1. Introduction
The hydrodynamic injection (HI) mouse model has been explored in different studies in hepatitis B virus (HBV) research [
1]. It has been proven that the HBV mouse model based on HI could be used to study viral replication and persistence in order to analyze the immunological factors required for HBV clearance and to evaluate novel antiviral therapy strategies [
1,
2,
3,
4,
5,
6,
7,
8,
9,
10]. HI of plasmid pAAV/HBV1.2 (containing a 1.2-fold overlength HBV genome) was found in a previous study to cause HBV replication and HBV surface antigen (HBsAg) expression that persisted for more than six months in approximately 40% of the injected mice [
3]. It was speculated that the outcome after HI was also determined by the backbone of plasmids and the genetic background of injected mice. In our previous studies, we explored pAAV/HBV1.3 (containing a 1.3-fold overlength HBV genome) in C57BL/6 mice. We found that high levels of serum HBsAg and HBV DNA were detected at seven days post injection (dpi), and they declined to undetectable levels at 28 dpi, which indicated temporary HBV replication and antigen expression [
2,
7,
11]. Therefore, we speculated that in addition to the backbone of plasmids and the genetic background of the injected specimen, viral characteristics could also be an independent determinant for viral replication and persistence in the HI mouse model.
Woodchuck hepatitis virus (WHV), like HBV, is a member of the family
Hepadnaviridae. WHV and HBV share a remarkable similarity in genome organization and replication strategy [
12]. Their nucleotide (nt) sequences were found to have a homology of 62%–70% [
13]. Early studies revealed that the HBV capsid could encapsidate WHV polymerase-epsilon complexes and vice versa [
14]. Likewise, defective HBV polymerase (POL) could be complemented by WHV counterparts, and vice versa [
15]. Furthermore, HBV core antigen (HBcAg) and WHV core antigen (WHcAg) could interact with each other to produce chimeric capsids [
15]. In our previous studies, we constructed two plasmids, pBS-HBV1.3 (pHBV1.3, containing a 1.3-fold overlength HBV genome) and pBS-WHV1.3 (pWHV1.3, containing a 1.3-fold overlength WHV genome) in pBluescript II SK(+) vector. After HI of pHBV1.3 in BALB/c mice, serum HBV antigen and HBV DNA peaked at seven dpi and normally disappeared at 28 dpi. However, we found that WHV viral DNA (vDNA) and antigens could persist up to 45 weeks after HI with pWHV1.3. The reason for WHV persistence in BALB/c mice was unknown and therefore, we wished to explore it.
In the present study, we constructed a series of chimeric WHV-HBV genomes based on pHBV1.3, in which different fragments of the HBV genome were substituted by the counterparts from WHV. We studied whether the chimeric WHV-HBV genomes would persist or be cleared in HI mice model and attempted to determine the viral genomic fragments that assisted in the persistence of the chimeric WHV-HBV genomes.
2. Materials and Methods
2.1. Ethics Statement
Female BALB/c mice (six to eight weeks old) used in this study were purchased from SJA Co., Ltd. (Changsha, China). Mice were kept in specific-pathogen-free (SPF) conditions with free access to water and foods. Guidelines for laboratory animal experiments were strictly followed. This study was conducted under the Permit Number 2010-361 from the Institutional Animal Care and Use Committee of Tongji Medical College (Wuhan, China).
