubMed]
- Moratelli, R.; Calisher, C.H. Bats and zoonotic viruses: Can we confidently link bats with emerging deadly viruses? Mem. Inst. Oswaldo Cruz 2015, 110, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Han, H.J.; Wen, H.L.; Zhou, C.M.; Chen, F.F.; Luo, L.M.; Liu, J.W.; Yu, X.J. Bats as reservoirs of severe emerging infectious diseases. Virus Res. 2015, 205, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.Y.; Li, J.L.; Yang, X.L.; Chmura, A.A.; Zhu, G.J.; Epstein, J.H.; Mazet, J.K.; Hu, B.; Zhang, W.; Peng, C.; et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 2013, 503, 535–538. [Google Scholar] [CrossRef] [PubMed]
- Halpin, K.; Young, P.L.; Field, H.E.; Mackenzie, J.S. Isolation of Hendra virus from pteropid bats: A natural reservoir of Hendra virus. J. Gen. Virol. 2000, 81, 1927–1932. [Google Scholar] [PubMed]
- Chua, K.B.; Koh, C.L.; Hooi, P.S.; Wee, K.F.; Khong, J.H.; Chua, B.H.; Chan, Y.P.; Lim, M.E.; Lam, S.K. Isolation of Nipah virus from Malaysian Island flying-foxes. Microbes Infect. 2002, 4, 145–151. [Google Scholar] [CrossRef]
- Towner, J.S.; Amman, B.R.; Sealy, T.K.; Carroll, S.A.; Comer, J.A.; Kemp, A.; Swanepoel, R.; Paddock, C.D.; Balinandi, S.; Khristova, M.L.; et al. Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathog. 2009, 5, e1000536. [Google Scholar] [CrossRef] [PubMed]
- O’Shea, T.J.; Cryan, P.M.; Cunningham, A.A.; Fooks, A.R.; Hayman, D.T.S.; Luis, A.D.; Peel, A.J.; Plowright, R.K.; Wood, J.L.N. Bat Flight and Zoonotic Viruses. Emerg. Infect. Dis. 2014, 20, 741–745. [Google Scholar] [CrossRef] [PubMed]
- Calisher, C.H.; Childs, J.E.; Field, H.E.; Holmes, K.V.; Schountz, T. Bats: Important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 2006, 19, 531. [Google Scholar] [CrossRef] [PubMed]
- George, D.B.; Webb, C.T.; Farnsworth, M.L.; O’Shea, T.J.; Bowen, R.A.; Smith, D.L.; Stanley, T.R.; Ellison, L.E.; Rupprecht, C.E. Host and viral ecology determine bat rabies seasonality and maintenance. Proc. Natl. Acad. Sci. USA 2011, 108, 10208–10213. [Google Scholar] [CrossRef] [PubMed]
- Hayman, D.T. As the bat flies. Science 2016, 354, 1099–1100. [Google Scholar] [CrossRef] [PubMed]
- Luis, A.D.; Hayman, D.T.S.; O’Shea, T.J.; Cryan, P.M.; Gilbert, A.T.; Pulliam, J.R.C.; Mills, J.N.; Timonin, M.E.; Willis, C.K.R.; Cunningham, A.A.; et al. A comparison of bats and rodents as reservoirs of zoonotic viruses: Are bats special? Proc. R. Soc. Lond. B Biol. Sci. 2013, 280, 20122753. [Google Scholar] [CrossRef] [PubMed]
- Hoar, B.R.; Chomel, B.B.; Rodriguez, F.D.A.; Colley, P.A. Zoonoses and potential zoonoses transmitted by bats. J. Am. Vet. Med. Assoc. 1998, 212, 1714–1720. [Google Scholar] [PubMed]
- Plowright, R.K.; Foley, P.; Field, H.E.; Dobson, A.P.; Foley, J.E.; Eby, P.; Daszak, P. Urban habituation, ecological connectivity and epidemic dampening: The emergence of Hendra virus from flying foxes (Pteropus spp.). Proc. R. Soc. Lond. B Biol. Sci. 2011, 278, 3703–3712. [Google Scholar] [CrossRef] [PubMed]
- Kuzmin, I.V.; Bozick, B.; Guagliardo, S.A.; Kunkel, R.; Shak, J.R.; Tong, S.; Rupprecht, C.E. Bats, emerging infectious diseases, and the rabies paradigm revisited. Emerg. Health Threats J. 2011, 4, 7159. [Google Scholar] [CrossRef] [PubMed]
