TMPRSS2 and MSPL Facilitate Trypsin-Independent Porcine Epidemic Diarrhea Virus Replication in Vero Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmids and Primers
2.2. Cell and Virus Culture
2.3. Expression of TTSPs in Transfected Vero Cells
2.4. Quantitative Real-Time PCR Analysis
2.5. Determination of Viral Titer of PEDV Propagated in Vero Cells Expressing TTSPs
2.6. Determination of Effects of TTSPs and TTSP Inhibitor on Viral Replication
2.7. Analysis of PEDV and TTSP Co-Localization
2.8. Cleavage of PEDV S Protein by TTSPs
2.9. Analysis of TTSP Activation of PEDV for Cell–Cell Fusion
2.10. Quantitative Analysis of TTSP Expression in the Normal and PEDV-Infected Piglet Small Intestine/IECs
2.11. Adaptation of PEDV Isolated from Clinical Samples to Vero Cells Transiently Expressing TTSPs
2.12. Statistical Analysis
3. Results
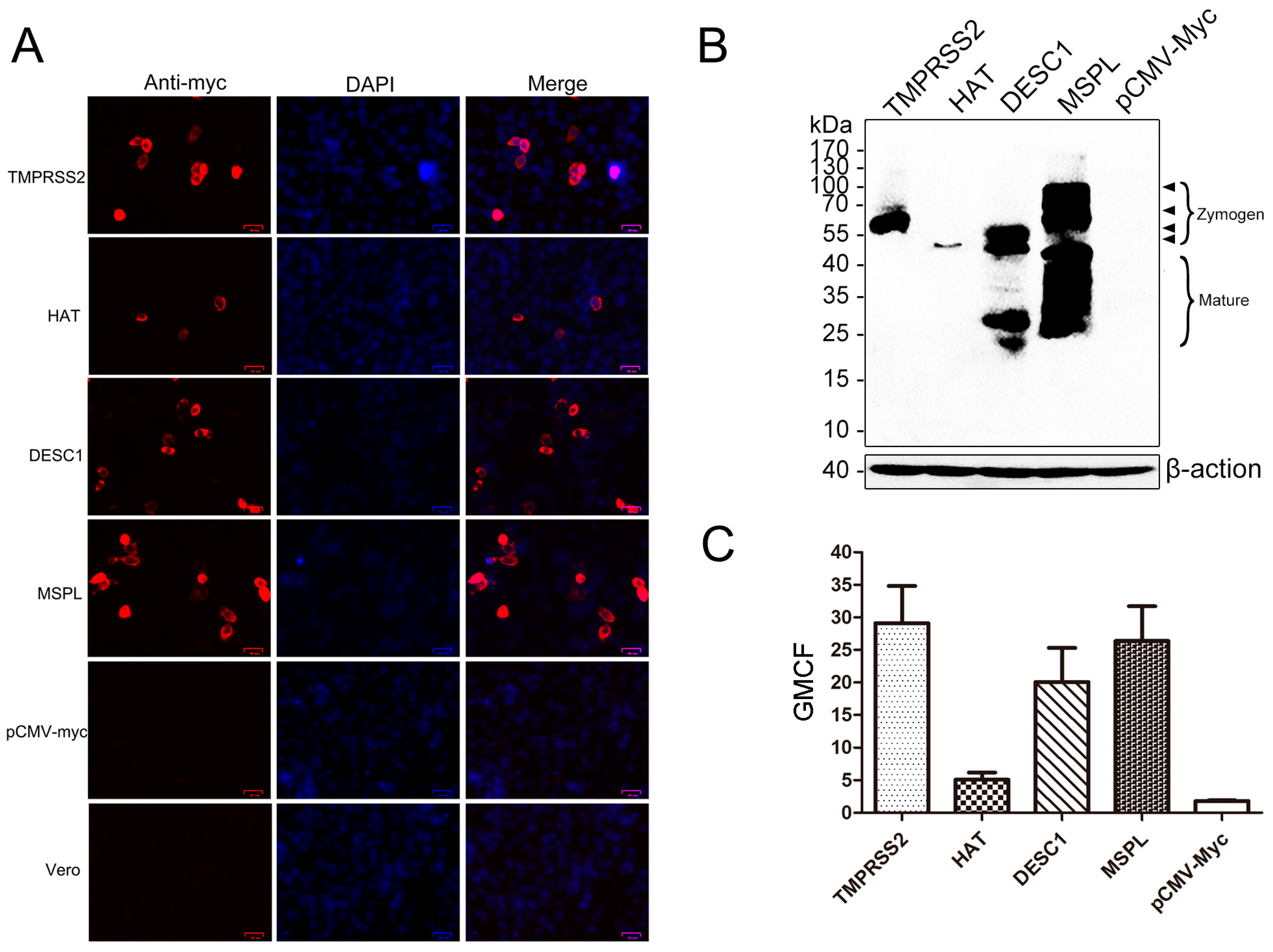
3.1. Expression of TTSPs in Transfected Vero Cells
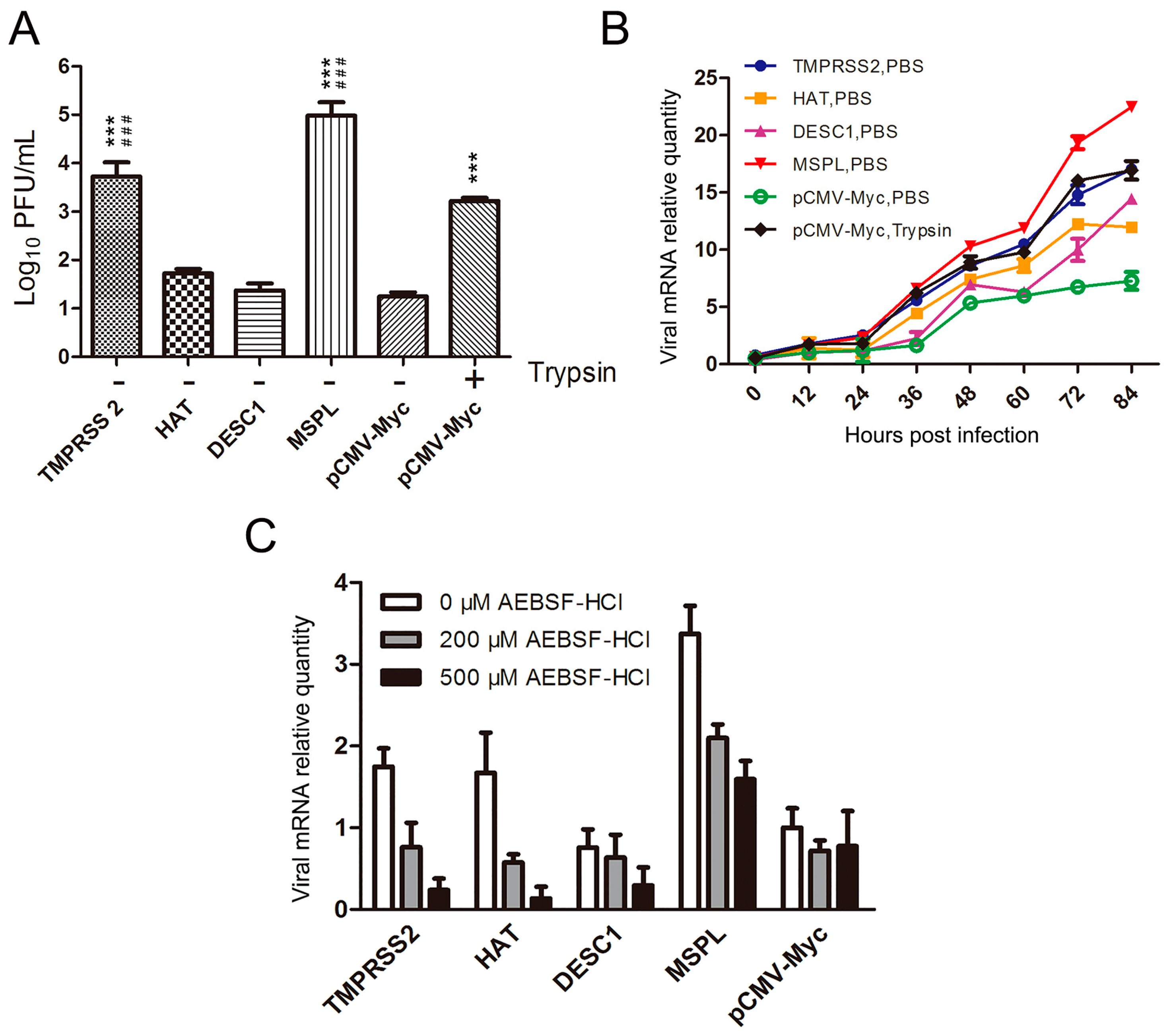
3.2. Effects of TTSPs and TTSP Inhibitor on Viral Replication
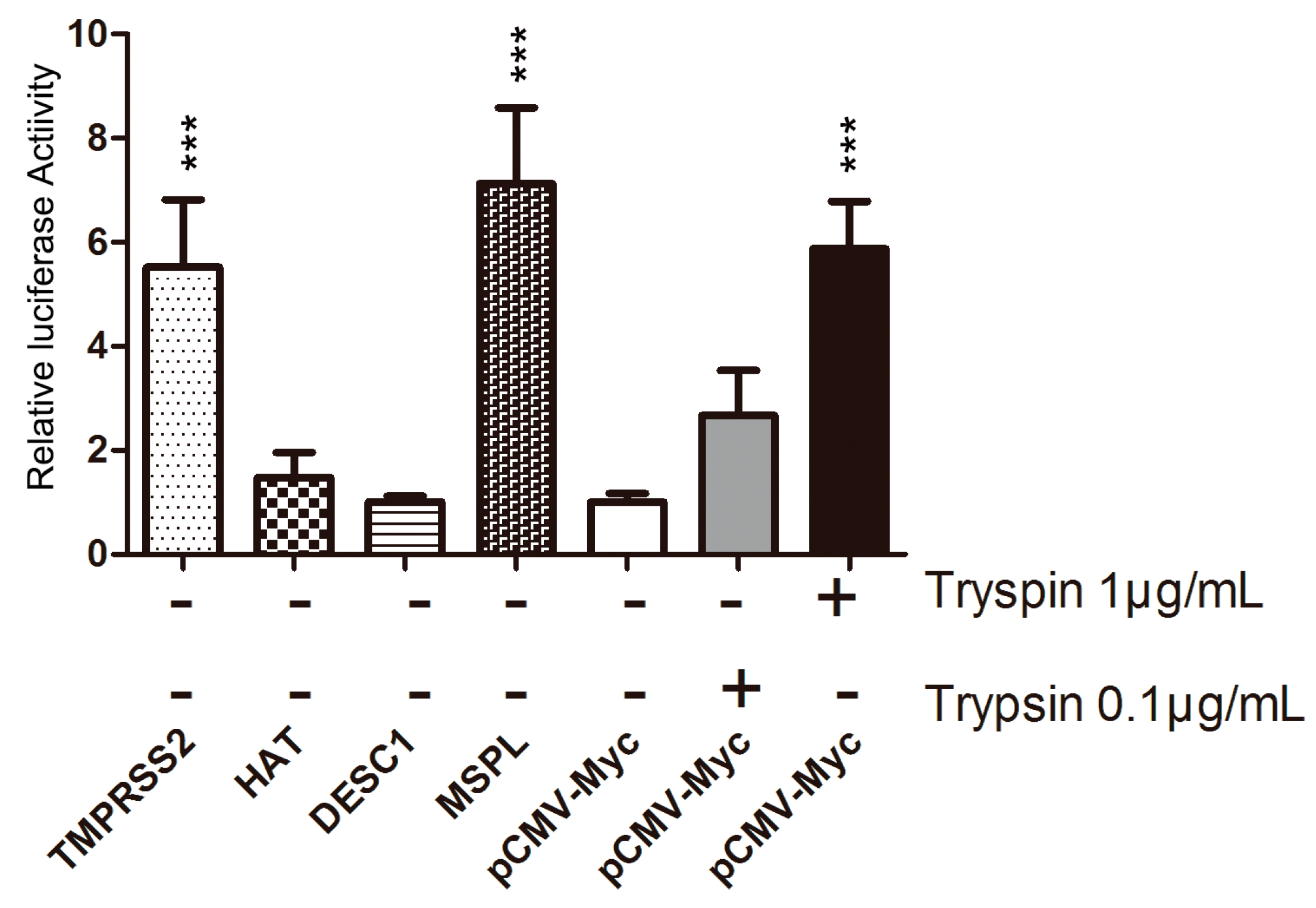
3.3. TTSP Activation of PEDV for Cell–Cell Fusion
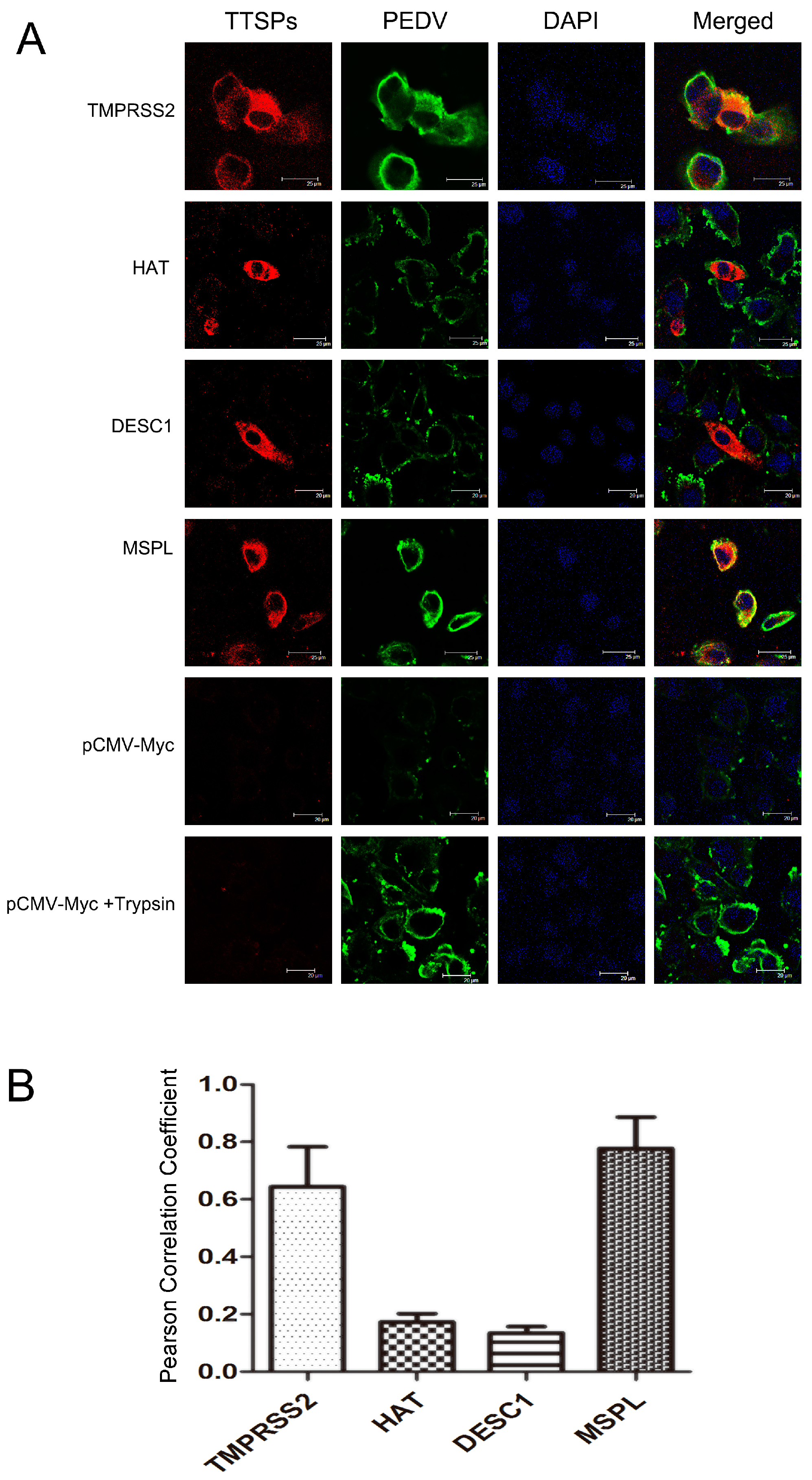
3.4. Co-Localization of TTSPs and PEDV
3.5. Effects of TTSPs on PEDV S Protein Cleavage
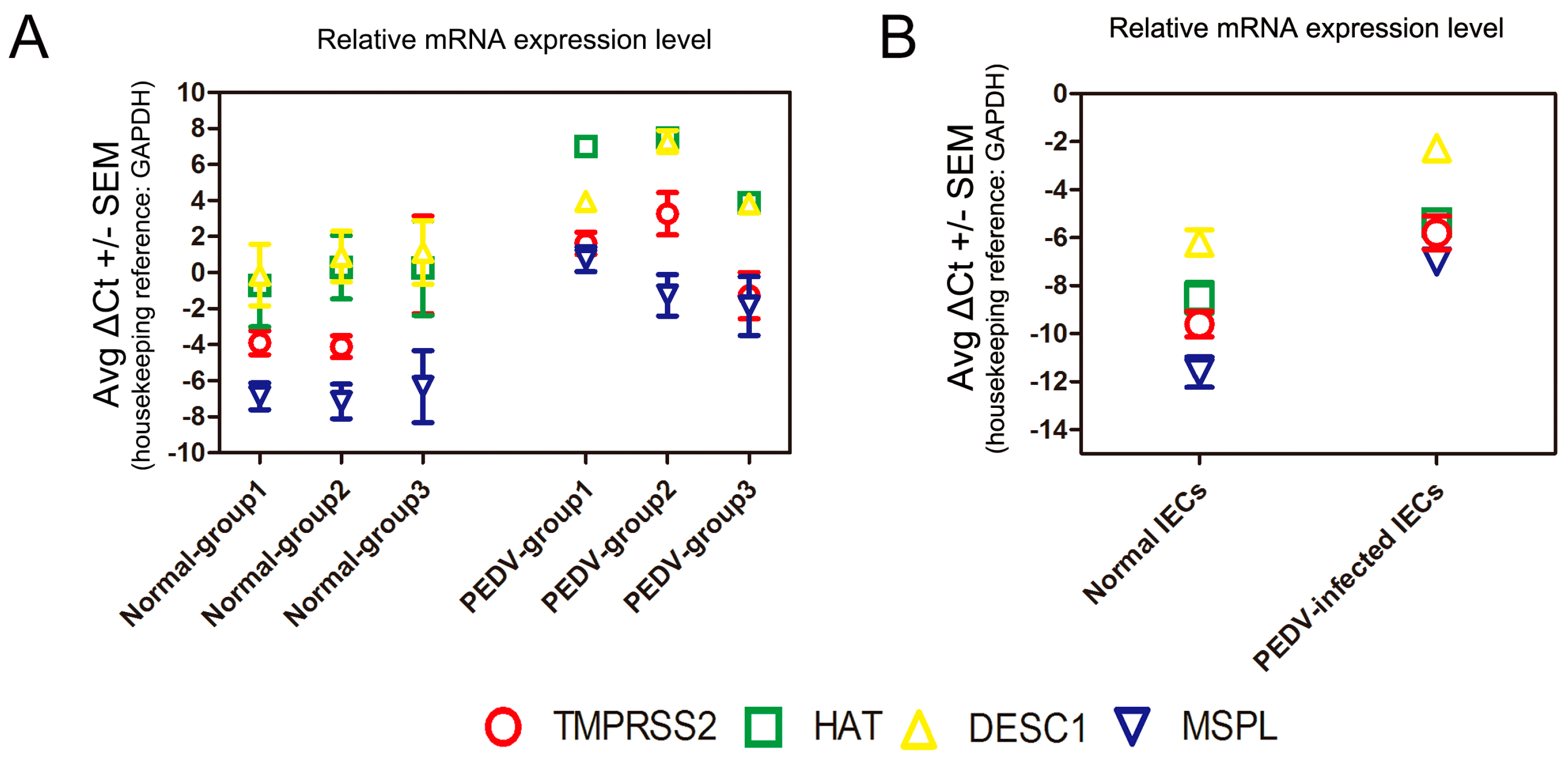
3.6. Determination of TTSP Expression in the Normal and PEDV-Infected Piglet Small Intestine/IECs
3.7. TTSPs Facilitate Propagation of PEDV Isolates in Vero Cells
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chen, Q.; Li, G.; Stasko, J.; Thomas, J.T.; Stensland, W.R.; Pillatzki, A.E.; Gauger, P.C.; Schwartz, K.J.; Madson, D.; Yoon, K.-J. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J. Clin. Microbiol. 2014, 52, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Debouck, P.; Pensaert, M. Experimental infection of pigs with a new porcine enteric coronavirus, CV 777. Am. J. Vet. Res. 1980, 41, 219–223. [Google Scholar] [PubMed]
- Kim, S.H.; Kim, I.J.; Pyo, H.M.; Tark, D.S.; Song, J.Y.; Hyun, B.H. Multiplex real-time RT-PCR for the simultaneous detection and quantification of transmissible gastroenteritis virus and porcine epidemic diarrhea virus. J. Virol. Methods 2007, 146, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Ojkic, D.; Hazlett, M.; Fairles, J.; Marom, A.; Slavic, D.; Maxie, G.; Alexandersen, S.; Pasick, J.; Alsop, J.; Burlatschenko, S. The first case of porcine epidemic diarrhea in Canada. Can. Vet. J. 2015, 56, 149–152. [Google Scholar] [PubMed]
