Global Transmission Dynamics of Measles in the Measles Elimination Era
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequence Data
2.2. Phylogeographic Analysis
3. Results
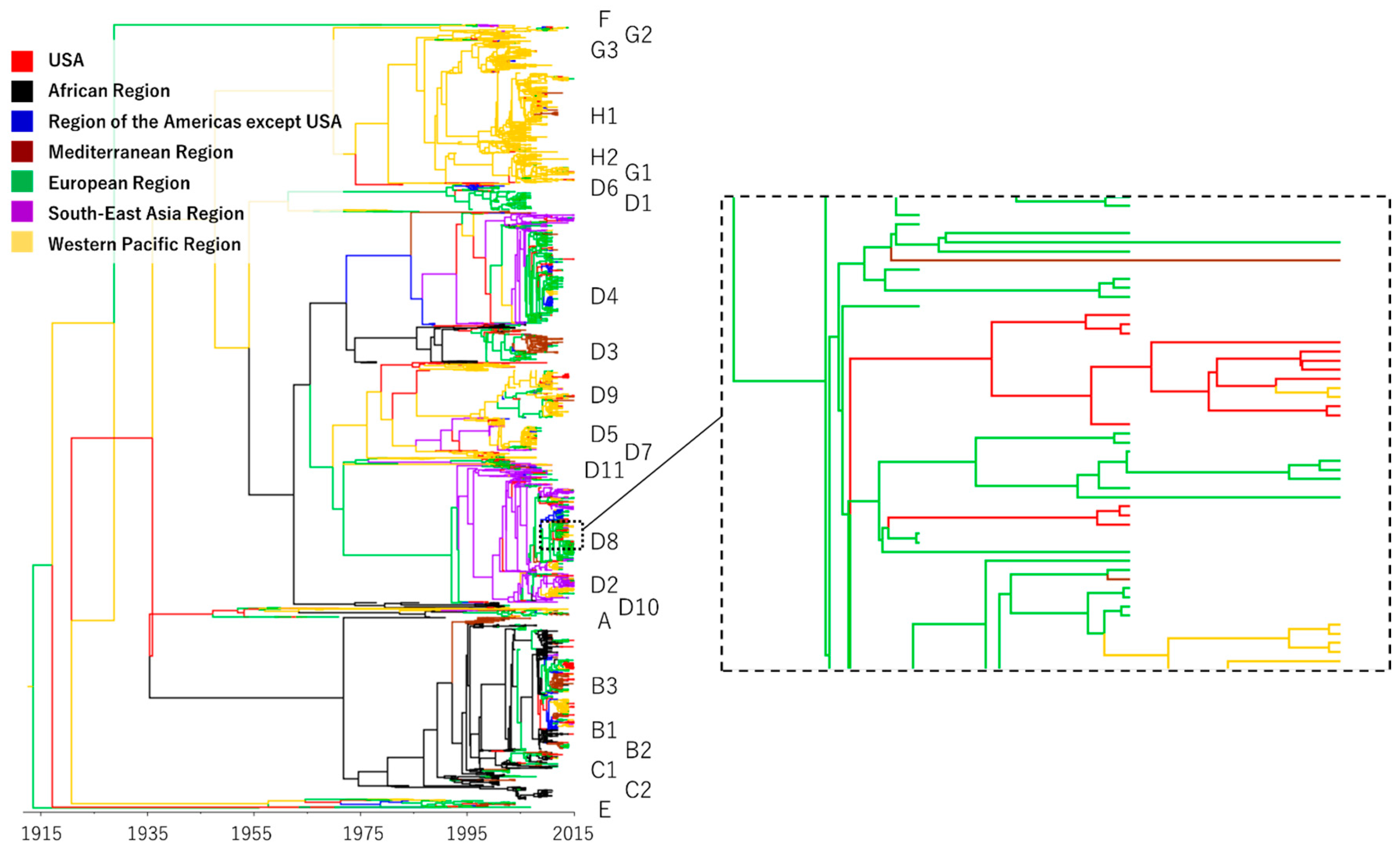
