Nutrients and Other Environmental Factors Influence Virus Abundances across Oxic and Hypoxic Marine Environments
Abstract
:1. Introduction
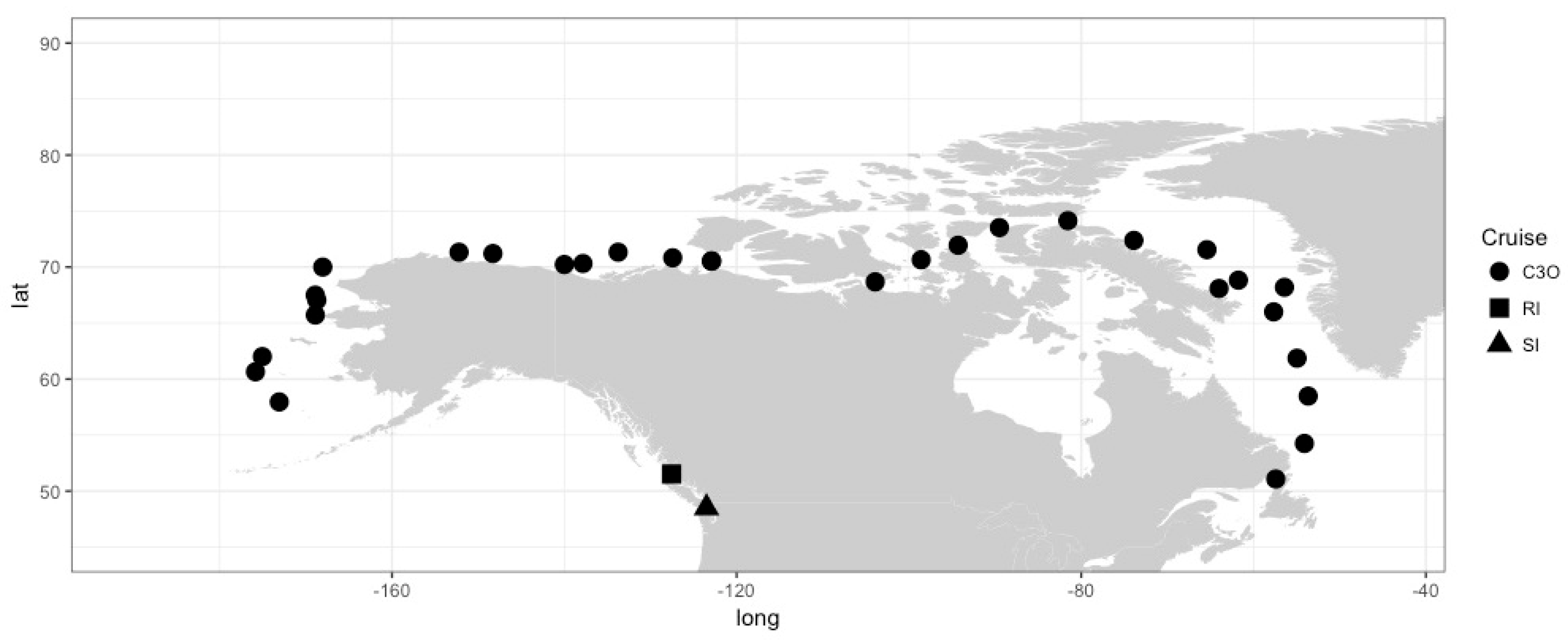
2. Materials and Methods
3. Results
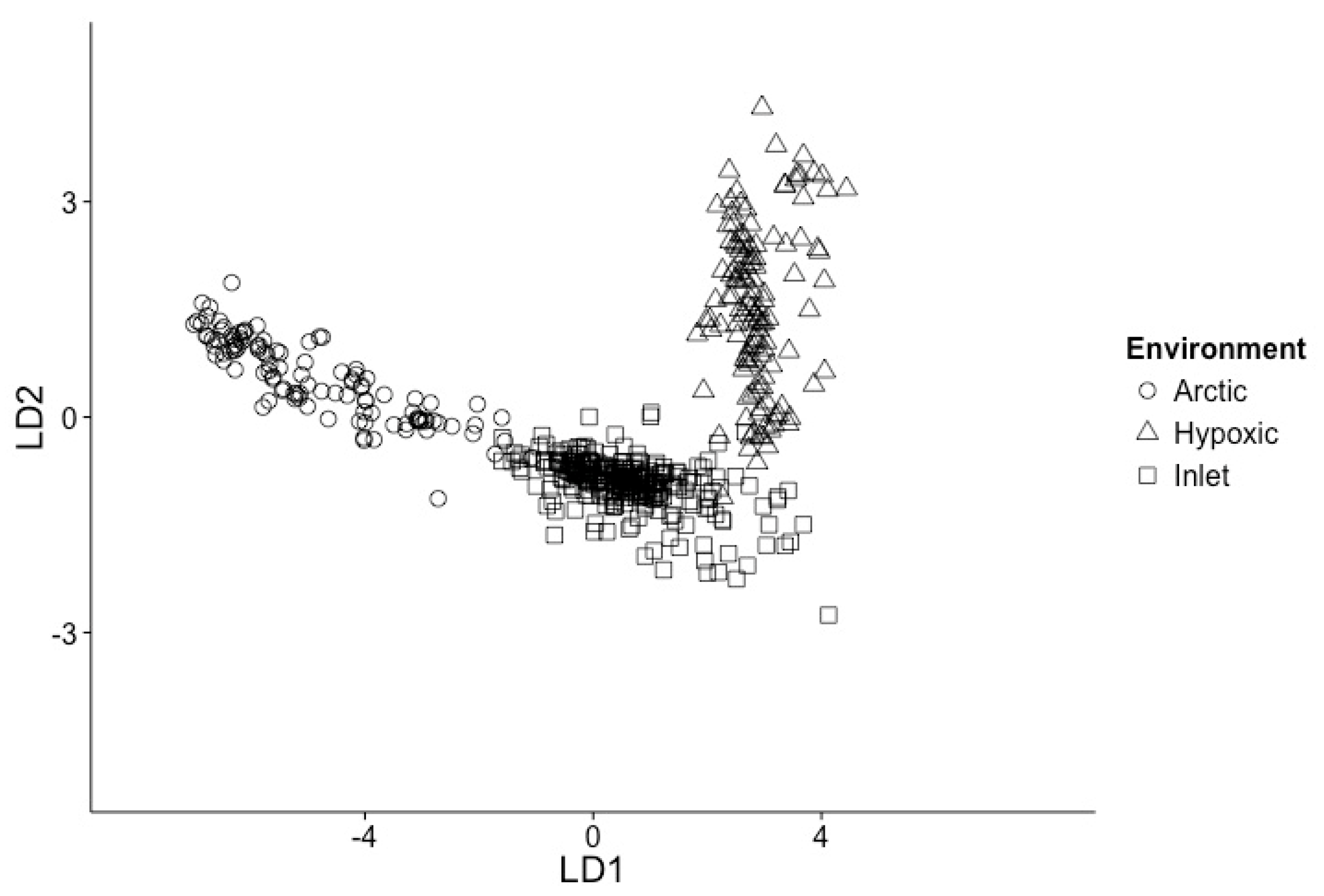
3.1. Samples Can Be Classified into Environments
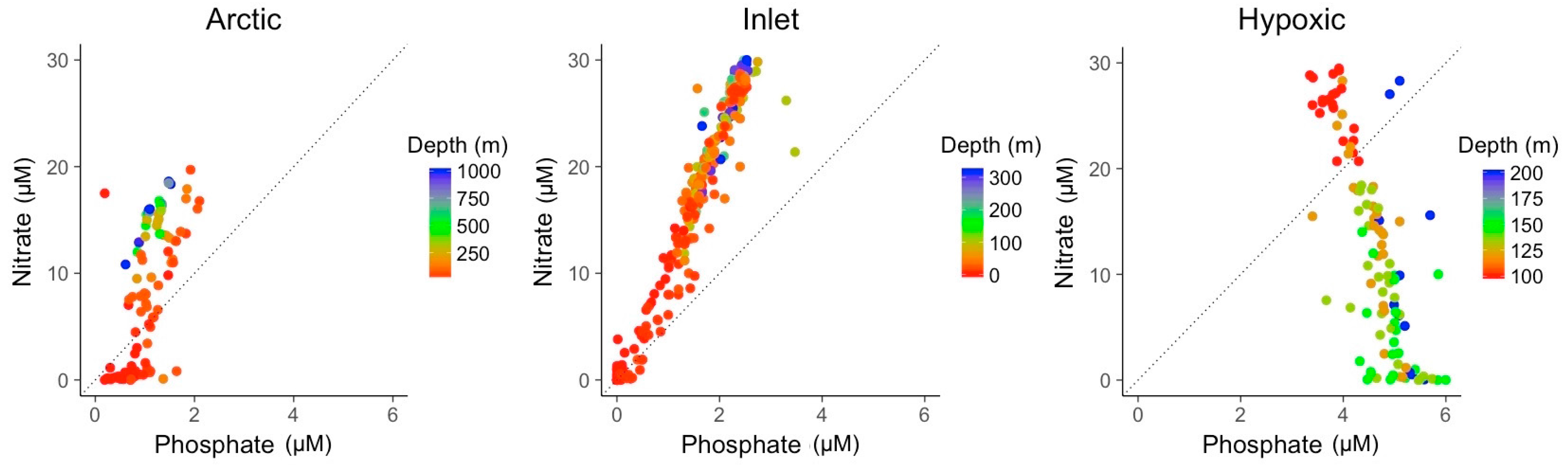
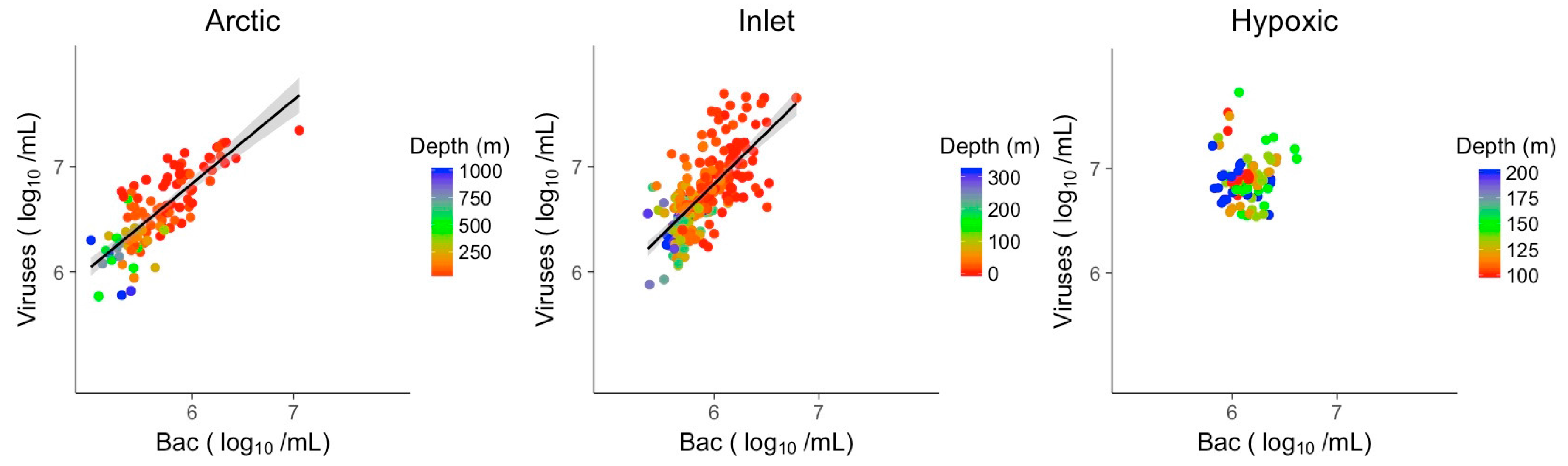
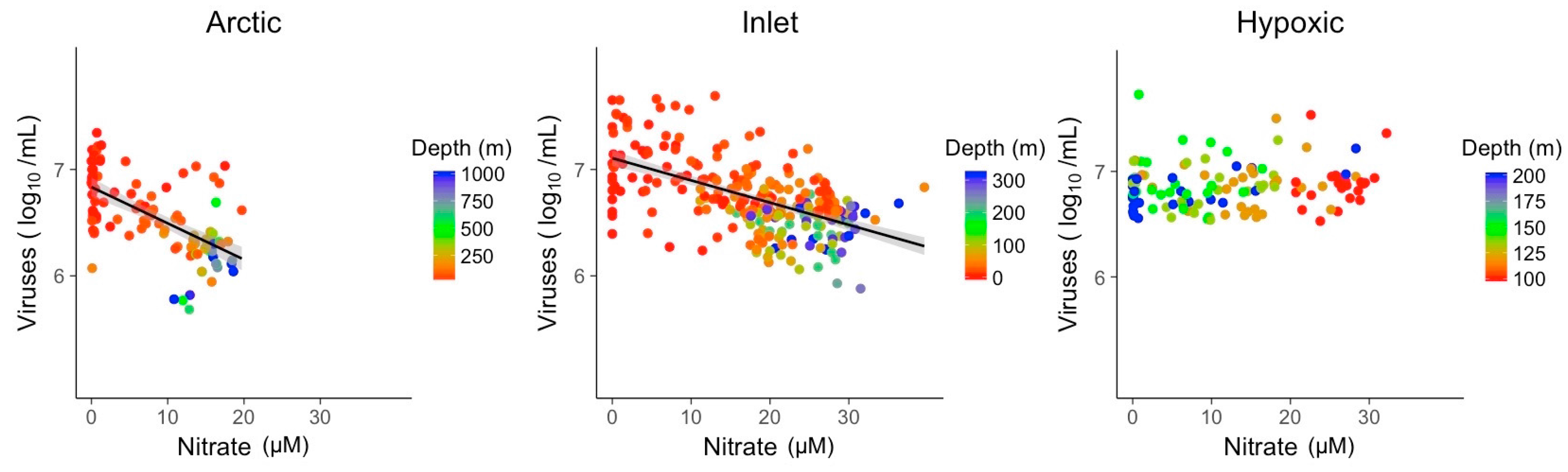
3.2. Explanatory Power of Single Variable Linear Models
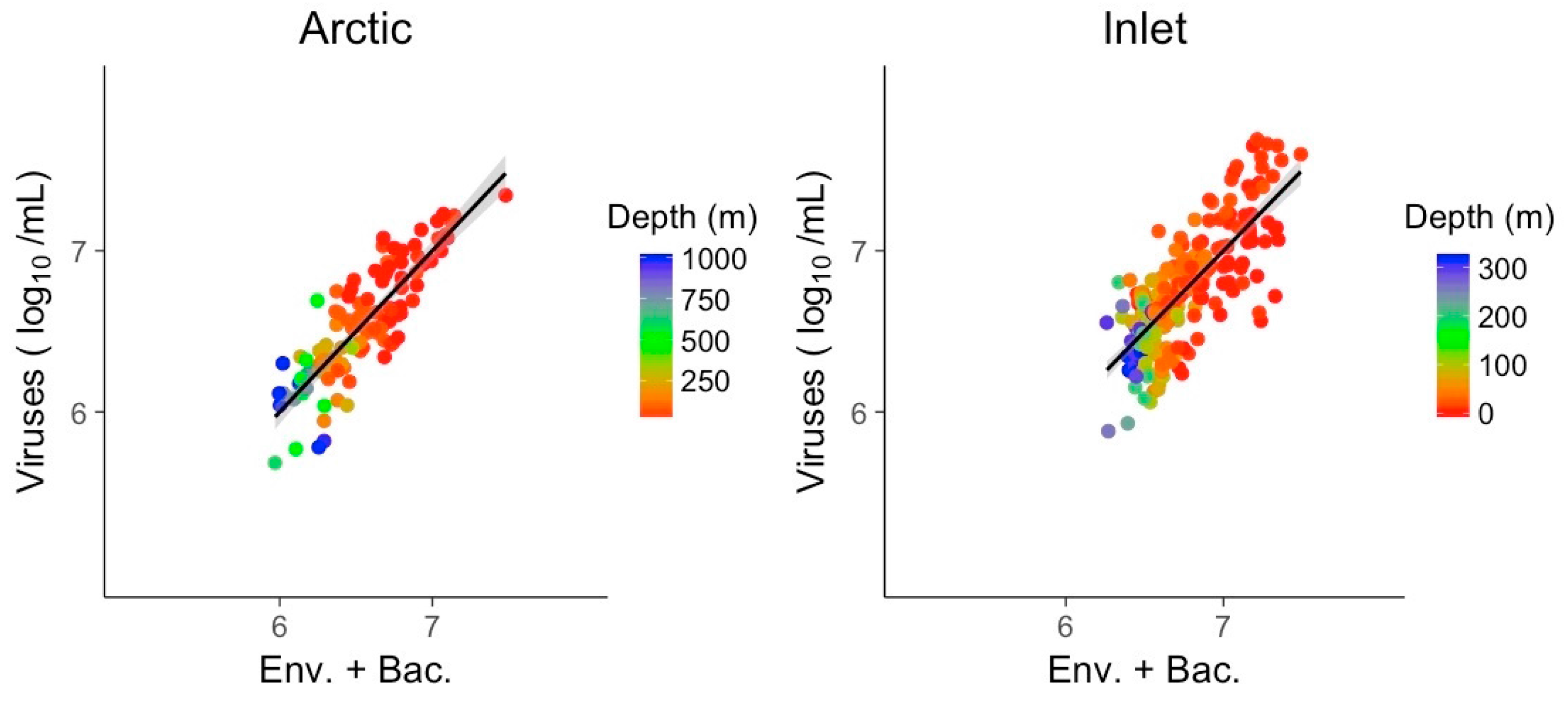
3.3. Multivariate Models Show Increased Explanatory Power
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wilhelm, S.W.; Suttle, C.A. Viruses and nutrient cycles in the sea. Bioscience 1999, 49, 781–788. [Google Scholar] [CrossRef]
- Ortmann, A.C.; Suttle, C.A. High abundances of viruses in a deep-sea hydrothermal vent system indicates viral mediated microbial mortality. Deep Sea Res Part I 2005, 52, 1515–1527. [Google Scholar] [CrossRef]
- Paul, J.H.; Rose, J.B.; Jiang, S.C.; Kellogg, C.A.; Dickson, L. Distribution of viral abundance in the reef environment of Key Largo, Florida. Appl. Environ. Microbiol. 1993, 59, 718–724. [Google Scholar] [PubMed]
- Proctor, L.M.; Fuhrman, J.A. Viral mortality of marine bacteria and cyanobacteria. Nature 1990, 343, 60–62. [Google Scholar] [CrossRef]
- Murray, A.G.; Jackson, G.A. Viral dynamics: A model of the effects of size, shape, motion and abundance of single-celled planktonic organisms and other particles. Mar. Ecol. Prog. Ser. 1992, 89, 103–116. [Google Scholar] [CrossRef]
- Mann, N.H. Phages of the marine cyanobacterial picophytoplankton. FEMS Microbiol. Rev. 2003, 27, 17–34. [Google Scholar] [CrossRef]
- Wommack, K.E.; Colwell, R.R. Virioplankton: Viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 2000, 64, 69–114. [Google Scholar] [CrossRef] [PubMed]