- Drexler, J.F.; Corman, V.M.; Wegner, T.; Tateno, A.F.; Zerbinati, R.M.; Gloza-Rausch, F.; Seebens, A.; Muller, M.A.; Drosten, C. Amplification of Emerging Viruses in a Bat Colony. Emerg. Infect. Dis. 2011, 17, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Dufkova, L.; Strakova, P.; Sirmarova, J.; Salat, J.; Moutelikova, R.; Chrudimsky, T.; Bartonicka, T.; Nowotny, N.; Ruzek, D. Detection of Diverse Novel Bat Astrovirus Sequences in the Czech Republic. Vector Borne Zoonotic Dis. 2015, 15, 518–521. [Google Scholar] [CrossRef] [PubMed]
- Rougeron, V.; Suquet, E.; Maganga, G.D.; Jiolle, D.; Mombo, I.M.; Bourgarel, M.; Motsch, P.; Arnathau, C.; Durand, P.; Drexler, F.; et al. Characterization and phylogenetic analysis of new bat astroviruses detected in Gabon, Central Africa. Acta Virol. 2016, 60, 386–392. [Google Scholar] [CrossRef] [PubMed]
- Wikimedia Commons. Available online: https://commons.wikimedia.org/wiki/File:A_large_blank_world_map_with_oceans_marked_in_blue.svg (accessed on 29 December 2016).
- Chu, D.K.W.; Chin, A.W.H.; Smith, G.J.; Chan, K.H.; Guan, Y.; Peiris, J.S.M.; Poon, L.L.M. Detection of novel astroviruses in urban brown rats and previously known astroviruses in humans. J. Gen. Virol. 2010, 91, 2457–2462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouzalas, I.G.; Wuthrich, D.; Selimovic-Hamza, S.; Drogemuller, C.; Bruggmann, R.; Seuberlich, T. Full-genome based molecular characterization of encephalitis-associated bovine astroviruses. Infect. Genet. Evol. 2016, 44, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Selimovic-Hamza, S.; Bouzalas, I.G.; Vandevelde, M.; Oevermann, A.; Seuberlich, T. Detection of astrovirus in historical cases of European sporadic bovine encephalitis, Switzerland 1958–1976. Front. Vet. Sci. 2016, 3, 91. [Google Scholar] [CrossRef] [PubMed]
- Bouzalas, I.G.; Wuthrich, D.; Walland, J.; Drogemuller, C.; Zurbriggen, A.; Vandevelde, M.; Oevermann, A.; Bruggmann, R.; Seuberlich, T. Neurotropic astrovirus in cattle with nonsuppurative encephalitis in Europe. J. Clin. Microbiol. 2014, 52, 3318–3324. [Google Scholar] [CrossRef] [PubMed]
- Seuberlich, T.; Wuthrich, D.; Selimovic-Hamza, S.; Drogemuller, C.; Oevermann, A.; Bruggmann, R.; Bouzalas, I. Identification of a second encephalitis-associated astrovirus in cattle. Emerg. Microbes Infect. 2016, 5, e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Diab, S.; McGraw, S.; Barr, B.; Traslavina, R.; Higgins, R.; Talbot, T.; Blanchard, P.; Rimoldi, G.; Fahsbender, E.; et al. Divergent astrovirus associated with neurologic disease in cattle. Emerg. Infect. Dis. 2013, 19, 1385–1392. [Google Scholar] [CrossRef] [PubMed]
- Pfaff, F.; Schlottau, K.; Scholes, S.; Courtenay, A.; Hoffmann, B.; Höper, D.; Beer, M. A novel astrovirus associated with enephalitis and ganlionitis in domestic sheep. Transbound. Emerg. Dis. 2017, 1–6. [Google Scholar]