- Pensaert, M.B.; de Bouck, P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978, 58, 243–247. [Google Scholar] [CrossRef] [PubMed]
- Trujillo-Ortega, M.E.; Beltran-Figueroa, R.; Garcia-Hernandez, M.E.; Juarez-Ramirez, M.; Sotomayor-Gonzalez, A.; Hernandez-Villegas, E.N.; Becerra-Hernandez, J.F.; Sarmiento-Silva, R.E. Isolation and characterization of porcine epidemic diarrhea virus associated with the 2014 disease outbreak in Mexico: Case report. BMC Vet. Res. 2016, 12, 132. [Google Scholar] [CrossRef] [PubMed]
- Wood, E.N. An apparently new syndrome of porcine epidemic diarrhoea. Vet. Rec. 1977, 100, 243–244. [Google Scholar] [CrossRef] [PubMed]
- Oldham, J. Letter to the editor. Pig Farming 1972, 10, 72–73. [Google Scholar] [CrossRef]
- Hofmann, M.; Wyler, R. Propagation of the virus of porcine epidemic diarrhea in cell culture. J. Clin. Microbiol. 1988, 26, 2235–2239. [Google Scholar] [PubMed]
- Park, J.E.; Cruz, D.J.; Shin, H.J. Receptor-bound porcine epidemic diarrhea virus spike protein cleaved by trypsin induces membrane fusion. Arch. Virol. 2011, 156, 1749. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.M.; Cavanagh, D. The molecular biology of coronaviruses. Adv. Virus Res. 1997, 48, 1–100. [Google Scholar] [PubMed]
- Bosch, B.J.; van der Zee, R.; de Haan, C.A.; Rottier, P.J. The coronavirus spike protein is a class I virus fusion protein: Structural and functional characterization of the fusion core complex. J. Virol. 2003, 77, 8801–8811. [Google Scholar] [CrossRef] [PubMed]
- White, J.M.; Delos, S.E.; Brecher, M.; Schornberg, K. Structures and mechanisms of viral membrane fusion proteins: Multiple variations on a common theme. Crit. Rev. Biochem. Mol. 2008, 43, 189–219. [Google Scholar] [CrossRef] [PubMed]
- Kawase, M.; Shirato, K.; Matsuyama, S.; Taguchi, F. Protease-mediated entry via the endosome of human coronavirus 229E. J. Virol. 2009, 83, 712–721. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, S.; Taguchi, F. Two-step conformational changes in a coronavirus envelope glycoprotein mediated by receptor binding and proteolysis. J. Virol. 2009, 83, 11133–11141. [Google Scholar] [CrossRef] [PubMed]
- Simmons, G.; Reeves, J.D.; Rennekamp, A.J.; Amberg, S.M.; Piefer, A.J.; Bates, P. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. USA 2004, 101, 4240–4245. [Google Scholar] [CrossRef] [PubMed]