2.2. Chimeric Woodchuck Hepatitis Virus and Hepatitis B Virus (WHV-HBV) Genomes
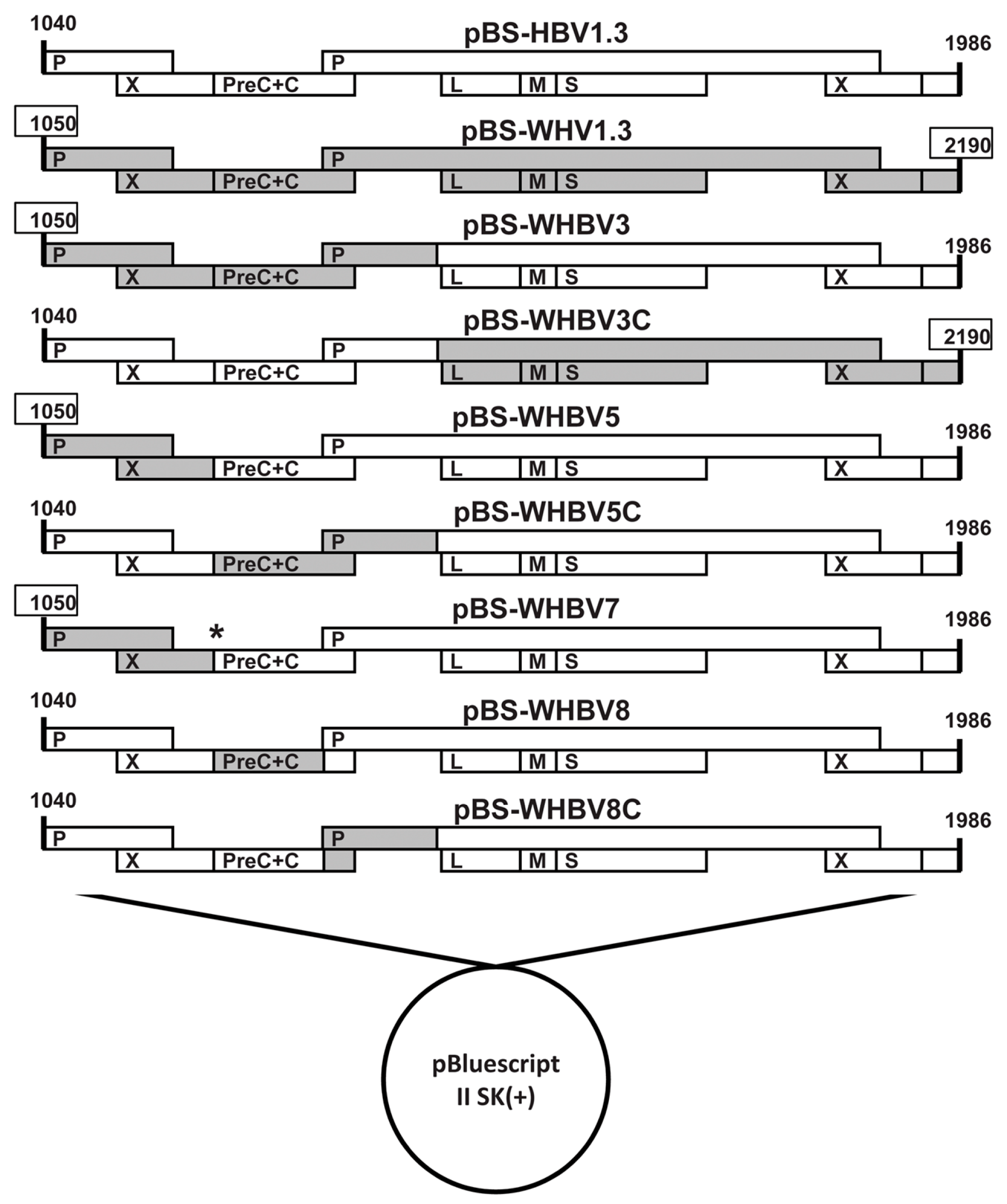
pWHV1.3 contained 1.3 copies of WHV genome (nt 1050–2190, GenBank J04514). pHBV1.3 contained 1.3 copies of the HBV genome (nt 1040–1986, GenBank AY220698). Both pWHV1.3 and pHBV1.3 were constructed based on the pBluescript II SK(+) vector. We constructed a series of chimeric genomes based on pHBV1.3 and pWHV1.3, in which the individual HBV sequences of pHBV1.3 were substituted by the corresponding WHV sequences.
First, we constructed the chimeric plasmid pWHBV3, in which an HBV fragment (nt 1040-2817) was substituted by its counterpart WHV fragment (nt 1050–2950) (
Figure 1,
Figure S1 and
Table S1). The WHV fragment (nt 1050–2950) was amplified from pWHV1.3 using high-fidelity polymerase chain reaction (PCR) with primers WHBV3F and WHBV3R (
Table S2). Both pHBV1.3 and the cloned WHV fragment (nt 1050–2950) were double-digested with the
KpnI and
BstEII and then fused together. pWHBV3 contains the intact HBV
S gene and WHV
C gene, the chimeric
P gene and two
X genes.