- Laurin, M.A.; Dastor, M.; L’Homme, Y. Detection and genetic characterization of a novel pig astrovirus: Relationship to other astroviruses. Arch. Virol. 2011, 156, 2095–2099. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.T.; Gimenez-Lirola, L.G.; Gerber, P.F.; Jiang, Y.H.; Halbur, P.G.; Opriessnig, T. Identification and characterization of novel porcine astroviruses (PAstVs) with high prevalence and frequent co-infection of individual pigs with multiple PAstV types. J. Gen. Virol. 2013, 94, 570–582. [Google Scholar] [CrossRef] [PubMed]
- Woode, G.N.; Bridger, J.C. Isolation of small viruses resembling astroviruses and caliciviruses from acute enteritis of calves. J. Med. Microbiol. 1978, 11, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Aroonprasert, D.; Fagerland, J.A.; Kelso, N.E.; Zheng, S.; Woode, G.N. Cultivation and partial characterization of bovine astrovirus. Vet. Microbiol. 1989, 19, 113–125. [Google Scholar] [CrossRef]
- Woode, G.N.; Gourley, N.E.; Pohlenz, J.F.; Liebler, E.M.; Mathews, S.L.; Hutchinson, M.P. Serotypes of bovine astrovirus. J. Clin. Microbiol. 1985, 22, 668–670. [Google Scholar] [PubMed]
- Nagai, M.; Omatsu, T.; Aoki, H.; Otomaru, K.; Uto, T.; Koizumi, M.; Minami-Fukuda, F.; Takai, H.; Murakami, T.; Masuda, T.; et al. Full genome analysis of bovine astrovirus from fecal samples of cattle in Japan: Identification of possible interspecies transmission of bovine astrovirus. Arch. Virol. 2015, 160, 2491–2501. [Google Scholar] [CrossRef] [PubMed]
- Blomstrom, A.L.; Widen, F.; Hammer, A.S.; Belak, S.; Berg, M. Detection of a novel astrovirus in brain tissue of mink suffering from shaking mink syndrome by use of viral metagenomics. J. Clin. Microbiol. 2010, 48, 4392–4396. [Google Scholar] [CrossRef] [PubMed]
- Brnic, D.; Prpic, J.; Keros, T.; Roic, B.; Staresina, V.; Jemersic, L. Porcine astrovirus viremia and high genetic variability in pigs on large holdings in Croatia. Infect. Genet. Evol. 2013, 14, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Geyer, A.; Steele, A.D.; Peenze, I.; Lecatsas, G. Astrovirus-like particles, adenoviruses and rotaviruses associated with diarrhoea in piglets. J. S. Afr. Vet. Assoc. 1994, 65, 164–166. [Google Scholar] [PubMed]
- Shan, T.; Li, L.; Simmonds, P.; Wang, C.; Moeser, A.; Delwart, E. The fecal virome of pigs on a high-density farm. J. Virol. 2011, 85, 11697–11708. [Google Scholar] [CrossRef] [PubMed]
- Ulloa, J.C.; Gutierrez, M.F. Genomic analysis of two ORF2 segments of new porcine astrovirus isolates and their close relationship with human astroviruses. Can. J. Microbiol. 2010, 56, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.T.; Luo, Z.; Lv, S.L.; Opriessnig, T.; Li, R.C.; Yu, X.L. Identification and characterization of multiple porcine astrovirus genotypes in Hunan province, China. Arch Virol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Brnic, D.; Jemersic, L.; Keros, T.; Prpic, J. High prevalence and genetic heterogeneity of porcine astroviruses in domestic pigs. Vet. J. 2014, 202, 390–392. [Google Scholar] [CrossRef] [PubMed]
- Indik, S.; Valicek, L.; Smid, B.; Dvorakova, H.; Rodak, L. Isolation and partial characterization of a novel porcine astrovirus. Vet. Microbiol. 2006, 117, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Machnowska, P.; Ellerbroek, L.; Johne, R. Detection and characterization of potentially zoonotic viruses in faeces of pigs at slaughter in Germany. Vet. Microbiol. 2014, 168, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, J.B.; Lee, T.W. Human astrovirus serotypes. Lancet 1984, 2, 1405. [Google Scholar] [CrossRef]