- Spaan, W.; Cavanagh, D.; Horzinek, M.C. Coronaviruses: Structure and genome expression. J. Gen. Virol. 1988, 69, 2939–2952. [Google Scholar] [CrossRef] [PubMed]
- Zamolodchikova, T.S. Serine proteases of small intestine mucos—Localization, functional properties, and physiological role. Biochemistry 2012, 77, 820–829. [Google Scholar] [PubMed]
- Qiu, Z.; Hingley, S.T.; Simmons, G.; Yu, C.; Das Sarma, J.; Bates, P.; Weiss, S.R. Endosomal proteolysis by cathepsins is necessary for murine coronavirus mouse hepatitis virus type 2 spike-mediated entry. J. Virol. 2006, 80, 5768–5776. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.K.; Takimoto, K.; Yabe, M.; Taguchi, F. Acquired fusion activity of a murine coronavirus MHV-2 variant with mutations in the proteolytic cleavage site and the signal sequence of the S protein. Virology 1997, 227, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Yoshikura, H.; Tejima, S. Role of protease in mouse hepatitis virus-induced cell fusion. Studies with a cold-sensitive mutant isolated from a persistent infection. Virology 1981, 113, 503–511. [Google Scholar] [CrossRef]
- Bottcher, E.; Matrosovich, T.; Beyerle, M.; Klenk, H.D.; Garten, W.; Matrosovich, M. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J. Virol. 2006, 80, 9896–9898. [Google Scholar] [CrossRef] [PubMed]
- Bottcher, E.; Freuer, C.; Steinmetzer, T.; Klenk, H.D.; Garten, W. MDCK cells that express proteases TMPRSS2 and HAT provide a cell system to propagate influenza viruses in the absence of trypsin and to study cleavage of HA and its inhibition. Vaccine 2009, 27, 6324–6329. [Google Scholar] [CrossRef] [PubMed]
- Bottcher, E.; Freuer, C.; Sielaff, F.; Schmidt, S.; Eickmann, M.; Uhlendorff, J.; Steinmetzer, T.; Klenk, H.D.; Garten, W. Cleavage of influenza virus hemagglutinin by airway proteases TMPRSS2 and HAT differs in subcellular localization and susceptibility to protease inhibitors. J. Virol. 2010, 84, 5605–5614. [Google Scholar] [CrossRef] [PubMed]
- Bertram, S.; Heurich, A.; Lavender, H.; Gierer, S.; Danisch, S.; Perin, P.; Lucas, J.M.; Nelson, P.S.; Pohlmann, S.; Soilleux, E.J. Influenza and SARS-coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PLoS ONE 2012, 7, e35876. [Google Scholar] [CrossRef] [PubMed]