Next, we constructed the chimeric plasmids of pWHBV5 and pWHBV5C, in which the inserted WHV fragment (nt 1050–2950) in pWHBV3 was subdivided. For pWHBV5, the WHV fragment (nt 1050–1933) replaced the corresponding HBV fragment (nt 1040–1818), based on pHBV1.3 (
Figure 1,
Figure S1 and
Table S1). The WHV fragment (nt 1050–1933) was amplified from pWHV1.3 with primers WHBV4F and WHBV5FR, and the HBV fragment (nt 1819–2817) was amplified from pHBV1.3 with primers WHBV4R and WHBV5RF (
Table S2). Then, the cloned WHV fragment (nt 1050–1933) and HBV fragment (nt 1819–2817) were fused together by overlapping PCR. The fused WHV-HBV fragment and pHBV1.3 were both digested with the restriction enzymes
XhoI and
BstEII and then fused together, resulting in pWHBV5. pWHBV5C was constructed by the same strategy. For pWHBV5C, the HBV fragment (nt 1820–2817) was replaced by the corresponding WHV fragment (nt 1935–2950), based on pHBV1.3 (
Figure 1,
Figure S1 and
Table S1). The HBV fragment (nt 1040–1819) was amplified from pHBV1.3 with primers WHBV5CF and WHBV5CFR, and the WHV fragment (nt 1935–2950) was amplified from pWHV1.3 using primers WHBV5CRF and WHBV5CR (
Table S2). The cloned HBV fragment (nt 1040–1819) and WHV fragment (nt 1935–2950) were fused together by overlapping PCR. The fused HBV-WHV fragment and pHBV1.3 were both digested by the restriction enzymes
PstІ and
BstEII and then fused together, resulting in pWHBV5C.
Finally, the chimeric plasmids of pWHBV8 and pWHBV8C were constructed based on pWHBV5C, in which the inserted WHV fragment (nt 1935–2950) in pWHBV5C was further subdivided (
Figure 1,
Figure S1, and
Table S1). For pWHBV8, the HBV fragment (nt 1820–2331) was substituted by the corresponding WHV fragment (nt 1935–2449) based on pHBV1.3. The fragment (HBV nt 1040–1819, WHV nt 1935–2449) was digested from pWHBV5C by the restriction enzymes
PstIand
BspEI and then fused with the digested pHBV1.3 fragment using the same restriction enzymes, resulting in pWHBV8. For pWHBV8C, the HBV fragment (nt 2332–2817) was substituted by the corresponding WHV fragment (nt 2450–2950) based on pHBV1.3 (
Figure 1,
Figure S1 and
Table S1). Two HBV fragments (nt 1040–2031 and nt 2035–2331) were amplified from pHBV1.3 using primers WHBV5CF/WHBV8CFR and WHBV8CRF/WHBV4R, respectively (
Table S2), and were fused by overlapping PCR to change the redundant restriction enzyme
BspEI site (TCT to AGC). The fused HBV fragment (HBV nt 1040–2031, AGC, HBV nt 2035–2331) and pWHBV5C were both digested with
PstI and
BspEI and then fused together, resulting in pWHBV8C.
All of the chimeric HBV-WHV plasmids were sequenced by a commercial service (Beijing Genomics Institute, Shenzhen, China). We found that pWHBV5 had a stop codon at the beginning of the HBV preC region, leading to the termination of HBeAg production. Therefore, pWHBV7 was generated by site-directed mutagenesis using primers WHBV7FR and WHBV7RF to correct the second encoding codon from TAA to CAA in the HBV preC region (
Table S2).
2.3. Hydrodynamic Injection (HI) of BALB/c Mice
HI procedures were performed as previously described [
6]. Briefly, for each mouse, 10 μg of plasmids were diluted in normal saline (10% of mouse body weight) and injected via the tail vein within 8 s.
2.4. Serological Assays
Mouse sera were diluted (1:10), then enzyme-linked immunosorbent assay (ELISA; Kehua Tech, Xiamen, China) was used to measure viral antigens and related antibodies in sera (HBsAg, anti-HBs, HBeAg, anti-HBe, and anti-HBc). The results were indicated as optical density (OD) 450 nm, as measured on a microplate reader (cutoff = 0.1).
2.5. Measurementof Serum Viral DNA (vDNA)
At first, residual input DNA in sera was eliminated by DNase I (TaKaRa Bio Inc., Kusatsu, Japan). vDNA kits (Omega Bio-tek Inc., Norcross, GA, USA) were used to isolate DNA from viruses according to the manufacturer’s instructions. Primer QuantS and QuantAS (
Table S3) were employed to measure vDNA via real-time PCR (qPCR) [
16].
2.6. Measurementof WHV Core Antigen (WHcAg) and HBV Core Antigen (HBcAg) via Immunohistochemical Staining (IHC)
Mice were sacrificed and livers were collected. Then, livers were fixed with formalin and embedded in paraffin. WHcAg and HBcAg in paraffin sections were measured as previously described [
17].
