A Multiplex RT-PCR Assay to Detect and Discriminate Porcine Reproductive and Respiratory Syndrome Viruses in Clinical Specimens
Abstract
:1. Introduction
2. Materials and Methods
2.1. Viruses, Cells, and Reagents
2.2. Clinical Specimen Collection
2.3. Primers Design
2.4. Nucleic Acid Extractions
2.5. First Strand cDNA Synthesis
2.6. Single PCR and Plasmid Template Construction
2.7. Optimization of the MultiplexRT-PCR Assay
2.8. Specificity of the Proposed Multiplex RT-PCR Assay
2.9. Sensitivity of the Proposed Multiplex RT-PCR Assay
2.10. Detection of PRRSV in Clinical Specimens by the Multiplex PCR
3. Results
3.1. MultiplexRT-PCR Assay Conditions
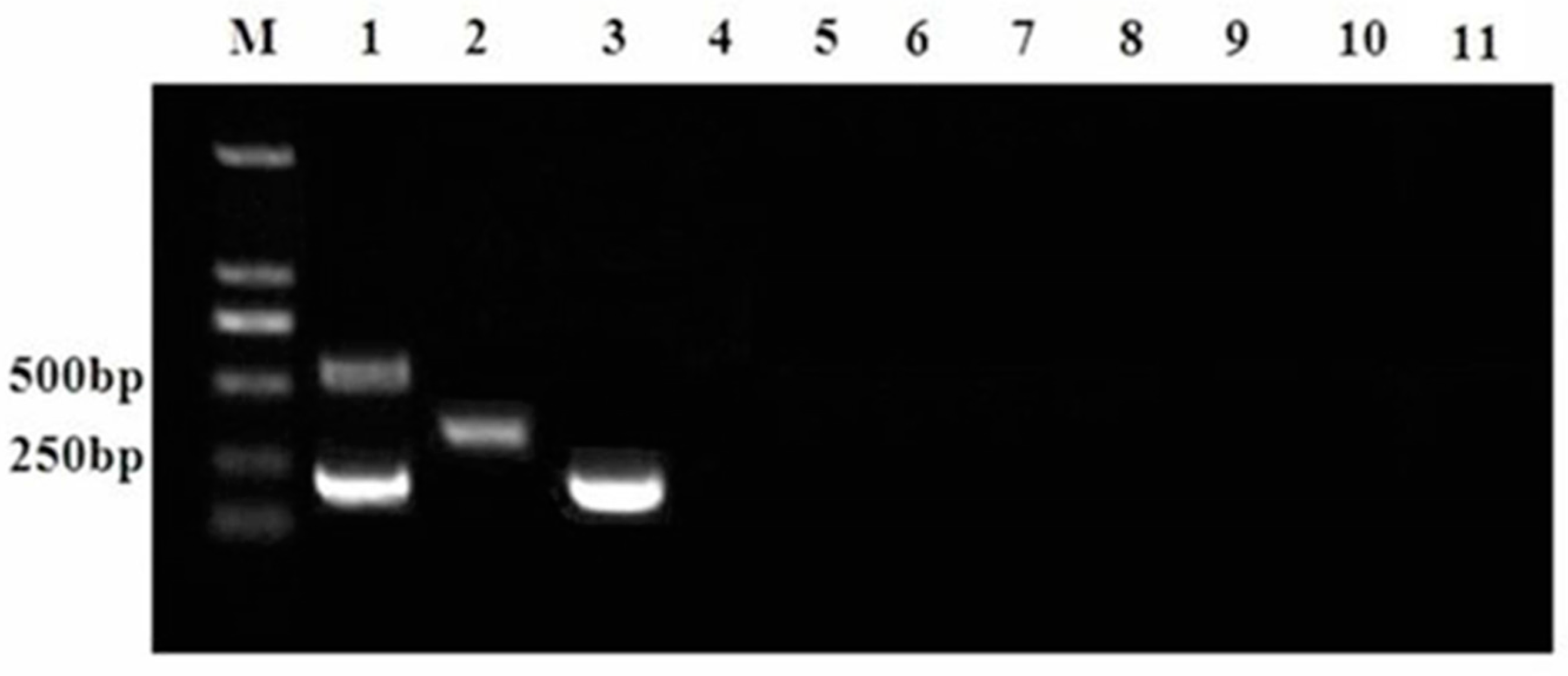
3.2. Specificity of the Proposed Multiplex RT-PCR Assay
3.3. Sensitivity of Multiplex RT-PCR
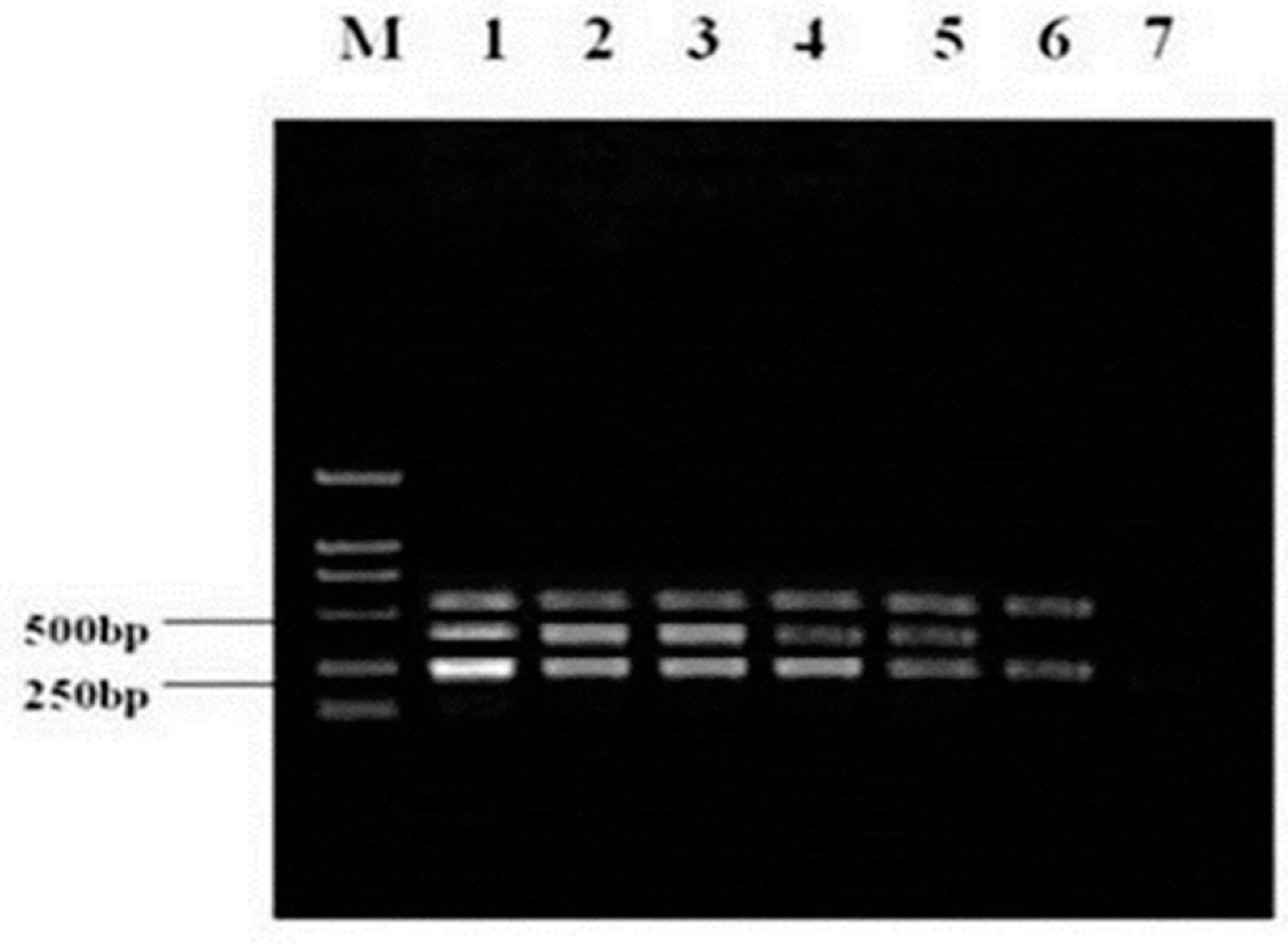
3.4. Detection of Viruses in Clinical Specimens
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| PRRSV | porcine reproductive and respiratory syndrome virus |
| CSFV | classical swine fever virus |
| PRV | pseudorabies virus |
| PCV2 | porcine circovirus type 2 |
| PPV | porcine parvovirus |
| JEV | Japanese encephalitis virus |
| RV | rotavirus |
| PEDV | porcine epidemic diarrhea virus |
References
- Renukaradhya, G.J.; Meng, X.J.; Calvert, J.G.; Roof, M.; Lager, K.M. Live porcine reproductive and respiratory syndrome virus vaccines: Current status and future direction. Vaccine 2015, 33, 4069–4080. [Google Scholar] [CrossRef] [PubMed]
- Loving, C.L.; Osorio, F.A.; Murtaugh, M.P.; Zuckermann, F.A. Innate and adaptive immunity against porcine reproductive and respiratory syndrome virus. Vet. Immunol. Immunopathol. 2015, 167, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Brar, M.S.; Shi, M.; Murtaugh, M.P.; Leung, F.C. Evolutionary diversification of type 2 porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 2015, 96, 1570–1580. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.Q.; Chen, Z.S.; Liu, W.X.; Cui, Y.Z. Isolation and identification of porcine reproductive and respiratory syndrome (PRRS) virus from aborted fetuses suspected of PRRS. Chin. J. Anim. Pollut. Infect. Dis. 1996, 26, 1–5. (In Chinese) [Google Scholar]