- Bertram, S.; Glowacka, I.; Blazejewska, P.; Soilleux, E.; Allen, P.; Danisch, S.; Steffen, I.; Choi, S.Y.; Park, Y.; Schneider, H.; et al. TMPRSS2 and TMPRSS4 facilitate trypsin-independent spread of influenza virus in Caco-2 cells. J. Virol. 2010, 84, 10016–10025. [Google Scholar] [CrossRef] [PubMed]
- Glowacka, I.; Bertram, S.; Muller, M.A.; Allen, P.; Soilleux, E.; Pfefferle, S.; Steffen, I.; Tsegaye, T.S.; He, Y.; Gnirss, K.; et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011, 85, 4122–4134. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, S.; Nagata, N.; Shirato, K.; Kawase, M.; Takeda, M.; Taguchi, F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010, 84, 12658–12664. [Google Scholar] [CrossRef] [PubMed]
- Shulla, A.; Heald-Sargent, T.; Subramanya, G.; Zhao, J.; Perlman, S.; Gallagher, T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 2011, 85, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Gierer, S.; Bertram, S.; Kaup, F.; Wrensch, F.; Heurich, A.; Kramer-Kuhl, A.; Welsch, K.; Winkler, M.; Meyer, B.; Drosten, C.; et al. The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J. Virol. 2013, 87, 5502–5511. [Google Scholar] [CrossRef] [PubMed]
- Shirato, K.; Kawase, M.; Matsuyama, S. Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J. Virol. 2013, 87, 12552–12561. [Google Scholar] [CrossRef] [PubMed]
- Zmora, P.; Blazejewska, P.; Moldenhauer, A.S.; Welsch, K.; Nehlmeier, I.; Wu, Q.; Schneider, H.; Pohlmann, S.; Bertram, S. DESC1 and MSPL activate influenza A viruses and emerging coronaviruses for host cell entry. J. Virol. 2014, 88, 12087–12097. [Google Scholar] [CrossRef] [PubMed]
- Shirato, K.; Matsuyama, S.; Ujike, M.; Taguchi, F. Role of proteases in the release of porcine epidemic diarrhea virus from infected cells. J. Virol. 2011, 85, 7872–7880. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Jia, S.; Zhao, H.; Yin, J.; Wang, X.; Yu, M.; Ma, S.; Wu, Y.; Chen, Y.; Fan, W.; et al. Novel approach for isolation and identification of Porcine epidemic diarrhea virus (PEDV) strain NJ using porcine intestinal epithelial cells. Viruses 2017, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hu, G.; Lin, Z.; He, L.; Xu, L.; Zhang, Y. Characteristic and functional analysis of a newly established porcine small intestinal epithelial cell line. PLoS ONE 2014, 9, e110916. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, G.; Liang, W.; Kang, K.; Guo, K.; Zhang, Y. Integrin beta3 is required in infection and proliferation of classical swine fever virus. PLoS ONE 2014, 9, e110911. [Google Scholar]