- Fuhrman, J.A.; Suttle, C.A. Viruses in marine planktonic systems. Oceanography 1993, 6, 51–63. [Google Scholar] [CrossRef]
- Knowles, B.; Silveira, C.B.; Bailey, B.A.; Barott, K.; Coutinho, F.H.; Dinsdale, E.A.; Felts, B.; Furby, K.A.; George, E.E.; Green, K.T.; et al. Lytic to temperate switching of viral communities. Nature 2016, 531, 466–470. [Google Scholar] [CrossRef] [PubMed]
- Wigington, C.H.; Sonderegger, D.; Brussaard, C.P.D.; Buchan, A.; Finke, J.F.; Fuhrman, J.A.; Lennon, J.T.; Middleboe, M.; Suttle, C.A.; Stock, C.; et al. Marine virus and microbial cell abundances. Nat. Microbiol. 2016, 1, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Parikka, K.J.; Le Romancer, M.; Wauters, N.; Jacquet, S. Deciphering the virus-to-prokaryote ratio (VPR): Insights into virus-host relationships in a variety of ecosystems. Biol. Rev. Camb. Philos. Soc. 2017, 92, 1081–1100. [Google Scholar] [CrossRef] [PubMed]
- Clasen, J.L.; Brigden, S.M.; Payet, J.P.; Suttle, C.A. Evidence that viral abundance across oceans and lakes is driven by different biological factors. Freshw. Biol. 2008, 53, 1090–1100. [Google Scholar] [CrossRef]
- Baudoux, A.C.; Brussaard, C.P.D. Characterization of different viruses infecting the marine harmful algal bloom species Phaeocystis globosa. Virology 2005, 341, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Nagasaki K, YM. Effect of temperature on the algicidal activity and the stability of HaV (Heterosigma akashiwo virus). Aquat. Microb. Ecol. 1998, 15, 211–216. [Google Scholar] [CrossRef]
- Hardies, S.C.; Hwang, Y.J.; Hwang, C.Y.; Jang, G.I.; Cho, B.C. Morphology, physiological characteristics, and complete sequence of marine bacteriophage RIO-1 infecting Pseudoalteromonas marina. J. Virol. 2013, 87, 9189–9198. [Google Scholar] [CrossRef] [PubMed]