- Gao, Z.Q.; Guo, X.; Yang, H.C. Genomic characterization of two Chinese isolates of porcine respiratory and reproductive syndrome virus. Arch. Virol. 2004, 149, 1341–1351. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, T.; Zhu, C.G.; Jin, Y.F.; Zhang, Y.Z. Genetic variation of Chinese PRRSV strains based on ORF5 sequence. Biochem. Genet. 2006, 44, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Tian, K.G.; Yu, X.L.; Zhao, T.Z.; Feng, Y.J.; Cao, Z.; Wang, C.B.; Hu, Y.; Chen, X.Z.; Hu, D.M.; Tian, X.S.; et al. Emergence of fatal PRRSV variants: Unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS ONE 2007, 2, e526. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Ye, C.; Chang, X.B.; Jiang, C.G.; Wang, S.J.; Cai, X.H.; Tong, G.Z.; Tian, Z.J.; Shi, M.; An, T.Q. Importation and recombination are responsible for the latest emergence of highly pathogenic porcine reproductive and respiratory syndrome virus in China. J. Virol. 2015, 89, 10712–10716. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Ni, J.; Cao, Z.; Han, X.; Xia, Y.; Zi, Z.; Ning, K.; Liu, Q.; Cai, L.; Qiu, P.; et al. The epidemic status and genetic diversity of 14 highly pathogenic porcine reproductive and respiratory syndrome virus (HP-PRRSV) isolates from China in 2009. Vet. Microbiol. 2011, 150, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Wernike, K.; Hoffmann, B.; Dauber, M.; Lange, E.; Schirrmeier, H.; Beer, M. Detection and typing of highly pathogenic porcine reproductive and respiratory syndrome virus by multiplex real-time RT-PCR. PLoS ONE 2012, 7, e38251. [Google Scholar] [CrossRef] [PubMed]
- Chai, Z.; Ma, W.J.; Fu, F.; Lang, Y.K.; Wang, W.; Tong, G.Z.; Liu, Q.F.; Cai, X.H.; Li, X. A SYBR Green-based real-time RT-PCR assay for simple and rapid detection and differentiation of highly pathogenic and classical type 2 porcine reproductive and respiratory syndrome virus circulating in China. Arch. Virol. 2013, 158, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.L.; Li, Y.H.; Duan, Z.Y.; Guo, R.; Liu, Z.W.; Zhou, D.N.; Yuan, F.Y.; Tian, Y.X. A one step RT-PCR assay to detect and discriminate porcine reproductive and respiratory syndrome viruses in clinical specimens. Gene 2013, 531, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Reed, L.J.; Muench, H. A simple method of estimating fifty percent end points. Am. J. Hyg. 1938, 27, 709–716. [Google Scholar]