- Xu, X.; Zhang, H.; Zhang, Q.; Dong, J.; Liang, Y.; Huang, Y.; Liu, H.J.; Tong, D. Porcine epidemic diarrhea virus E protein causes endoplasmic reticulum stress and up-regulates interleukin-8 expression. Virol. J. 2013, 10, 26. [Google Scholar] [CrossRef] [PubMed]
- Junwei, G.; Baoxian, L.; Lijie, T.; Yijing, L. Cloning and sequence analysis of the N gene of porcine epidemic diarrhea virus LJB/03. Virus Genes 2006, 33, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Jinghui, F.; Yijing, L. Cloning and sequence analysis of the M gene of porcine epidemic diarrhea virus LJB/03. Virus Genes 2005, 30, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.Y.; Zhang, G.H.; Ge, J.W.; Jiang, Y.P.; Qiao, X.Y.; Cui, W.; Li, Y.J. Isolation and characteristics of virus culture of porcine epidemic diarrhea virus LJB/03. Chin. J. Virol. 2010, 26, 483–489. [Google Scholar]
- Park, J.E.; Cruz, D.J.; Shin, H.J. Clathrin- and serine proteases-dependent uptake of porcine epidemic diarrhea virus into Vero cells. Virus Res. 2014, 191, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Bolte, S.; Cordelieres, F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc.-Oxf. 2006, 224, 213–232. [Google Scholar] [CrossRef] [PubMed]
- Afar, D.E.; Vivanco, I.; Hubert, R.S.; Kuo, J.; Chen, E.; Saffran, D.C.; Raitano, A.B.; Jakobovits, A. Catalytic cleavage of the androgen-regulated TMPRSS2 protease results in its secretion by prostate and prostate cancer epithelia. Cancer Res. 2001, 61, 1686–1692. [Google Scholar] [PubMed]
- Miyake, Y.; Yasumoto, M.; Tsuzuki, S.; Fushiki, T.; Inouye, K. Activation of a membrane-bound serine protease matriptase on the cell surface. J. Biochem. 2009, 146, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Chasey, D.; Cartwright, S.F. Virus-like particles associated with porcine epidemic diarrhoea. Res. Vet. Sci. 1978, 25, 255–256. [Google Scholar] [PubMed]
- Nagy, B.; Nagy, G.; Meder, M.; Mocsari, E. Enterotoxigenic Escherichia coli, rotavirus, porcine epidemic diarrhoea virus, adenovirus and calici-like virus in porcine postweaning diarrhoea in Hungary. Acta Vet. Hung. 1996, 44, 9–19. [Google Scholar] [PubMed]
- Van Reeth, K.; Pensaert, M. Prevalence of infections with enzootic respiratory and enteric viruses in feeder pigs entering fattening herds. Vet. Rec. 1994, 135, 594–597. [Google Scholar] [PubMed]