- Williamson, S.J.; Paul, J.H. Environmental factors that influence the transition from lysogenic to lytic existence in the HSIC/Listonella pelagia marine phage-host system. Microb. Ecol. 2006, 52, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Kukkaro, P.; Bamford, D.H. Virus-host interactions in environments with a wide range of ionic strengths. Environ. Microbiol. Rep. 2009, 1, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Keynan, A.; Nealson, K.; Sideropoulos, H.; Hastings, J.W. Marine transducing bacteriophage attacking a luminous bacterium. J. Virol. 1974, 14, 333–340. [Google Scholar] [PubMed]
- Bettarel, Y.; Bouvier, T.; Bouvier, C.; Carr, C.; Desnues, A.; Domaizon, I.; Jacquet, S.; Robin, A.; Sime-Ngando, T. Ecological traits of planktonic viruses and prokaryotes along a full-salinity gradient. FEMS Micriobiol. Ecol. 2011, 76, 360–372. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Shan, J.; Millard, A.; Clokie, M.R.J.; Mann, N.H. Light-dependent adsorption of photosynthetic cyanophages to Synechococcus sp. WH7803. FEMS Microbiol. Lett. 2010, 310, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Baudoux, A.C.; Brussaard, C.P.D. Influence of irradiance on virus-algal host interactions. J. Phycol. 2008, 44. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.M.; Campbell, D.A.; Lawrence, J.E. Resource dynamics during infection of Micromonas pusilla by virus MpV-Sp1. Environ. Microbiol. 2007, 9, 2720–2727. [Google Scholar] [CrossRef] [PubMed]
- Juneau, P.; Lawrence, J.E.; Suttle, C.A.; Harrison, P.J. Effects of viral infection on photosynthetic processes in the bloom-forming alga Heterosigma akashiwo. Aquat. Microb. Ecol. 2003, 31, 9–17. [Google Scholar] [CrossRef]
- Lawrence, J.E.; Suttle, C.A. Effect of viral infection of sinking rates of Heterosigma akashiwo and its implications for bloom termination. Aquat. Microb. Ecol. 2004, 37, 1–7. [Google Scholar] [CrossRef]