- Zhou, L.; Yang, H.C.; Lunney, J.K.; Rowland, R.R.R. Porcine reproductive and respiratory syndrome in China. Virus Res. 2010, 154, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Lunney, J.K.; Fang, Y.; Ladinig, A.; Chen, N.; Li, Y.; Rowland, B.; Renukaradhya, G.J. Porcine reproductive and respiratory syndrome virus (PRRSV): Pathogenesis and interaction with the immune system. Annu. Rev. Anim. Biosci. 2016, 4, 129–154. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Chen, N.; Wan, L.; Wu, J.; Zhou, Z.; Ni, J.; Li, X.; Zhai, X.; Shi, J.; Tian, K. New genomic characteristics of highly pathogenic porcine reproductive and respiratory syndrome viruses do not lead to significant changes in pathogenicity. Vet. Microbiol. 2012, 158, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Mengeling, W.L.; Lager, K.M. A brief review of procedures and potential problems associated with the diagnosis of porcine reproductive and respiratory syndrome. Vet. Res. 2000, 31, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Kleiboeker, S.B.; Schommer, S.K.; Lee, S.M.; Watkins, S.; Chittick, W.; Polson, D. Simultaneous detection of North American and European porcine reproductive and respiratory syndrome virus using real-time quantitative reverse transcriptase-PCR. J. Vet. Diagn. Investig. 2005, 17, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Lurchachaiwong, W.; Payungporn, S.; Srisatidnarakul, U.; Mungkundar, C.; Theamboonlers, A.; Poovorawan, Y. Rapid detection and strain identification of porcine reproductive and respiratory syndrome virus (PRRSV) by real-time RT-PCR. Lett. Appl. Microbiol. 2008, 46, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.L.; Wu, H.; Yu, Y.G.; Cheng, B.Z.; Yang, X.G.; Chen, G.; Liu, D.M.; Li, X.F. Rapid detection of a highly virulent Chinese-type isolate of Porcine Reproductive and Respiratory Syndrome virus by real-time reverse transcriptase PCR. J. Virol. Methods 2008, 149, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Markoulatos, P.; Siafakas, N.; Moncany, M. Multiplex polymerase chain reaction: A practical approach. J. Clin. Lab. Anal. 2002, 16, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.G.; Chen, G.D.; Huang, Y.; Ding, L.; Li, Z.C.; Chang, C.D.; Wang, C.Y.; Tong, D.W.; Liu, H.J. Development of multiplex PCR for simultaneous detection of six swine DNA and RNA viruses. J. Virol. Methods 2012, 183, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.K.; Wei, C.H.; Yang, X.Y.; Dai, A.L.; Li, X.H. Multiplex PCR for the simultaneous detection of porcine reproductive and respiratory syndrome virus, classical swine fever virus, and porcine circovirus in pigs. Mol. Cell. Probes 2013, 27, 149–152. [Google Scholar] [CrossRef] [PubMed]