2.7. Measurement of Cytotoxic T Lymphocyte (CTL) Responses
Cytotoxic T lymphocyte (CTL) responses were measured using peptide stimulation and flow cytometry as previously described [
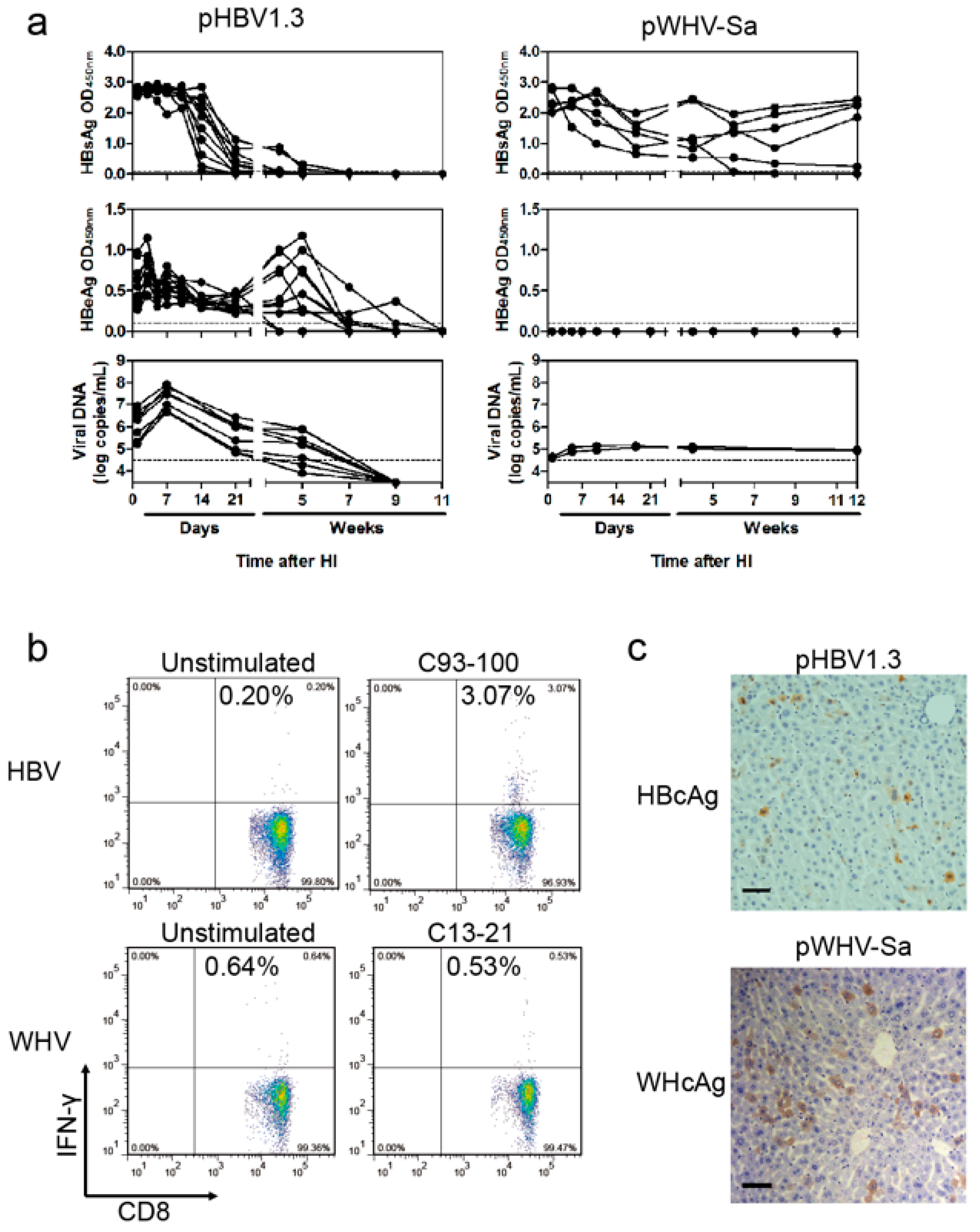
18]. Briefly, at four weeks after HI with pHBV1.3 or pWHV-Sa (WHV) in C57BL/6 mice, the splenocytes were stimulated for six hours with the HBcAg peptide C93-100 (MGLKFRQL) or WHcAg peptide C13-21 (YQLLNFLPL; Sangon Biotech, Shanghai, China), respectively. For cell staining, anti-mIFN-γ-APC and anti-mCD8-PE (BD Biosciences, San Jose, CA, USA) were used. Dead cells were excluded using the LIVE/DEAD fixable dead cell stain kit (Invitrogen, Waltham, MA, USA). Cells were measured on FACSCalibur and analyzed using FlowJo software (Ashland, OR, USA).