- Takahashi, K.; Okada, K.; Ohshima, K. An outbreak of swine diarrhea of a new-type associated with coronavirus-like particles in Japan. Nihon juigaku zasshi. Jpn. J. Vet. Sci. 1983, 45, 829–832. [Google Scholar] [CrossRef]
- Xuan, H.; Xing, D.; Wang, D.; Zhu, W.; Zhao, F.; Gong, H. Study on the culture of porcine epidemic diarrhea virus adapted to fetal porcine intestine primary cell monolayer. Chin. J. Vet. Sci. 1984, 4, 202–208. [Google Scholar]
- Kweon, C.H.; Kwon, B.J.; Jung, T.S.; Kee, Y.J.; Hur, D.H.; Hwang, E.K.; Rhee, J.C.; An, S.H. Isolation of porcine epidemic diarrhea virus (PEDV) in Korea. Korean J. Vet. Res. 1993, 33, 249–254. [Google Scholar]
- Puranaveja, S.; Poolperm, P.; Lertwatcharasarakul, P.; Kesdaengsakonwut, S.; Boonsoongnern, A.; Urairong, K.; Kitikoon, P.; Choojai, P.; Kedkovid, R.; Teankum, K.; et al. Chinese-like strain of porcine epidemic diarrhea virus, Thailand. Emerg. Infect. Dis. 2009, 15, 1112–1115. [Google Scholar] [CrossRef] [PubMed]
- Panel, E.A. Scientific Opinion on porcine epidemic diarrhoea and emerging pig deltacoronavirus. EFSA J. 2014, 12, 3877. [Google Scholar]
- Song, D.; Moon, H.; Kang, B. Porcine epidemic diarrhea: A review of current epidemiology and available vaccines. Clin. Exp. Vaccine Res. 2015, 4, 166–176. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.Q.; Cai, R.J.; Chen, Y.Q.; Liang, P.S.; Chen, D.K.; Song, C.X. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg. Infect. Dis. 2012, 18, 161–163. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Zhang, J.; Deng, X.; Ye, Y.; Liao, M.; Fan, H. Complete genome sequence of a highly prevalent isolate of porcine epidemic diarrhea virus in South China. J. Virol. 2012, 86, 9551. [Google Scholar] [CrossRef] [PubMed]
- Dhanasekaran, S.M.; Barrette, T.R.; Ghosh, D.; Shah, R.; Varambally, S.; Kurachi, K.; Pienta, K.J.; Rubin, M.A.; Chinnaiyan, A.M. Delineation of prognostic biomarkers in prostate cancer. Nature 2001, 412, 822–826. [Google Scholar] [CrossRef] [PubMed]
- Kitamoto, Y.; Yuan, X.; Wu, Q.; McCourt, D.W.; Sadler, J.E. Enterokinase, the initiator of intestinal digestion, is a mosaic protease composed of a distinctive assortment of domains. Proc. Natl. Acad. Sci. USA 1994, 91, 7588–7592. [Google Scholar] [CrossRef] [PubMed]
- Yamaoka, K.; Masuda, K.I.; Ogawa, H.; Takagi, K.I.; Umemoto, N.; Yasuoka, S. Cloning and characterization of the cDNA for human airway trypsin-like protease. J. Biol. Chem. 1998, 273, 11895–11901. [Google Scholar] [CrossRef] [PubMed]