- Bosch, A.; Guix, S.; Krishna, N.K.; Brown, M.; Davison, A.J. Astroviruses. In Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Elsevier: San Diego, CA, USA, 2012. [Google Scholar]
- Kapoor, A.; Li, L.; Victoria, J.; Oderinde, B.; Mason, C.; Pandey, P.; Zaidi, S.Z.; Delwart, E. Multiple novel astrovirus species in human stool. J. Gen. Virol. 2009, 90, 2965–2972. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.R.; Morfopoulou, S.; Hubb, J.; Emmett, W.A.; Ip, W.; Shah, D.; Brooks, T.; Paine, S.M.; Anderson, G.; Virasami, A.; et al. Astrovirus VA1/HMO-C: An increasingly recognized neurotropic pathogen in immunocompromised patients. Infect. Dis. Soc. 2015, 60, 881–888. [Google Scholar] [CrossRef] [PubMed]
- Fremond, M.L.; Perot, P.; Muth, E.; Cros, G.; Dumarest, M.; Mahlaoui, N.; Seilhean, D.; Desguerre, I.; Hebert, C.; Corre-Catelin, N.; et al. Next-Generation Sequencing for Diagnosis and Tailored Therapy: A Case Report of Astrovirus-Associated Progressive Encephalitis. J. Pediatr. Infect. Dis. Soc. 2015, 4, e53–e57. [Google Scholar] [CrossRef] [PubMed]
- Walter, J.E.; Mitchell, D.K.; Guerrero, M.L.; Berke, T.; Matson, D.O.; Monroe, S.S.; Pickering, L.K.; Ruiz-Palacios, G. Molecular epidemiology of human astrovirus diarrhea among children from a periurban community of Mexico City. J. Infect. Dis. 2001, 183, 681–686. [Google Scholar] [CrossRef] [PubMed]
- Wolfaardt, M.; Kiulia, N.M.; Mwenda, J.M.; Taylor, M.B. Evidence of a recombinant wild-type human astrovirus strain from a Kenyan child with gastroenteritis. J. Clin. Microbiol. 2011, 49, 728–731. [Google Scholar] [CrossRef] [PubMed]
- Marshall, J.A.; Kennett, M.L.; Rodger, S.M.; Studdert, M.J.; Thompson, W.L.; Gust, I.D. Virus and virus-like particles in the faeces of cats with and without diarrhoea. Aust. Vet. J. 1987, 64, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Krishna, N.K. Identification of structural domains involved in astrovirus capsid biology. Viral Immunol. 2005, 18, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Brinker, J.P.; Blacklow, N.R.; Herrmann, J.E. Human astrovirus isolation and propagation in multiple cell lines. Arch. Virol. 2000, 145, 1847–1856. [Google Scholar] [CrossRef] [PubMed]
| Family | Species | Region | Tested Animals | Positive Results (%) | Reference |
|---|---|---|---|---|---|
| Vespertilionidae | Barbastella barbastellus | Hungary | 13 | 0 (0%) | [20] |
| Eptesicus nilssonii | Czech Republic | 1 | 0 (0%) | [39] | |
| Eptesicus serotinus | Hungary | 3 | 0 (0%) | [21] | |
| Hungary | 7 | 0 (0%) | [20] | ||
| Czech Republic | 1 | 1 (100%) | [39] | ||
| Hesperoptenus sp. | Cambodia | 1 | 0 (0%) | [15] | |
| Hypsugo savii | Czech Republic | 4 | 1 (25%) | [39] | |
| Ia io | China | 11 | 4 (36.4%) | [22] | |
| Lao PDR | 32 | 1 (3.1%) | [15] | ||
| Miniopterus inflatus | Gabon | 155 | 16 (10.3%) | [40] | |
| Miniopterus magnater | China (Hong Kong) | 122 | 67 (54.9%) | [11] | |
| Miniopterus pusillus | China (Hong Kong) | 73 | 31 (42.5%) | [11] | |