3.1. Phylogeography of Measles Virus
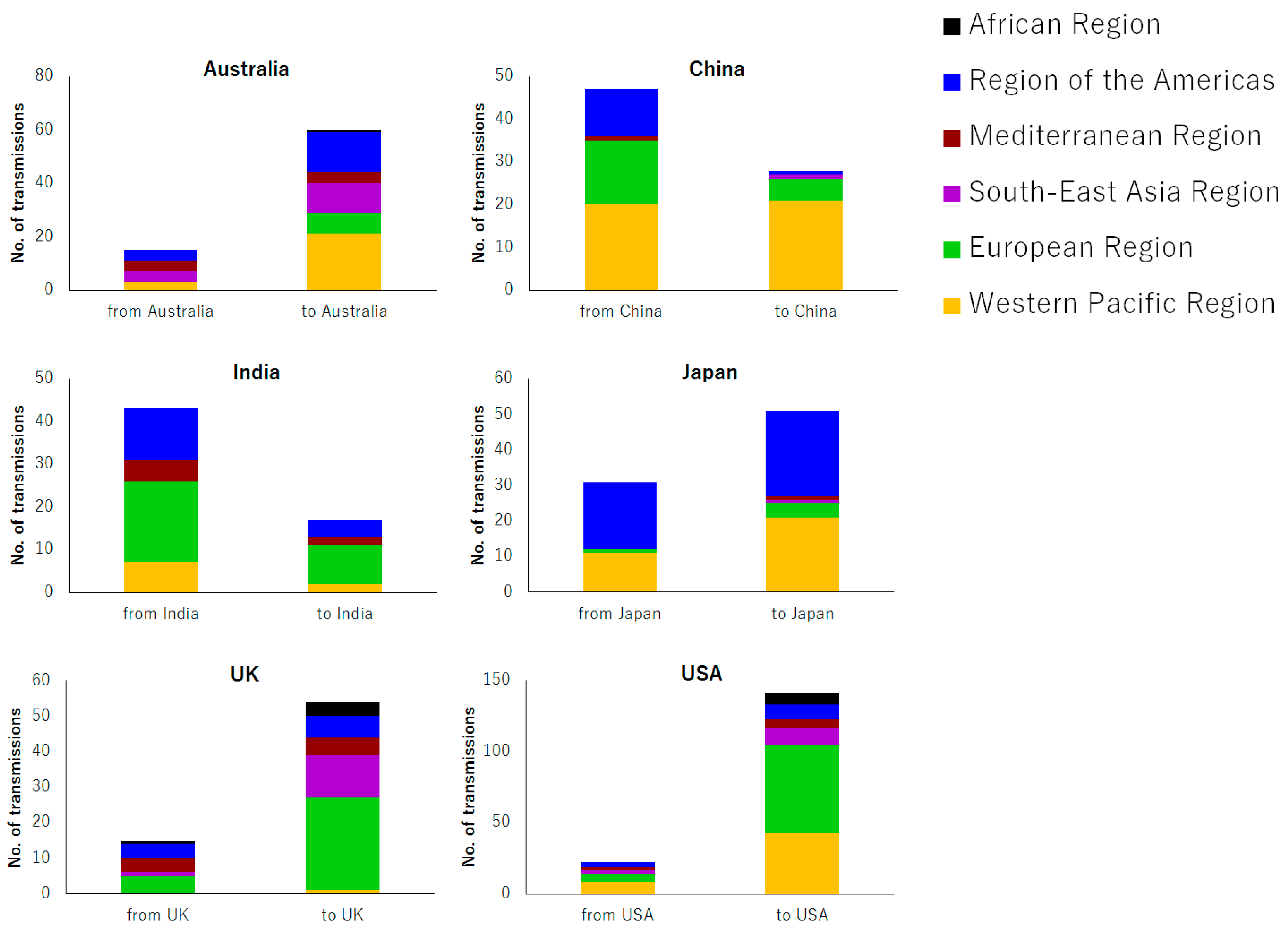
3.2. Geographical Trend in Measles Transmission
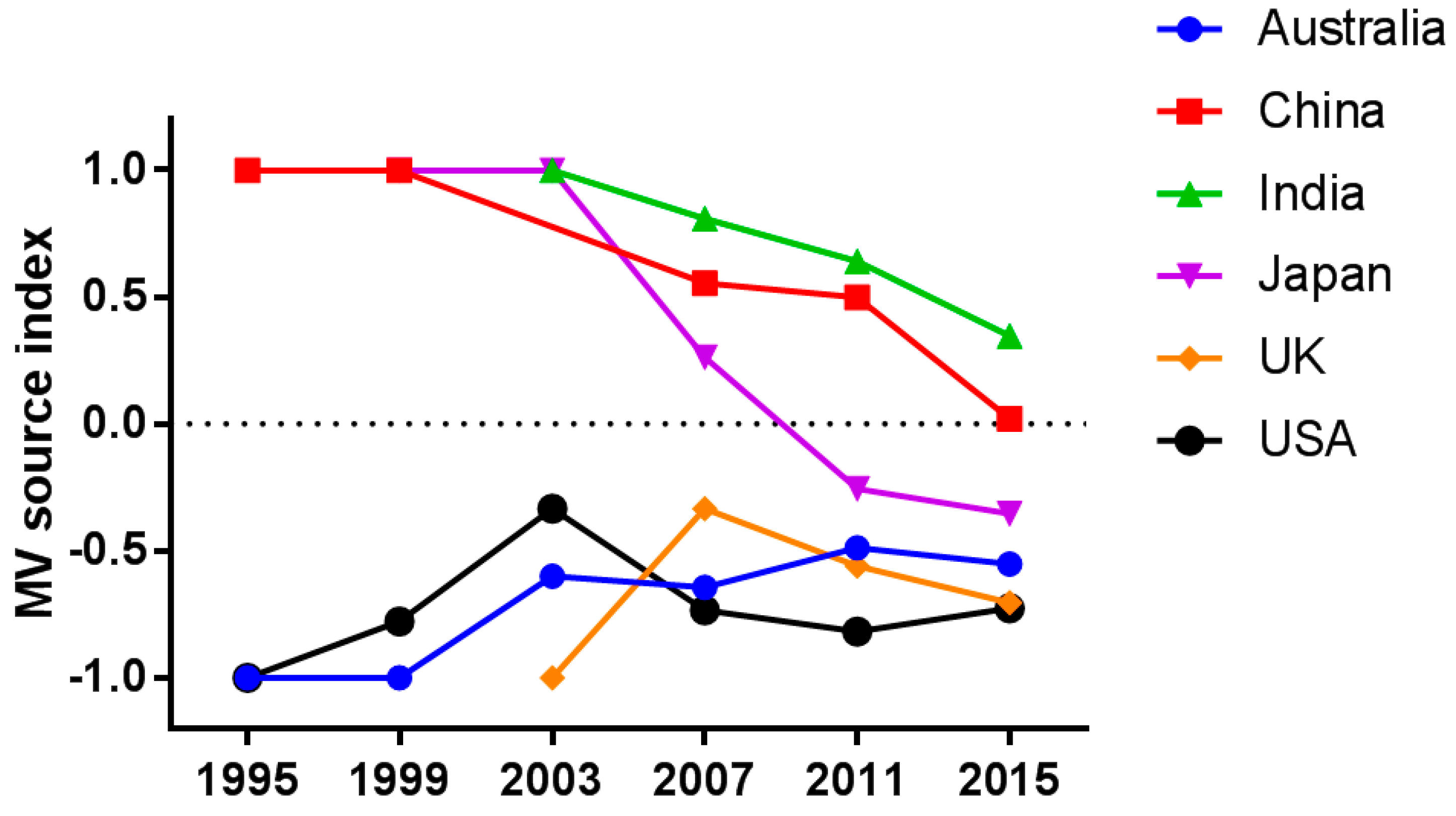
3.3. Temporal Trend of Measles Transmission
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- WHO Measles. Available online: http://www.who.int/mediacentre/factsheets/fs286/en/ (accessed on 21 March 2017).