- Mojica, K.D.A.; Brussaard, C.P.D. Factors affecting virus dynamics and microbial host-virus interactions in marine environments. FEMS Microbiol. Ecol. 2014, 89, 495–515. [Google Scholar] [CrossRef] [PubMed]
- Cottrell, M.T.; Suttle, C.A. Dynamics of a lytic virus infecting the photosynthetic marine picoflagellate Micromonas pusilla. Limnol. Oceanogr. 1995, 40, 730–739. [Google Scholar] [CrossRef]
- Noble, R.T.; Fuhrman, J.A. Virus decay and its causes in coastal waters. Appl. Environ. Microbiol. 1997, 63, 77–83. [Google Scholar] [PubMed]
- Garza, D.R.; Suttle, C.A. The effect of cyanophages on the mortality of Synechoccocus spp. and selection for UV resistant viral communities. Microb. Ecol. 1998, 36, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Jover, L.F.; Effler, T.C.; Buchan, A.; Wilhelm, S.W.; Weitz, J.S. The elemental composition of virus particles: Implications for marine biogeochemical cycles. Nat. Rev. Microbiol. 2014, 12, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Suttle, C.A. Marine viruses-major players in the global ecosystem. Nat. Rev. Microbiol. 2007, 5, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Redfield, A.C.; Ketchum, B.H.; Richards, F.A. The composition of seawater: Comparative and descriptive oceanography. In The Sea: Ideas and Observations on Progress in the Study of the Seas; Hill, N.M., Ed.; Interscience: New York, NY, USA, 1963; pp. 26–77. [Google Scholar]
- Bratbak, G.; Jacobsen, A.; Heldal, M.; Nagasaki, K.; Thingstad, F. Virus production in Phaeocystis pouchetii and its relation to host cell growth and nutrition. Aquat. Microb. Ecol. 1998, 16, 1–9. [Google Scholar] [CrossRef]
- Maat, D.S.; Crawfurd, K.J.; Timmermans, K.R.; Brussaard, C.P.D. Elevated CO2 and phosphate limitation favor Micromonas pusilla through stimulated growth and reduced viral impact. Appl. Environ. Microbiol. 2014, 80, 3119–3127. [Google Scholar] [CrossRef] [PubMed]
