Therapeutic Potency of Nanoformulations of siRNAs and shRNAs in Animal Models of Cancers
Abstract
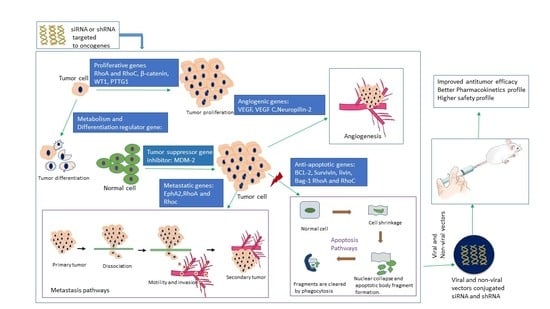
:1. Introduction
2. Challenges and Breakthrough of siRNA Delivery: From Concept to Clinical Trial
3. In-Vivo Delivery of siRNAs and shRNAs Directed against Different Cancer-Causing Genes in Various Cancer Models
3.1. Silencing of Bcl-2 Gene
3.2. Silencing of VEGF Gene
3.3. Silencing of EGF Receptor Genes
3.4. Silencing of Survivin Gene
3.5. Silencing of Cyclin-B1 Gene
3.6. Silencing of RhoA and RhoC Gene
3.7. Silencing of β-Catenin Gene
3.8. Silencing of EphA2 Gene
3.9. Silencing of MDM-2 Gene
3.10. Silencing of IGF-1R Gene
3.11. Silencing of Livin Gene
3.12. Silencing of WT1 Gene
3.13. Miscellaneous
4. Future Directions
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Naldini, L. Gene therapy returns to centre stage. Nature 2015, 526, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Zamore, P.D. RNA interference: Big applause for silencing in Stockholm. Cell 2006, 127, 1083–1086. [Google Scholar] [CrossRef] [PubMed]
- Hannon, G.J. RNA interference. Nature 2002, 418, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Williams, B. Role of the Double-Stranded RNA-Activated Protein Kinase (PKR) in Cell Regulation; Portland Press Limited: London, UK, 1997. [Google Scholar]
- Elbashir, S.M.; Harborth, J.; Lendeckel, W.; Yalcin, A.; Weber, K.; Tuschl, T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001, 411, 494–498. [Google Scholar] [CrossRef] [PubMed]
- Hannon, G.J.; Rossi, J.J. Unlocking the potential of the human genome with RNA interference. Nature 2004, 431, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Meister, G.; Tuschl, T. Mechanisms of gene silencing by double-stranded RNA. Nature 2004, 431, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, K.A.; Langer, R.; Anderson, D.G. Knocking down barriers: Advances in siRNA delivery. Nat. Rev. Drug Discov. 2009, 8, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Matranga, C.; Tomari, Y.; Shin, C.; Bartel, D.P.; Zamore, P.D. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 2005, 123, 607–620. [Google Scholar] [CrossRef] [PubMed]
- Rand, T.A.; Petersen, S.; Du, F.; Wang, X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 2005, 123, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Hutvágner, G.; Zamore, P.D. A microRNA in a multiple-turnover RNAi enzyme complex. Science 2002, 297, 2056–2060. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, D.W.; Davis, M.E. Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging. Nucl. Acids Res. 2006, 34, 322–333. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.M.; Li, M.Z.; Chang, K.; Ge, W.; Golding, M.C.; Rickles, R.J.; Siolas, D.; Hu, G.; Paddison, P.J.; Schlabach, M.R.; et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat. Genet. 2005, 37, 1281. [Google Scholar] [CrossRef] [PubMed]
- Rao, D.D.; Vorhies, J.S.; Senzer, N.; Nemunaitis, J. siRNA vs. shRNA: Similarities and differences. Adv. Drug Deliv. Rev. 2009, 61, 746–759. [Google Scholar] [CrossRef] [PubMed]
- Cullen, B.R. RNAi the natural way. Nat. Genet. 2005, 37, 1163. [Google Scholar] [CrossRef] [PubMed]
- Karim, E.; Rosli, R.; Chowdhury, E.H. Systemic Delivery of Nanoformulations of Anti-cancer Drugs with Therapeutic Potency in Animal Models of Cancer. Curr. Cancer Ther. Rev. 2016, 12, 204–220. [Google Scholar] [CrossRef]