- Rota, J.S.; Heath, J.L.; Rota, P.A.; King, G.E.; Celma, M.L.; Carabaña, J.; Fernandez-Muñoz, R.; Brown, D.; Jin, L.; Bellini, W.J. Molecular epidemiology of measles virus: Identification of pathways of transmission and implications for measles elimination. J. Infect. Dis. 1996, 173, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Bellini, W.J.; Rota, P.A. Genetic diversity of wild-type measles viruses: Implications for global measles elimination programs. Emerg. Infect. Dis. 1998, 4, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Cutts, F.T.; Lessler, J.; Metcalf, C.J. Measles elimination: Progress, challenges and implications for rubella control. Expert Rev. Vaccines 2013, 12, 917–932. [Google Scholar] [CrossRef] [PubMed]
- Rota, P.A.; Brown, K.; Mankertz, A.; Santibanez, S.; Shulga, S.; Muller, C.P.; Hübschen, J.M.; Siqueira, M.; Beirnes, J.; Ahmed, H.; et al. Global distribution of measles genotypes and measles molecular epidemiology. J. Infect. Dis. 2011, 204, S514–S523. [Google Scholar] [CrossRef] [PubMed]
- Vukshich Oster, N.; Harpaz, R.; Redd, S.B.; Papania, M.J. International importation of measles virus-United States, 1993–2001. J. Infect. Dis. 2004, 189, S48–S53. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhang, Y.; Huang, F.; Wang, H.; Liu, D.; Li, J.; Rodewald, L.; Wu, J.; Deng, Y.; Xu, W. Endemic and imported measles virus-associated outbreaks among adults, Beijing, China, 2013. Emerg. Infect. Dis. 2015, 21, 477–479. [Google Scholar] [CrossRef] [PubMed]
- Lemey, P.; Rambaut, A.; Drummond, A.J.; Suchard, M.A. Bayesian phylogeography finds its roots. PLoS Comput. Biol. 2009, 5, e1000520. [Google Scholar] [CrossRef] [PubMed]
- Paraskevis, D.; Pybus, O.; Magiorkinis, G.; Hatzakis, A.; Wensing, A.M.; van de Vijver, D.A.; Albert, J.; Angarano, G.; Asjö, B.; Balotta, C.; et al. Tracing the HIV-1 subtype B mobility in Europe: A phylogeographic approach. Retrovirology 2009, 6, 49. [Google Scholar] [CrossRef] [PubMed]
- Magiorkinis, G.; Magiorkinis, E.; Paraskevis, D.; Ho, S.Y.W.; Shapiro, B.; Pybus, O.G.; Allain, J.-P.; Hatzakis, A. The global spread of hepatitis C virus 1a and 1b: A phylodynamic and phylogeographic analysis. PLoS Med. 2009, 6, e1000198. [Google Scholar] [CrossRef] [PubMed]
- Okoro, C.K.; Kingsley, R.A.; Connor, T.R.; Harris, S.R.; Parry, C.M.; Al-Mashhadani, M.N.; Kariuki, S.; Msefula, C.L.; Gordon, M.A.; de Pinna, E.; et al. Intracontinental spread of human invasive Salmonella Typhimurium pathovariants in sub-Saharan Africa. Nat. Genet. 2012, 44, 1215–1221. [Google Scholar] [CrossRef] [PubMed]