- Yasuoka, S.; Ohnishi, T.; Kawano, S.; Tsuchihashi, S.; Ogawara, M.; Masuda, K.; Yamaoka, K.; Takahashi, M.; Sano, T. Purification, characterization, and localization of a novel trypsin-like protease found in the human airway. Am. J. Respir. Cell Mol. 1997, 16, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Hobson, J.P.; Netzel-Arnett, S.; Szabo, R.; Rehault, S.M.; Church, F.C.; Strickland, D.K.; Lawrence, D.A.; Antalis, T.M.; Bugge, T.H. Mouse DESC1 is located within a cluster of seven DESC1-like genes and encodes a type II transmembrane serine protease that forms serpin inhibitory complexes. J. Biol. Chem. 2004, 279, 46981–46994. [Google Scholar] [CrossRef] [PubMed]
- Lang, J.C.; Schuller, D.E. Differential expression of a novel serine protease homologue in squamous cell carcinoma of the head and neck. Br. J. Cancer 2001, 84, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Bugge, T.H.; Antalis, T.M.; Wu, Q. Type II transmembrane serine proteases. J. Biol. Chem. 2009, 284, 23177–23181. [Google Scholar] [CrossRef] [PubMed]
- Szabo, R.; Bugge, T.H. Membrane-anchored serine proteases in vertebrate cell and developmental biology. Annu. Rev. Cell Dev. Biol. 2011, 27, 213–235. [Google Scholar] [CrossRef] [PubMed]
- Wicht, O.; Li, W.; Willems, L.; Meuleman, T.J.; Wubbolts, R.W.; van Kuppeveld, F.J.; Rottier, P.J.; Bosch, B.J. Proteolytic activation of the porcine epidemic diarrhea coronavirus spike fusion protein by trypsin in cell culture. J. Virol. 2014, 88, 7952–7961. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; (Northeast Agricultural University, Harbin, China). TMPRSS2 and MSPL promote PEDV replication in a dose-dependent manner at the stage of virus entry. in preparations. 2017. [Google Scholar]
- Liu, C.; Ma, Y.; Yang, Y.; Zheng, Y.; Shang, J.; Zhou, Y.; Jiang, S.; Du, L.; Li, J.; Li, F. Cell entry of porcine epidemic diarrhea coronavirus is activated by lysosomal proteases. J. Biol. Chem. 2016, 291, 24779–24786. [Google Scholar] [CrossRef] [PubMed]
| Primers | Primer Sequence (5′→3′) | Targets (ID) |
|---|---|---|
| Primers for the construction of TTSPs plasmids | ||
| TMPRSS2-F | CCGGAATTCGGATGGCTTTGAACTCAGGG | TMPRSS2 |
| TMPRSS2-R | GGAAGATCTTTAGCCGTCTGCCCTCAT | (BC051839) |
| HAT-F | CCGGAATTCGGATGTATAGGCCAGCACG | HAT |
| HAT-R | GGAAGATCTCTAGATCCCAGTTTGTTG | (BC125195) |
| DESC1-F | CCGGAATTCGGATGATGTATCGGCCAGATG | DESC1 |