- Davis, M.E. Non-viral gene delivery systems. Curr. Opin. Biotechnol. 2002, 13, 128–131. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, A.; Miyata, K.; Kataoka, K. Recent progress in development of siRNA delivery vehicles for cancer therapy. Adv. Drug Deliv. Rev. 2016, 104, 61–77. [Google Scholar] [CrossRef] [PubMed]
- Pahle, J.; Walther, W. Vectors and strategies for nonviral cancer gene therapy. Expert Opin. Biol. Ther. 2016, 16, 443–461. [Google Scholar] [CrossRef] [PubMed]
- Wirth, T.; Parker, N.; Ylä-Herttuala, S. History of gene therapy. Gene 2013, 525, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, N.; Wu, J.; Xu, X.; Kamaly, N.; Farokhzad, O.C. Cancer nanotechnology: The impact of passive and active targeting in the era of modern cancer biology. Adv. Drug Deliv. Rev. 2014, 66, 2–25. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Liu, X.; Zhu, D.; Wang, Y.; Zhang, Z.; Zhou, X.; Qiu, N.; Chen, X.; Shen, Y. Nonviral cancer gene therapy: Delivery cascade and vector nanoproperty integration. Adv. Drug Deliv. Rev. 2017, 115, 115–154. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.D.; Siegel, R.L.; Lin, C.C.; Mariotto, A.B.; Kramer, J.L.; Rowland, J.H.; Stein, K.D.; Alteri, R.; Jemal, A. Cancer treatment and survivorship statistics, 2016. CA Cancer J. Clin. 2016, 66, 271–289. [Google Scholar] [CrossRef] [PubMed]
- Fitzmaurice, C.; Allen, C.; Barber, R.M.; Barregard, L.; Bhutta, Z.A.; Brenner, H.; Dicker, D.J.; Chimed-Orchir, O.; Dandona, R.; Dandona, L. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: A systematic analysis for the global burden of disease study. JAMA Oncol. 2017, 3, 524–548. [Google Scholar] [PubMed]
- Bitko, V.; Musiyenko, A.; Shulyayeva, O.; Barik, S. Inhibition of respiratory viruses by nasally administered siRNA. Nat. Med. 2005, 11, 50–55. [Google Scholar] [CrossRef] [PubMed]
- DiFiglia, M.; Sena-Esteves, M.; Chase, K.; Sapp, E.; Pfister, E.; Sass, M.; Yoder, J.; Reeves, P.; Pandey, R.K.; Rajeev, K.G. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc. Natl. Acad. Sci. USA 2007, 104, 17204–17209. [Google Scholar] [CrossRef] [PubMed]
- Pecot, C.V.; Calin, G.A.; Coleman, R.L.; Lopez-Berestein, G.; Sood, A.K. RNA interference in the clinic: Challenges and future directions. Nat. Rev. Cancer 2011, 11, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Bumcrot, D.; Manoharan, M.; Koteliansky, V.; Sah, D.W. RNAi therapeutics: A potential new class of pharmaceutical drugs. Nat. Chem. Biol. 2006, 2, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Alexis, F.; Pridgen, E.; Molnar, L.K.; Farokhzad, O.C. Factors affecting the clearance and biodistribution of polymeric nanoparticles. Mol. Pharm. 2008, 5, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Scherphof, G. In vivo behavior of liposomes: Interactions with the mononuclear phagocyte system and implications for drug targeting. In Targeted Drug Delivery; Springer: Berlin, Germany, 1991; pp. 285–327. [Google Scholar]
- Zamecnik, J.; Vargova, L.; Homola, A.; Kodet, R.; Sykova, E. Extracellular matrix glycoproteins and diffusion barriers in human astrocytic tumours. Neuropathol. Appl. Neurobiol. 2004, 30, 338–350. [Google Scholar] [CrossRef] [PubMed]
- Decuzzi, P.; Causa, F.; Ferrari, M.; Netti, P. The effective dispersion of nanovectors within the tumor microvasculature. Ann. Biomed. Eng. 2006, 34, 633–641. [Google Scholar] [CrossRef] [PubMed]