- Kalaycioglu, A.T.; Baykal, A.; Guldemir, D.; Bakkaloglu, Z.; Korukluoglu, G.; Coskun, A.; Torunoglu, M.A.; Ertek, M.; Durmaz, R. Molecular characterization of measles viruses in Turkey (2010–2011): First report of genotype D9 involved in an outbreak in 2011. J. Med. Virol. 2013, 85, 2128–2135. [Google Scholar] [CrossRef] [PubMed]
- Harvala, H.; Wiman, Å.; Wallensten, A.; Zakikhany, K.; Englund, H.; Brytting, M. Role of Sequencing the Measles Virus Hemagglutinin Gene and Hypervariable Region in the Measles Outbreak Investigations in Sweden During 2013–2014. J. Infect. Dis. 2016, 213, 592–599. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Sun, J.; Zhang, Y.; Wang, C.; Song, Y.; Song, L.; Xu, Q.; Li, B.; Chen, P.; Li, H.; et al. The first measles outbreak caused by imported genotype D9 measles virus in Shandong Province, China, 2013. Jpn. J. Infect. Dis. 2014, 67, 300–303. [Google Scholar] [CrossRef] [PubMed]
- O’Riordan, B.; Carr, M.J.; Connell, J.; Dunford, L.; Hall, W.W.; Hassan, J. Seroepidemiology and phylogenetic characterisation of measles virus in Ireland, 2004–2013. J. Clin. Virol. 2014, 60, 374–380. [Google Scholar] [CrossRef] [PubMed]
- MeaNS. Available online: http://www.who-measles.org/Public/Web_Front/main.php (accessed on 25 February 2015).
- Expanded Programme on Immunization (EPI). Standardization of the nomenclature for describing the genetic characteristics of wild-type measles viruses. Wkly. Epidemiol. Rec. 1998, 73, 265–269. [Google Scholar]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [PubMed]
- Genetic diversity of wild-type measles viruses and the global measles nucleotide surveillance database (MeaNS). Wkly. Epidemiol. Rec. 2015, 90, 373–380.
- WHO Regional Offices. Available online: http://www.who.int/about/regions/en/ (accessed on 3 February 2017).
- Furuse, Y.; Suzuki, A.; Oshitani, H. Origin of measles virus: Divergence from rinderpest virus between the 11th and 12th centuries. Virol. J. 2010, 7, 52. [Google Scholar] [CrossRef] [PubMed]
- Pomeroy, L.W.; Bjørnstad, O.N.; Holmes, E.C. The Evolutionary and Epidemiological Dynamics of the Paramyxoviridae. J. Mol. Evol. 2008, 66, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Saito, H. Measles exportation from Japan to the United States, 1994 to 2006. J. Travel Med. 2008, 15, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Gomi, H.; Takahashi, H. Why is measles still endemic in Japan? Lancet 2004, 364, 328–329. [Google Scholar] [CrossRef]
- Brunei Darussalam, Cambodia, Japan Verified as Achieving Measles Elimination. Available online: http://www.wpro.who.int/mediacentre/releases/2015/20150327/en/ (accessed on 12 March 2016).