- Jacquet, S.; Heldal, M.; Iglesias-Rodriguez, D.; Larsen, A.; Wilson, W.H.; Bratbak, G. Flow cytometric analysis of an Emiliania huxleyi bloom terminated by viral infection. Aquat. Microb. Ecol. 2002, 27, 111–124. [Google Scholar] [CrossRef]
- Motegi, C.; Kaiser, K.; Benner, R.; Weinbauer, M.G. Effect of P-limitation on prokaryotic and viral production in surface waters of the Northwestern Mediterranean Sea. J. Plankton Res. 2015, 37, 16–20. [Google Scholar] [CrossRef]
- Bratbak, G.; Egge, J.K.; Heldal, M. Viral mortality of the marine alga Emiliania huxleyi (Haptophyceae) and termination of algal blooms. Mar. Ecol. Prog. Ser. 1993, 93, 39–48. [Google Scholar] [CrossRef]
- Chow, C.T.; Kim, D.Y.; Sachdeva, R.; Caron, D.A. Top-down controls on bacterial community structure: Microbial network analysis of bacteria, T4-like viruses and protists. ISME J. 2013, 8, 816–829. [Google Scholar] [CrossRef] [PubMed]
- Tommasi, D.; Hunt, B.P.V.; Pakhomov, E.A.; Mackas, D.L. Mesozooplankton community seasonal succession and its drivers: Insights from a British Columbia, Canada, fjord. J. Mar. Syst. 2013, 116, 10–32. [Google Scholar] [CrossRef]
- Carmack, E.C.; Mclaughlin, F.A.; Vagle, S.; Melling, H.; Williams, W.J. Structures and property distributions in the three oceans surrounding Canada in 2007: A basis for a long-term ocean climate monitoring strategy. Atmosphere-Ocean 2010, 48, 211–224. [Google Scholar] [CrossRef]
- Brussaard, C.P.D. Optimization of procedures for counting viruses by flow cytometry. Appl. Environ. Microbiol. 2004, 70, 1506–1513. [Google Scholar] [CrossRef] [PubMed]
- Vaulot, D. CYTOPC: Processing software for flow cytometric data. Signal Noise 1989, 2, 292. [Google Scholar]