| Miniopterus schreibersii | China (Hong Kong) | 3 | 3 (100%) | [11] | |
| China | 19 | 12 (63.2%) | [22] | ||
| China | 187 | 22 (11.8%) | [23] | ||
| Hungary | 15 | 12 (80%) | [20] | ||
| Myotis alcathoe | Hungary | 16 | 0 (0%) | [20] | |
| Myotis bechsteinii | Hungary | 22 | 1 (4.5%) | [21] | |
| Hungary | 125 | 5 (4%) | [20] | ||
| Czech Republic | 1 | 0 (0%) | [39] | ||
| Germany | 321 | 35 (10.9%) | [19] | ||
| Myotis brandtii | Hungary | 3 | 0 (0%) | [20] | |
| Myotis blythii | Hungary | 2 | 0 (0%) | [21] | |
| Hungary | 10 | 0 (0%) | [20] | ||
| Myotis chinensis | China (Hong Kong) | 9 | 3 (33.3%) | [11] | |
| Myotis dasycneme | Hungary | 11 | 0 (0%) | [20] | |
| Myotis daubentonii | Hungary | 7 | 3 (42.9%) | [21] | |
| Hungary | 81 | 6 (7.4%) | [20] | ||
| Czech Republic | 3 | 0 (0%) | [39] | ||
| Germany | 47 | 30 (63.8%) | [19] | ||
| Myotis dasycheme | Hungary | 4 | 0 (0%) | [21] | |
| Myotis emarginatus | Hungary | 5 | 1 (20%) | [20] | |
| Czech Republic | 1 | 1 (100%) | [39] | ||
| Myotis horsfieldii | Cambodia | 47 | 20 (42.6%) | [15] | |
| Myotis myotis | Hungary | 6 | 0 (0%) | [21] | |
| Hungary | 29 | 0 (0%) | [20] | ||
| Myotis mystacinus | Hungary | 1 | 0 (0%) | [20] | |
| Czech Republic | 1 | 1 (100%) | [39] | ||
| Myotis nattereri | Hungary | 4 | 0 (0%) | [21] | |
| Hungary | 37 | 1 (2.7%) | [20] | ||
| Germany | 248 | 99 (39.9%) | [19] | ||
| Myotis pilosus | China (Hong Kong) | 12 | 10 (83.3%) | [11] | |
| China | 16 | 2 (12.5%) | [22] | ||
| China | 1 | 0 (0%) | [23] | ||
| Myotis spp. | China | 5 | 3 (60%) | [22] | |
| Nyctalus leisleri | Hungary | 6 | 0 (0%) | [20] | |
| Nyctalus noctula | Hungary | 14 | 4 (28.6%) | [20] | |
| Czech Republic | 7 | 1 (14.3%) | [39] | ||
| Nyctalus plancyi velutinus | China | 1 | 0 (0%) | [22] | |
| Pipistrellus abramus | China (Hong Kong) | 2 | 1 (50%) | [11] | |
| China | 20 | 1 (5%) | [22] | ||
| Pipistrellus nathusii | Hungary | 3 | 0 (0%) | [20] | |
| Czech Republic | 1 | 0 (0%) | [39] | ||
| Germany | 22 | 6 (27.3%) | [19] | ||
| Pipistrellus pipistrellus | Hungary | 1 | 0 (0%) | [21] | |
| Hungary | 12 | 0 (0%) | [20] | ||
| Czech Republic | 12 | 1 (8.3%) | [39] | ||
| Germany | 7 | 0 (0%) | [19] | ||
| Pipistrellus pygmaeus | Hungary | 6 | 1 (16.7%) | [20] | |
| Czech Republic | 1 | 1 (100%) | [39] | ||
| Germany | 12 | 6 (50%) | [19] | ||
| Pipistrellus spp. | China | 5 | 0 (0%) | [22] | |
| Cambodia | 29 | 0 (0%) | [15] | ||
| Plecotus auritus | Hungary | 11 | 1 (9.1%) | [21] | |
| Hungary | 29 | 1 (3.4%) | [20] | ||
| Czech Republic | 2 | 0 (0%) | [39] | ||
| Germany | 118 | 24 (20.3%) | [19] | ||
| Plecotus austriacus | Hungary | 3 | 0 (0%) | [20] | |
| Czech Republic | 2 | 0 (0%) | [39] | ||
| Scotophilus kuhlii | China | 38 | 6 (15.8%) | [23] | |
| China | 2 | 0 (0%) | [22] | ||
| Scotophilus spp. | Cambodia | 524 | 39 (7.4%) | [15] | |
| Tylonycteris pachypus | China | 2 | 0 (0%) | [22] | |
| Tylonycteris sp. | Cambodia | 1 | 0 (0%) | [15] | |
| Vespertilio murinus | Hungary | 3 | 0 (0%) | [20] | |
| Czech Republic | 5 | 1 (20%) | [39] | ||
| TOTAL | 2468 | 469 (19%) | |||
| Rhinolophidae | Rhinolophus affinis | China | 2 | 0 (0%) | [22] |