- Ramsay, M.E.; Jin, L.; White, J.; Litton, P.; Cohen, B.; Brown, D. The elimination of indigenous measles transmission in England and Wales. J. Infect. Dis. 2003, 187, S198–S207. [Google Scholar] [CrossRef] [PubMed]
- Orenstein, W.A.; Papania, M.J.; Wharton, M.E. Measles elimination in the United States. J. Infect. Dis. 2004, 189, S1–S3. [Google Scholar] [CrossRef] [PubMed]
- Heywood, A.E.; Gidding, H.F.; Riddell, M.A.; McIntyre, P.B.; MacIntyre, C.R.; Kelly, H.A. Elimination of endemic measles transmission in Australia. Bull. World Health Organ. 2009, 87, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Wilson, S.E.; Khan, K.; Gilca, V.; Miniota, J.; Deeks, S.L.; Lim, G.; Eckhardt, R.; Bolotin, S.; Crowcroft, N.S. Global travel patterns and risk of measles in Ontario and Quebec, Canada: 2007–2011. BMC Infect. Dis. 2015, 15, 341. [Google Scholar] [CrossRef] [PubMed]
- Pegorie, M.; Shankar, K.; Welfare, W.S.; Wilson, R.W.; Khiroya, C.; Munslow, G.; Fiefield, D.; Bothra, V.; McCann, R. Measles outbreak in Greater Manchester, England, October 2012 to September 2013: Epidemiology and control. Euro. Surveill. 2014, 19. [Google Scholar] [CrossRef]
- Cohuet, S.; Bukasa, A.; Heathcock, R.; White, J.; Brown, K.; Ramsay, M.; Fraser, G. A measles outbreak in the Irish traveller ethnic group after attending a funeral in England, March–June 2007. Epidemiol. Infect. 2009, 137, 1759–1765. [Google Scholar] [CrossRef] [PubMed]
- Jost, M.; Luzi, D.; Metzler, S.; Miran, B.; Mutsch, M. Measles associated with international travel in the region of the Americas, Australia and Europe, 2001–2013: A systematic review. Travel Med. Infect. Dis. 2015, 13, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Hanratty, B.; Holt, T.; Duffell, E.; Patterson, W.; Ramsay, M.; White, J.M.; Jin, L.; Litton, P. UK measles outbreak in non-immune anthroposophic communities: The implications for the elimination of measles from Europe. Epidemiol. Infect. 2000, 125, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Slade, T.A.; Klekamp, B.; Rico, E.; Mejia-Echeverry, A. Measles outbreak in an unvaccinated family and a possibly associated international traveler-Orange County, Florida, December 2012–January 2013. MMWR. Morb. Mortal. Wkly. Rep. 2014, 63, 781–784. [Google Scholar] [PubMed]
- Abad, C.L.; Safdar, N. The Reemergence of Measles. Curr. Infect. Dis. Rep. 2015, 17, 51. [Google Scholar] [CrossRef] [PubMed]
- Rocha, H.A.L.; Correia, L.L.; Campos, J.S.; Silva, A.C.; Andrade, F.O.; Silveira, D.I.; Machado, M.M.; Leite, Á.J.; Cunha, A.J.L.A. Factors associated with non-vaccination against measles in northeastern Brazil: Clues about causes of the 2015 outbreak. Vaccine 2015, 33, 4969–4974. [Google Scholar] [CrossRef] [PubMed]
- Phadke, V.K.; Bednarczyk, R.A.; Salmon, D.A.; Omer, S.B. Association Between Vaccine Refusal and Vaccine-Preventable Diseases in the United States: A Review of Measles and Pertussis. JAMA 2016, 315, 1149–1158. [Google Scholar] [CrossRef] [PubMed]
- Muscat, M. Who gets measles in Europe? J. Infect. Dis. 2011, 204, 1293–1294. [Google Scholar] [CrossRef] [PubMed]