2.8. Statistical Analysis
In this study, GraphPad Prism (GraphPad Software Inc., La Jolla, CA, USA) was applied to perform all of the statistical analyses. Student’s t-test was utilized to analyze any differences between the two independent groups. A statistical significance was set at p < 0.05. Results were indicated as means ± SD.
4. Discussion
In this study, we created a series of chimeric WHV-HBV genomes based on a commonly used vector and explored hydrodynamic injection of the chimeric plasmids in BALB/c mice to study genomic factors that determine HBsAg persistence. Consistent with our previous results [
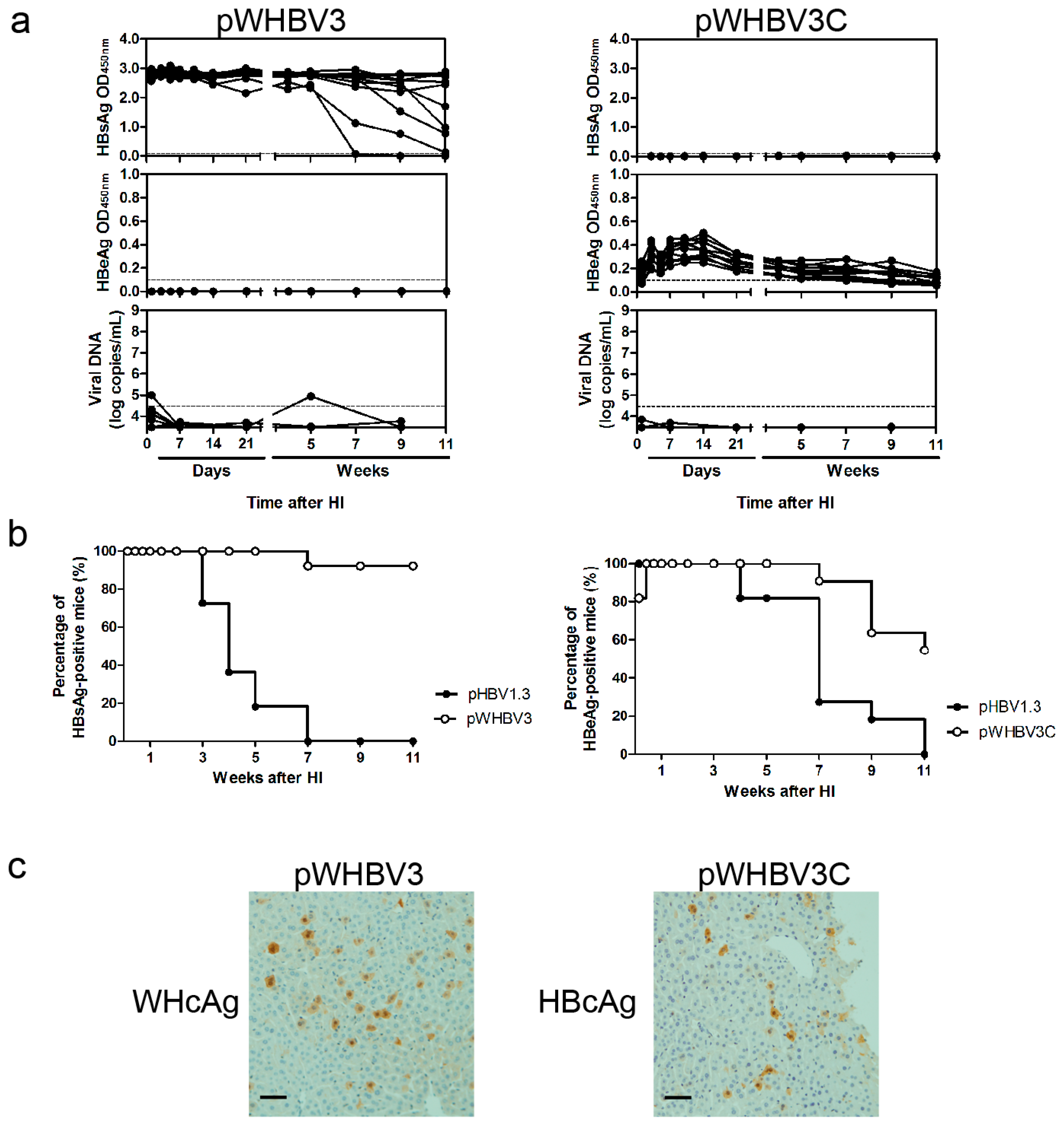
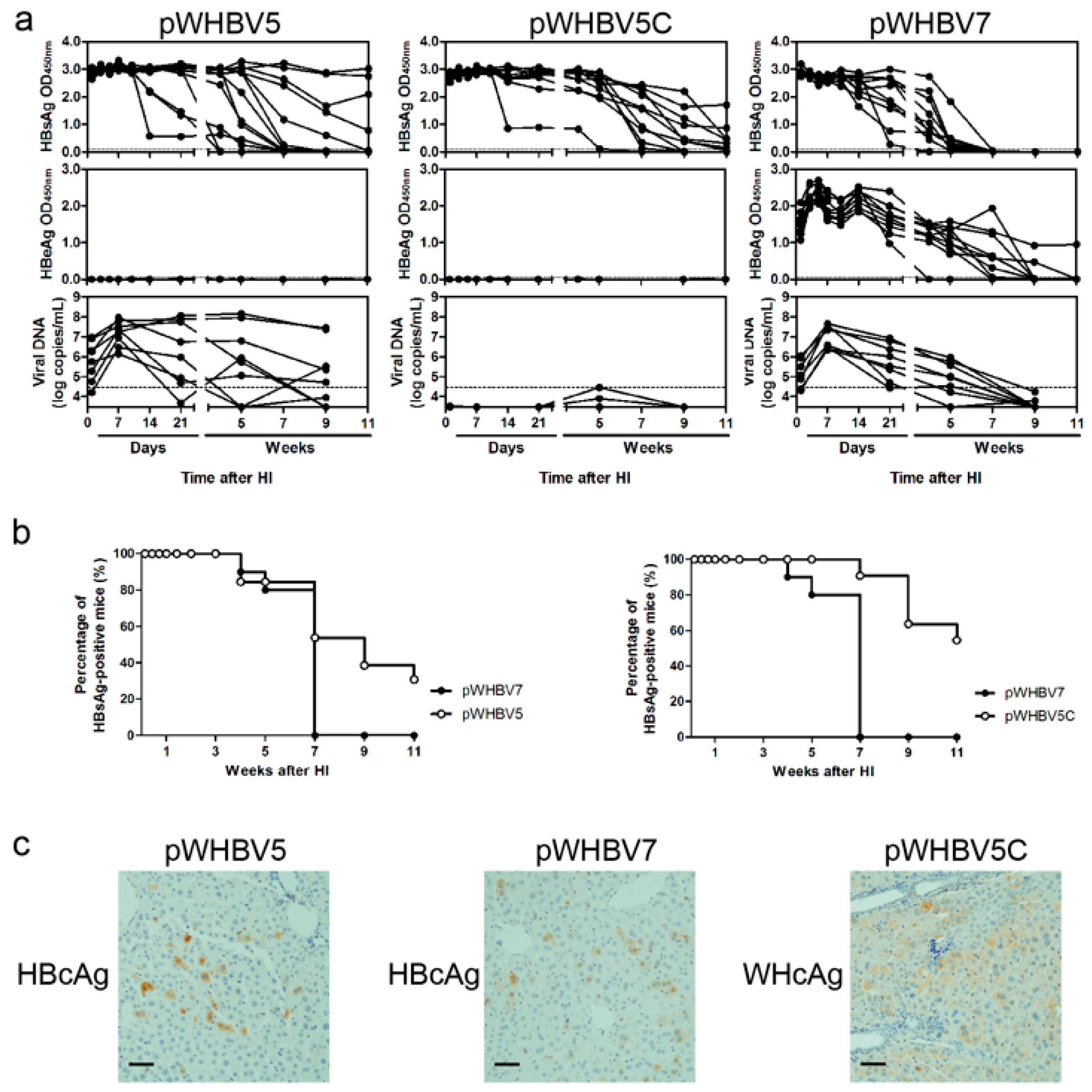
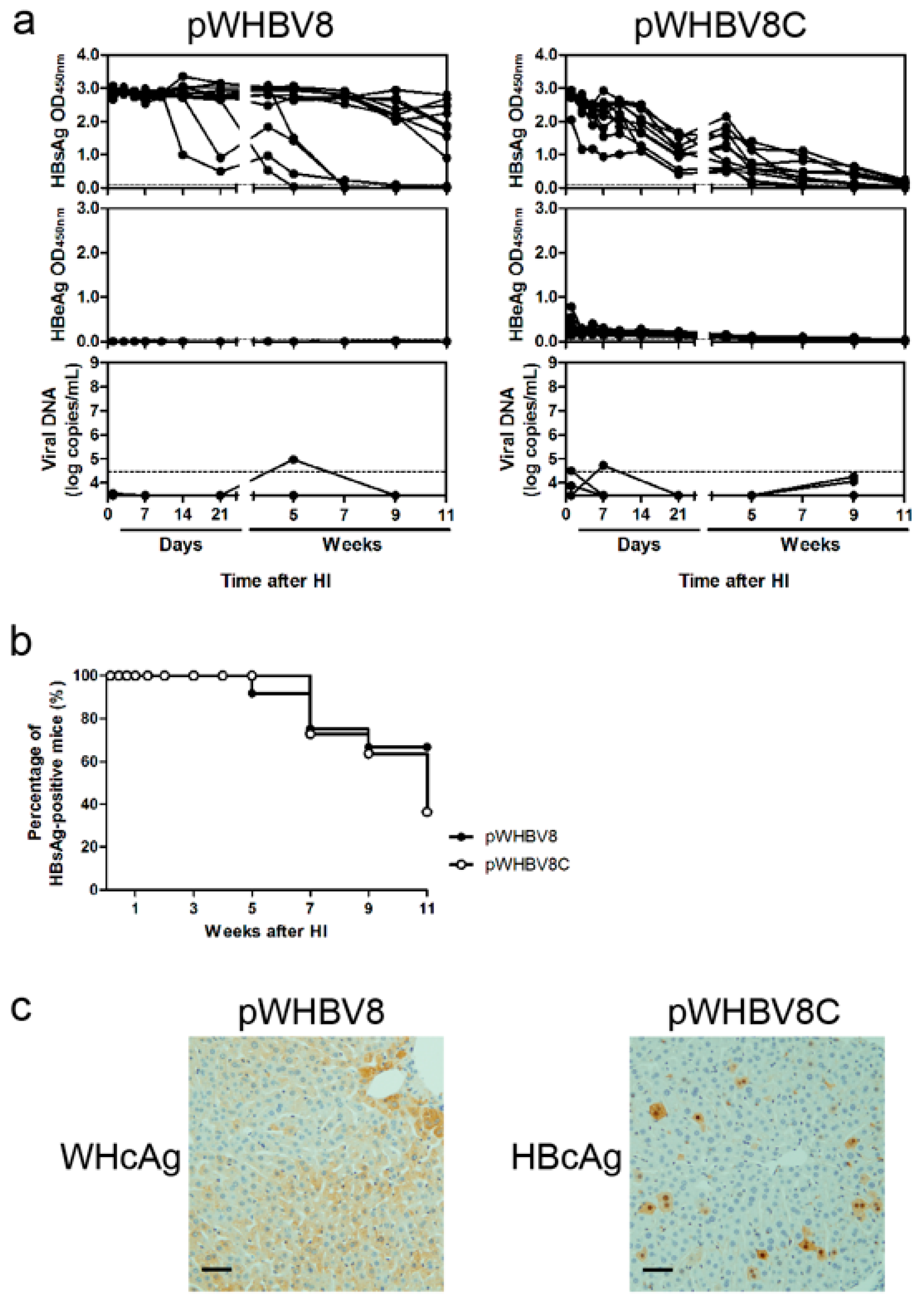
19], a part of such constructs (pWHBV3, pWHBV5C, and pWHBV8) was able to persist in mice and continuously produce HBsAg. For example, pWHBV3 containing a WHV fragment (nt 