- Armstrong, F.A.J.; Stearns, C.R.; Strickland, J.D.H. The measurement of upwelling and subsequent biological processes by means of the Technicon AutoAnalyzerTM and associated equipment. Deep Sea Res. Oceanogr. Abstr. 1967, 14, 381–389. [Google Scholar] [CrossRef]
- Murphey, J.; Riley, J.P. A modified single solution method for the determination of phosphate in natural waters. Anal. Chim. Acta 1962, 27, 31–36. [Google Scholar] [CrossRef]
- Moffitt, S.E.; Moffitt, R.A.; Sauthoff, W.; Davis, C.V.; Hewett, K.; Hill, T.M. Paleoceanographic insights on recent oxygen minimum zone expansion: Lessons for modern oceanography. PLoS ONE 2015, 10, e0115246. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2015. [Google Scholar]
- Venables, W.N.; Ripley, B.D. Modern Applied Statistics with S-Plus, 4th ed.; Springer: New York, NY, USA, 2002. [Google Scholar]
- Beaujean, A.A. BaylorEdPsych. 2012.
- Zaikova, E.; Walsh, D.; Stilwell, C.P.; Mohn, W.W.; Tortell, P.D.; Hallam, S.J. Microbial community dynamics in a seasonally anoxic fjord: Saanich Inlet, British Columbia. Environ. Microbiol. 2010, 12, 172–191. [Google Scholar] [CrossRef] [PubMed]
- Tyrrell, T. The relative influences of nitrogen and phosphorus on oceanic primary production. Nature 1999, 400, 525–531. [Google Scholar] [CrossRef]
- Somes, C.J.; Schmittner, A.; Galbraith, E.D.; Lehmann, M.F.; Altabet, M.A.; Montoya, J.P.; Letelier, R.M.; Mix, A.C.; Bourbonnais, A.; Eby, M. Simulating the global distribution of nitrogen isotopes in the ocean. Glob. Biogeochem. Cycles 2010, 24, 1–16. [Google Scholar] [CrossRef]
- Moore, C.M.; Mills, M.M.; Arragio, K.R.; Berman-Frank, I.; Bopp, L.; Boyd, P.W.; Galbraith, E.D.; Geider, R.J.; Jaccard, S.L.; Jickells, T.D.; et al. Processes and patterns of oceanic nutrient limitation. Nat. Geosci. 2013, 6, 701–710. [Google Scholar] [CrossRef]