| Rhinolophus euryale | Hungary | 3 | 0 (0%) | [20] | |
| Rhinolophus ferrumequinum | China | 7 | 0 (0%) | [23] | |
| China | 4 | 2 (50%) | [22] | ||
| Hungary | 12 | 0 (0%) | [20] | ||
| Rhinolophus hipposideros | Hungary | 3 | 0 (0%) | [20] | |
| Czech Republic | 2 | 1 (50%) | [39] | ||
| Rhinolophus lepidus | China | 11 | 0 (0%) | [23] | |
| Rhinolophus macrotis | China | 2 | 0 (0%) | [23] | |
| China | 1 | 0 (0%) | [22] | ||
| Rhinolophus pearsonii | China | 1 | 1 (100%) | [22] | |
| Rhinolophus rouxii | China (Hong Kong) | 8 | 1 (12.5%) | [11] | |
| Rhinolophus sinicus | China | 1 | 0 (0%) | [22] | |
| Rhinolophus sp. | Cambodia | 53 | 1 (1.9%) | [15] | |
| Lao PDR | 102 | 4 (3.9%) | [15] | ||
| TOTAL | 212 | 10 (4.7%) | |||
| Hipposideridae | Aselliscus stoliczkanus | China | 1 | 0 (0%) | [22] |
| Aselliscus sp. | Lao PDR | 7 | 0 (0%) | [15] | |
| Hipposideros armiger | China (Hong Kong) | 10 | 0 (0% | [11] | |
| China | 109 | 21 (19.3%) | [22] | ||
| Hipposideros gigas | Gabon | 226 | 7 (3.1%) | [40] | |
| Hipposideros larvatus | China | 29 | 4 (13.8%) | [22] | |
| China | 1 | 0 (0%) | [23] | ||
| Hipposideros pomona | China | 95 | 13 (13.7%) | [22] | |
| China | 15 | 0 (0%) | [23] | ||
| Hipposideros ruber | Gabon | 394 | 17 (4.3%) | [40] | |
| Hipposideros spp. | Cambodia | 4 | 1 (25%) | [15] | |
| Lao PDR | 26 | 1 (3.8%) | [15] | ||
| TOTAL | 917 | 64 (7.0%) | |||
| Pteropodidae | Cynopterus sphinx | China (Hong Kong) | 11 | 0 (0%) | [11] |
| Cynopterus spp. | Cambodia | 321 | 0 (0%) | [15] | |
| Lao PDR | 19 | 0 (0%) | [15] | ||
| Eonycteris sp. | Cambodia | 28 | 0 (0%) | [15] | |
| Lao PDR | 51 | 3 (5.9%) | [15] | ||
| Macroglossus sp. | Cambodia | 21 | 0 (0%) | [15] | |
| Lao PDR | 1 | 0 (0%) | [15] | ||
| Megaerops sp. | Cambodia | 29 | 0 (0%) | [15] | |
| Lao PDR | 69 | 0 (0%) | [15] | ||
| Pteropus sp. | Cambodia | 10 | 0 (0%) | [15] | |
| Rousettus aegyptiacus | Gabon | 162 | 2 (1.2%) | [40] | |
| Rousettus leschenaultia | China | 59 | 1 (1.7%) | [23] | |
| Rousettus sp. | Cambodia | 11 | 1 (9.1%) | [15] | |
| Lao PDR | 322 | 23 (7.1%) | [15] | ||
| TOTAL | 1114 | 30 (2.7%) | |||
| Emballonuridae | Coleura afra | Gabon | 25 | 2 (8%) | [40] |
| Taphozous melanopogon | China | 172 | 160 (93%) | [22] | |
| Taphozous spp. | Cambodia | 147 | 4 (2.7%) | [15] | |
| TOTAL | 344 | 166 (48.3%) | |||
| Megadermatidae | Megaderma lyra | China | 1 | 1 (100%) | [22] |
| Cambodia | 21 | 2 (9.5%) | [15] | ||
| TOTAL | 22 | 3 (13.6%) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fischer, K.; Pinho dos Reis, V.; Balkema-Buschmann, A. Bat Astroviruses: Towards Understanding the Transmission Dynamics of a Neglected Virus Family. Viruses 2017, 9, 34. https://doi.org/10.3390/v9020034
Fischer K, Pinho dos Reis V, Balkema-Buschmann A. Bat Astroviruses: Towards Understanding the Transmission Dynamics of a Neglected Virus Family. Viruses. 2017; 9(2):34. https://doi.org/10.3390/v9020034
Chicago/Turabian StyleFischer, Kerstin, Vinícius Pinho dos Reis, and Anne Balkema-Buschmann. 2017. "Bat Astroviruses: Towards Understanding the Transmission Dynamics of a Neglected Virus Family" Viruses 9, no. 2: 34. https://doi.org/10.3390/v9020034