- Spear, A.; Faaberg, K.S. Development of a genome copy specific RT-qPCR assay for divergent strains of type 2 porcine reproductive and respiratory syndrome virus. J. Virol. Methods 2015, 218, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Pileri, E.; Gibert, E.; Martín-Valls, G.E.; Nofrarias, M.; López-Soria, S.; Martín, M.; Díaz, I.; Darwich, L.; Mateu, E. Transmission of Porcine reproductive and respiratory syndrome virus 1 to and from vaccinated pigs in a one-to-one model. Vet. Microbiol. 2017, 201, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Charpin, C.; Mahé, S.; Keranflec’h, A.; Belloc, C.; Cariolet, R.; Le Potier, M.F.; Rose, N. Infectiousness of pigs infected by the Porcine Reproductive and Respiratory Syndrome virus (PRRSV) is time-dependent. Vet. Res. 2012, 43, 69. [Google Scholar] [CrossRef] [PubMed]
- Pileri, E.; Gibert, E.; Soldevila, F.; García-Saenz, A.; Pujols, J.; Diaz, I.; Darwich, L.; Casal, J.; Martín, M.; Mateu, E. Vaccination with a genotype 1 modified live vaccine against porcine reproductive and respiratory syndrome virus significantly reduces viremia, viral shedding and transmission of the virus in aquasi-natural experimental model. Vet. Microbiol. 2015, 175, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Rose, N.; Renson, P.; Andraud, M.; Paboeuf, F.; Le Potier, M.F.; Bourry, O. Porcine reproductive and respiratory syndrome virus (PRRSv) modified-live vaccine reduces virus transmission in experimental conditions. Vaccine 2015, 33, 2493–2499. [Google Scholar] [CrossRef] [PubMed]

| Primer | Primer Sequences (5′–3′) | Origin/Target Gene | Location | Products | Source |
|---|---|---|---|---|---|
| Nsp2-F | TGAYGGGCGACAATGTCC | PRRSV CH-1a (GenBank:AY032626)/Nsp2 | 2745–2762(AY032626) | 319 bp (AY032626) | Previous study [12] |
| 1406–1423(FJ495082.2) | |||||
| Nsp2-R | CGCAGACAAATCCAGAVG | PRRSV 07HBEZ (GenBank:FJ495082.2)/Nsp2 | 3064–3047(AY032626) | 229 bp (FJ495082.2) | |
| 1635–1618(FJ495082.2) | |||||