- Payet, J.P.; Suttle, C.A. Physical and biological correlates of virus dynamics in the southern Beaufort Sea and Amundsen Gulf. J. Mar. Syst. 2008, 74, 933–945. [Google Scholar] [CrossRef]
- Maranger, R.; Bird, D. Viral abundance in aquatic systems: A comparison between marine and fresh waters. Mar. Ecol. Prog. Ser. 1995, 121, 217–226. [Google Scholar] [CrossRef]
- Capotondi, A.; Alexander, M.A.; Bond, N.A.; Curchitser, E.N.; Scott, J.D. Enhanced upper ocean stratification with climate change in the CMIP3 models. J. Geophys. Res. Ocean 2012, 117, 1–23. [Google Scholar] [CrossRef]
- Hordoir, R.; Meier, H.E.M. Effect of climate change on the thermal stratification of the baltic sea: A sensitivity experiment. Clim. Dyn. 2012, 38, 1703–1713. [Google Scholar] [CrossRef]
- Keeling, R.E.; Körtzinger, A.; Gruber, N. Ocean deoxygenation in a warming world. Ann. Rev. Mar. Sci. 2010, 2, 199–229. [Google Scholar] [CrossRef] [PubMed]
- Carmack, E.; Mclaughlin, F.; Whiteman, G.; Homer-dixon, T. Detecting and coping with disruptive shocks in Arctic marine systems: A resilience approach to place and people. AMBIO 2012, 41, 56–65. [Google Scholar] [CrossRef] [PubMed]


| Variable | Min. | Max. | Mean | Unit |
|---|---|---|---|---|
| Temperature | −1.710 | 15 | 7 | °C |
| Salinity | 3.060 | 35 | 31 | PSU |
| Chlorophyll | 0.030 | 44 | 2 | mg·m−3 |
| Oxygen | 0.005 | 10 | 4 | mL·L−1 |
| PAR | 0.000 | 669 | 25 | µmol quanta m−2·s−1 |
| NO3 | 0.010 | 54 | 15 | µM |
| PO4 | 0.006 | 7 | 2 | µM |
| SiO4 | 0.070 | 141 | 43 | µM |
| Bacteria | 7.31 × 104 | 7.40 × 107 | 1.66 × 106 | Cells mL−1 |
| Viruses | 4.83 × 105 | 1.40 × 108 | 8.35 × 106 | Viruses mL−1 |
| Variable | Parameter | Arctic | Inlet |
|---|---|---|---|
| Bacteria (log10) | R2 | 0.66 | 0.48 |
| Slope | 0.80 | 0.97 | |
| p-value | 2.5 × 10−27 | 1.9 × 10−37 | |
| NO3 | R2 | 0.37 | 0.33 |
| Slope | −0.03 | −0.02 | |
| p-value | 1.4 × 10−12 | 1.2 × 10−24 | |