1050–2950) supported persistent expression of HBsAg in 92% of challenged mice at 11 wpi, while pWHBV5C containing the shorter WHV fragment (nt 1935–2950) supported HBsAg persistence in only 54.5% of mice. Furthermore, pWHBV8 containing the smallest WHV fragment (nt 1935–2449) showed persistent HBsAg antigenemia in 67% of mice. Thus, we assume that the vector backbone does not represent a determinant for the persistence of WHV or the chimeric WHV-HBV genome in the HI mouse model. Compared with pHBV1.3, HBsAg expression in pWHV-Sa challenged mouse sera persisted until week 12 (
Figure 2). We speculated that the low-level replication and the weak immune responses induced by pWHV-Sa might lead to persistent HBsAg expression. It is likely that the persistence is because the expression of viral antigens was below a critical level and did not provoke an effective host immune response before tolerance was established. pWHBV3-, pWHBV5C-, and pWHBV8-challenged mice consistently expressed intrahepatic WHcAg. Moreover, the complementary chimeric plasmids of pWHBV3C, pWHBV7, and pWHBV8C producing intrahepatic HBcAg could not support HBsAg persistence in BALB/c mice. It was reported that HBcAg played an important role in the clearance of HBV infection in the liver. Intrahepatic antiviral response may be triggered by HBcAg. In the HI mouse model, the intrahepatic antiviral response triggered by HBcAg is helpful not only for the clearance of infected hepatocytes but also for the input HBV DNA [
3,
4,
20]. Our results showed that the mice receiving pWHBV3, pWHBV5C, or pWHBV8 with intrahepatic WHcAg expression provoked weak immune responses and failed to clear the chimeric plasmids in the liver, leading to HBsAg persistence. We speculated that WHcAg might present weaker antigenicity than HBcAg in BALB/c mice.