| JXA1-F | ATTTGAATGTTCGCACGGTCTC | PRRSV 07HBEZ (GenBank:FJ495082.2)/GP4 | 13,380–13,401 | None (FJ495082.2) | This study |
| JXA1-R | CCGCTGAAACTCTGGTTAAAGG | PRRSV JXA1-P170 (GenBank:JQ804986.1)/GP5 | 13,879–13,900 | 620 bp (JQ804986.1) |
| Multiplex RT-PCR | Sequencing Method | |||||||
|---|---|---|---|---|---|---|---|---|
| Pig Farm | No. of Specimens | HP-PRRSV JXA1-R Positive (%) | HP-PRRSV Positive (%) | C-PRRSV Positive (%) | HP-PRRSV JXA1-R Positive (%) | HP-PRRSV Positive (%) | C-PRRSV Positive (%) | Concordance Rate (%) |
| 1 | 56 | 1 (1.79) | 6 (10.71) | 0 (0) | 1 (1.79) | 6 (10.71) | 0 (0) | 100 |
| 2 | 60 | 2 a (3.33) | 11 a (18.33) | 2 (3.33) | 2 a (3.33) | 11 a (18.33) | 2 (3.33) | 100 |
| 3 | 45 | 1 (2.22) | 5 (11.11) | 0 (0) | 1 (2.22) | 5 (11.11) | 0 (0) | 100 |
| 4 | 58 | 2 a (3.45) | 9 a (15.52) | 1 (1.72) | 2 a (3.45) | 9 a (15.52) | 1 (1.72) | 100 |
| 5 | 32 | 0 (0) | 3 (9.38) | 0 (0) | 0 (0) | 3 (9.38) | 0 (0) | 100 |
| 6 | 35 | 0 (0) | 5 (14.29) | 0 (0) | 0 (0) | 5 (14.29) | 0 (0) | 100 |
| 7 | 49 | 1 (2.04) | 2 b (4.08) | 1 b (2.04) | 1 (2.04) | 2 b (4.08) | 1 b (2.04) | 100 |
| 8 | 57 | 2 (3.51) | 6 (10.53) | 0 (0) | 2 (3.51) | 6 (10.53) | 0 (0) | 100 |
| 9 | 66 | 2 c (3.03) | 10 (15.15) | 2 c (3.03) | 2c (3.03) | 10 (15.15) | 2 c (3.03) | 100 |
| 10 | 58 | 1 (1.72) | 6 (10.34) | 0 (0) | 1 (1.72) | 6 (10.34) | 0 (0) | 100 |
| Total | 516 | 12 (2.33) | 63 (12.21) | 6 (1.16) | 12 (2.33) | 63 (12.21) | 6 (1.16) | 100 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, K.; Tian, Y.; Zhou, D.; Duan, Z.; Guo, R.; Liu, Z.; Yuan, F.; Liu, W. A Multiplex RT-PCR Assay to Detect and Discriminate Porcine Reproductive and Respiratory Syndrome Viruses in Clinical Specimens. Viruses 2017, 9, 205. https://doi.org/10.3390/v9080205
Yang K, Tian Y, Zhou D, Duan Z, Guo R, Liu Z, Yuan F, Liu W. A Multiplex RT-PCR Assay to Detect and Discriminate Porcine Reproductive and Respiratory Syndrome Viruses in Clinical Specimens. Viruses. 2017; 9(8):205. https://doi.org/10.3390/v9080205
Chicago/Turabian StyleYang, Keli, Yongxiang Tian, Danna Zhou, Zhengying Duan, Rui Guo, Zewen Liu, Fangyan Yuan, and Wei Liu. 2017. "A Multiplex RT-PCR Assay to Detect and Discriminate Porcine Reproductive and Respiratory Syndrome Viruses in Clinical Specimens" Viruses 9, no. 8: 205. https://doi.org/10.3390/v9080205