| PO4 | R2 | 0.12 | 0.28 |
| Slope | −0.31 | −0.23 | |
| p-value | 1.0 × 10−04 | 3.6 × 10−20 |
| Env. | Arctic | Inlet | Hypoxic |
|---|---|---|---|
| McFadden (R2) | 0.56 | 0.47 | 0.31 |
| Slope | 1.00 | 1.00 | 1.00 |
| n/df | 109/104 | 261/258 | 126/122 |
| Intercept | 5.545 | 3.75 | 13.721 |
| Temperature | 0.141 | 1.068 | −2.763 |
| Salinity | - | 0.199 | - |
| Chlorophyll | 0.191 | - | 0.384 |
| Oxygen | 0.112 | - | - |
| NO3 | −0.018 | −0.013 | - |
| PO4 | - | - | −0.078 |
| SiO4 | 0.009 | - | 0.004 |
| PAR | - | - | - |
| Signif. level | <0.01 | <0.05 | <0.1 |
| Env. + Bac. | Arctic | Inlet | ||
|---|---|---|---|---|
| McFadden (R2) | 0.73 | 0.59 | ||
| Slope | 1.00 | 1.00 | ||
| n/df | 109/105 | 252/249 | ||
| Intercept | 5.008 | 1.020 | ||
| Temperature | - | 0.774 | ||
| Salinity | −0.523 | - | ||
| Chlorophyll | 0.098 | - | ||
| Oxygen | - | - | ||
| NO3 | −0.010 | −0.003 | ||
| PO4 | - | - | ||
| SiO4 | - | - | ||
| PAR | - | - | ||
| Bacteria (log10) | 0.607 | 0.665 | ||
| Signif. level | <0.01 | <0.05 | <0.1 | |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Finke, J.F.; Hunt, B.P.V.; Winter, C.; Carmack, E.C.; Suttle, C.A. Nutrients and Other Environmental Factors Influence Virus Abundances across Oxic and Hypoxic Marine Environments. Viruses 2017, 9, 152. https://doi.org/10.3390/v9060152
Finke JF, Hunt BPV, Winter C, Carmack EC, Suttle CA. Nutrients and Other Environmental Factors Influence Virus Abundances across Oxic and Hypoxic Marine Environments. Viruses. 2017; 9(6):152. https://doi.org/10.3390/v9060152
Chicago/Turabian StyleFinke, Jan F., Brian P.V. Hunt, Christian Winter, Eddy C. Carmack, and Curtis A. Suttle. 2017. "Nutrients and Other Environmental Factors Influence Virus Abundances across Oxic and Hypoxic Marine Environments" Viruses 9, no. 6: 152. https://doi.org/10.3390/v9060152