| DESC1-R | GGAAGATCTTTAGATACCAGTTTTTG | (BC113412) |
| MSPL-F | CCGGAATTCGGATGGAGAGGGACAGCC | MSPL |
| MSPL-R | GGAAGATCTTTAGGATTTTCTGAATCG | (BC114928) |
| Primers for identification of PEDV by real-time PCR | ||
| PN-F | ACTGAGGGTGTTTTCTGGGTTGC | Nucleocapsid gene of PEDV |
| PN-R | GGTTCAACAATCTCAACTACACTGG | (DQ072726) |
| Beta-actin-F | AAGGATTCATATGTGGGCGATG | β-actin gene of Vero cells |
| Beta-actin-R | TCTCCATGTCGTCCCAGTTGGT | (AB004047) |
| Primers for identification of swine TTSPs mRNA by real-time PCR | ||
| sw-TMPRSS2-F | CACCCGAACTATGACCCCAAGACC | Swine-TMPRSS2 |
| sw-TMPRSS2-R | CATAGCGGCGTTCAGCACCTC | (XM_013982601) |
| sw-HAT-F | ACAACGCACAATAACTCCCTCTG | Swine-HAT |
| sw-HAT-R | GACATTGTTCTGTTGAAGGCTGG | (XM_013978756) |
| sw-DESC1-F | TGCTGCTGATTTTTAGATTTCGCTC | Swine-DESC1 |
| sw-DESC1-R | AGGGGGTCCTACAGCATCTTG | (XM_013978755) |
| sw-MSPL-F | CCCATAAGTGGCTTCCCGTC | Swine-MSPL |
| sw-MSPL-R | TGTAGATGCTCTCCTGGATGGTG | (XM_013989517) |
| sw-GAPDH-F | AAGGTCGGAGTGAACGGATTTG | Swine-GAPDH |
| sw-GAPDH-R | GCCTTGACTGTGCCGTGGAAC | (XM_013991162) |
| Primers for identification of TTSPs in Vero cells | ||
| m-TMPRSS2-F | ACCGCCAGGTGTTGGACCTTAC | m-TMPRSS2 |
| m-TMPRSS2-R | GACACGCCATCGCACCAGTTAG | (XM_007968781) |
| m-HAT-F | AGTGTGTGTCTCCCAGCTGCTAC | m-HAT |
| m-HAT-R | TCGGTAGGTTGTCACTCGGGTAT | (XM_007998573) |
| m-DESC1-F | GGTGGAACAGAAGTAGAAGAGGG | m-DESC1 |
| m-DESC1-R | CACATCACCTGGGTGAAACTC | (XM_007998564) |
| m-MSPL-F | TGACCCTGTCCGCTCACATCCAC | m-MSPL |
| m-MSPL-R | AAATCGCACCTCACTCTCCATCTTG | (XM_008021030) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, W.; Fan, W.; Bai, J.; Tang, Y.; Wang, L.; Jiang, Y.; Tang, L.; Liu, M.; Cui, W.; Xu, Y.; et al. TMPRSS2 and MSPL Facilitate Trypsin-Independent Porcine Epidemic Diarrhea Virus Replication in Vero Cells. Viruses 2017, 9, 114. https://doi.org/10.3390/v9050114
Shi W, Fan W, Bai J, Tang Y, Wang L, Jiang Y, Tang L, Liu M, Cui W, Xu Y, et al. TMPRSS2 and MSPL Facilitate Trypsin-Independent Porcine Epidemic Diarrhea Virus Replication in Vero Cells. Viruses. 2017; 9(5):114. https://doi.org/10.3390/v9050114
Chicago/Turabian StyleShi, Wen, Wenlu Fan, Jing Bai, Yandong Tang, Li Wang, Yanping Jiang, Lijie Tang, Min Liu, Wen Cui, Yigang Xu, and et al. 2017. "TMPRSS2 and MSPL Facilitate Trypsin-Independent Porcine Epidemic Diarrhea Virus Replication in Vero Cells" Viruses 9, no. 5: 114. https://doi.org/10.3390/v9050114