- Blumberg, S.; Worden, L.; Enanoria, W.; Ackley, S.; Deiner, M.; Liu, F.; Gao, D.; Lietman, T.; Porco, T. Assessing Measles Transmission in the United States Following a Large Outbreak in California. PLoS Curr. 2015, 7. [Google Scholar] [CrossRef] [PubMed]
| African Region | Region of the Americas | Mediterranean Region | European Region | South-East Asia Region | Western Pacific Region | Total | |
|---|---|---|---|---|---|---|---|
| Before 1955 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| 1956–1965 | 0 | 2 | 0 | 5 | 0 | 2 | 9 |
| 1966–1975 | 0 | 2 | 0 | 4 | 0 | 5 | 11 |
| 1976–1985 | 12 | 5 | 0 | 6 | 0 | 15 | 38 |
| 1986–1995 | 25 | 19 | 0 | 15 | 1 | 46 | 106 |
| 1996–2005 | 292 | 144 | 140 | 275 | 98 | 687 | 1636 |
| 2006–2015 | 546 | 803 | 522 | 1495 | 456 | 1793 | 5655 |
| Total | 875 | 976 | 662 | 1800 | 595 | 2548 | 7456 |
| Australia | China | India | Japan | UK | USA | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sequence | Transmission a | Sequence | Transmission a | Sequence | Transmission a | Sequence | Transmission a | Sequence | Transmission a | Sequence | Transmission a | |||||||
| from Australia | to Australia | from China | to China | from India | to India | from Japan | to Japan | from UK | to UK | from USA | to USA | |||||||
| Before 1955 | 0 | 0 (0) | 0 (0) | 0 | 0 (0) | 0 (0) | 0 | 0 (0) | 0 (0) | 0 | 0 (0) | 0 (0) | 0 | 0 (1) | 0 (1) | 1 | 0 (1) | 0 (2) |
| 1956–1965 | 0 | 0 (1) | 0 (1) | 2 | 0 (0) | 0 (1) | 0 | 0 (0) | 0 (0) | 0 | 0 (0) | 0 (1) | 1 | 0 (0) | 0 (0) | 2 | 0 (3) | 0 (2) |
| 1966–1975 | 4 | 0 (1) | 0 (1) | 0 | 0 (0) | 0 (1) | 0 | 0 (0) | 0 (0) | 1 | 0 (2) | 0 (1) | 2 | 0 (0) | 1 (3) | 2 | 0 (2) | 0 (2) |
| 1976–1985 | 10 | 0 (1) | 0 (0) | 0 | 0 (1) | 0 (0) | 0 | 0 (0) | 0 (0) | 5 | 0 (0) | 0 (1) | 2 | 0 (1) | 0 (0) | 4 | 0 (2) | 0 (3) |
| 1986–1995 | 13 | 0 (1) | 1 (4) | 31 | 1 (3) | 0 (4) | 0 | 0 (0) | 0 (3) | 0 | 0 (0) | 0 (2) | 2 | 0 (2) | 0 (0) | 15 | 0 (3) | 2 (9) |
| 1996–2005 | 54 | 4 (15) | 19 (8) | 419 | 14 (21) | 3 (8) | 71 | 5 (8) | 2 (9) | 80 | 8 (14) | 3 (11) | 16 | 4 (7) | 6 (13) | 98 | 6 (26) | 27 (56) |
| 2006–2015 | 276 | 11 (20) | 40 (34) | 842 | 32 (42) | 25 (34) | 446 | 38 (58) | 15 (32) | 408 | 23 (27) | 48 (65) | 141 | 11 (28) | 47 (67) | 456 | 16 (39) | 112 (140) |
| Total | 357 | 15 (39) | 60 (48) | 1294 | 47 (67) | 28 (48) | 517 | 43 (66) | 17 (44) | 494 | 31 (43) | 51 (81) | 164 | 15 (39) | 54 (84) | 578 | 22 (76) | 141 (214) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Furuse, Y.; Oshitani, H. Global Transmission Dynamics of Measles in the Measles Elimination Era. Viruses 2017, 9, 82. https://doi.org/10.3390/v9040082
Furuse Y, Oshitani H. Global Transmission Dynamics of Measles in the Measles Elimination Era. Viruses. 2017; 9(4):82. https://doi.org/10.3390/v9040082
Chicago/Turabian StyleFuruse, Yuki, and Hitoshi Oshitani. 2017. "Global Transmission Dynamics of Measles in the Measles Elimination Era" Viruses 9, no. 4: 82. https://doi.org/10.3390/v9040082