The C-terminal of HBcAg is rich in arginine residues and the C-terminal 39 residues of HBcAg are not required for capsid assembly, but function in pgRNA binding and vDNA synthesis [
21]. It was reported that the last 10 residues of HBcAg played a key role in the immune response induction in the hydrodynamic mouse model [
4]. The chimeric plasmid pWHBV8 encoded the recombinant core protein of the N-terminal 1–145 aa of WHcAg fused with the C-terminal 38 residues of HBcAg. We aligned the C-terminal 38 residues of HBcAg with WHcAg and found that this region was highly conserved. In our study, pWHBV8 persisted in mice with weak immune responses, though it contained only the C-terminal 10 residues of HBcAg. We speculated that the N-terminal 1–145 aa of HBcAg might be equally important for inducing strong immune responses. The weak immune response provoked by pWHBV8 might be due to the weak antigenicity of the assembled WHcAg particle by the N-terminal 1–145 aa of WHcAg in BALB/c mouse. Using the same strategy, we have constructed pWHV-HBV-Sa and pWHV-HBV-SaC145 based on pWHV1.3, in which WHV sequences were substituted by the HBV counterparts. In detail, pWHV-HBV-Sa contained HBsAg a-determinant (aa 121–147). On the other hand, pWHV-HBV-SaC145 was based on pWHV-HBV-Sa and contained one additional HBV fragment, which included the HBV preC region and the part of the C region encoding the N-terminal 1–145 aa of HBcAg. Of the BALB/c mice injected with pWHV-HBV-Sa, 83% of mice presented serum HBsAg positive at 12 wpi. However, the persistence rate of HBsAg antigenemia at 12 wpi in mice receiving pWHV-HBV-SaC145 was only approximately 33% (
Figure S6). These results confirmed that in the HI BALB/c mouse model, the WHV genome fragment (nt 1935–2449) encoding the preC and N-terminal 145 aa of WHcAg could facilitate serum HBsAg persistence for the weak antigenicity of WHcAg. On the other hand, the HBV genome fragment (nt 1814–2331) encoding the preC and N-terminal 145 aa of HBcAg might promote the clearance of chimeric genomes and serum HBsAg in BALB/c mice. In the future, our approach may be used to further study the requirement of viral persistence or clearance in the mouse model based on the HI method.
Interestingly, the chimeric HBV-WHV genomes were able to stably express viral antigens in the liver. These constructs could be further used to express hepadnaviral antigens to study their pathogenic potential; for example, whether the persistent expression of HBsAg or HBeAg may impair intrahepatic immune responses to other antigens. Constructs with mutated viral antigens, such as secretion-defective HBsAg, may also be created to examine their impact on cellular processes.
It is still not clear how the persistent HBsAg and HBeAg expression was achieved in the HI mouse model. We showed that the replication-defective pWHV-Sa genome was not able to persistently express viral antigens under the same conditions [
19], suggesting that viral replication is a prerequisite of continuous expression of viral antigens in the mouse liver. However, it is not yet possible to detect covalently closed circle DNA (cccDNA) in the mouse liver after HI due to the large quantity of input plasmids and replication intermediates. The copy numbers of cccDNA may be extremely low if their existence can even be confirmed. Thus, this will be an issue to be clarified in the future.
We speculated that HBeAg expression might act as an immune activator in BALB/c mice, especially in the innate immune response phase. Therefore, pWHBV7 with HBeAg expression could be cleared at seven wpi, though the viral replication level and the titer of anti-HBc were higher. Without HBeAg expression, HBsAg persisted for a longer time in pWHBV5-injected mice.