- Dominska, M.; Dykxhoorn, D.M. Breaking down the barriers: siRNA delivery and endosome escape. J. Cell Sci. 2010, 123, 1183–1189. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Rossi, J.J. Strategies for silencing human disease using RNA interference. Nat. Rev. Genet. 2007, 8, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Jackson, A.L.; Bartz, S.R.; Schelter, J.; Kobayashi, S.V.; Burchard, J.; Mao, M.; Li, B.; Cavet, G.; Linsley, P.S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003, 21, 635–637. [Google Scholar] [CrossRef] [PubMed]
- Jackson, A.L.; Burchard, J.; Schelter, J.; Chau, B.N.; Cleary, M.; Lim, L.; Linsley, P.S. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA 2006, 12, 1179–1187. [Google Scholar] [CrossRef] [PubMed]
- Petros, R.A.; DeSimone, J.M. Strategies in the design of nanoparticles for therapeutic applications. Nat. Rev. Drug Discov. 2010, 9, 615–627. [Google Scholar] [CrossRef] [PubMed]
- Kanasty, R.L.; Whitehead, K.A.; Vegas, A.J.; Anderson, D.G. Action and reaction: The biological response to siRNA and its delivery vehicles. Mol. Ther. 2012, 20, 513–524. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.E.; Ehrhardt, A.; Kay, M.A. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 2003, 4, 346–358. [Google Scholar] [CrossRef] [PubMed]
- Castanotto, D.; Rossi, J.J. The promises and pitfalls of RNA-interference-based therapeutics. Nature 2009, 457, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Walter, W.; Stein, U. Viral vectors for gene transfer a review of their use in the treatment of human disease. Drugs 2000, 60, 249–271. [Google Scholar] [CrossRef]
- Ylä-Herttuala, S. Glybera’s second act: The curtain rises on the high cost of therapy. Mol. Ther. 2015, 23, 217–218. [Google Scholar] [CrossRef] [PubMed]
- Kay, M.A. State-of-the-art gene-based therapies: The road ahead. Nat. Rev. Genet. 2011, 12, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Mintzer, M.A.; Simanek, E.E. Nonviral vectors for gene delivery. Chem. Rev. 2008, 109, 259–302. [Google Scholar] [CrossRef] [PubMed]
- Gilleron, J.; Querbes, W.; Zeigerer, A.; Borodovsky, A.; Marsico, G.; Schubert, U.; Manygoats, K.; Seifert, S.; Andree, C.; Stöter, M. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 2013, 31, 638–646. [Google Scholar] [CrossRef] [PubMed]
- Schaffer, D.V.; Fidelman, N.A.; Dan, N.; Lauffenburger, D.A. Vector unpacking as a potential barrier for receptor-mediated polyplex gene delivery. Biotechnol. Bioeng. 2000, 67, 598–606. [Google Scholar] [CrossRef]
- Lv, H.; Zhang, S.; Wang, B.; Cui, S.; Yan, J. Toxicity of cationic lipids and cationic polymers in gene delivery. J. Control. Release 2006, 114, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Loh, X.J.; Lee, T.-C.; Dou, Q.; Deen, G.R. Utilising inorganic nanocarriers for gene delivery. Biomater. Sci. 2016, 4, 70–86. [Google Scholar] [CrossRef] [PubMed]
- Shim, M.S.; Kwon, Y.J. Efficient and targeted delivery of siRNA in vivo. FEBS J. 2010, 277, 4814–4827. [Google Scholar] [CrossRef] [PubMed]
- Rosenblum, D.; Joshi, N.; Tao, W.; Karp, J.M.; Peer, D. Progress and challenges towards targeted delivery of cancer therapeutics. Nat. Commun. 2018, 9, 1410. [Google Scholar] [CrossRef] [PubMed]
- Stacker, S.A.; Williams, S.P.; Karnezis, T.; Shayan, R.; Fox, S.B.; Achen, M.G. Lymphangiogenesis and lymphatic vessel remodelling in cancer. Nat. Rev. Cancer 2014, 14, 159. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.K.; Stylianopoulos, T. Delivering nanomedicine to solid tumors. Nat. Rev. Clin. Oncol. 2010, 7, 653. [Google Scholar] [CrossRef] [PubMed]
- Danquah, M.K.; Zhang, X.A.; Mahato, R.I. Extravasation of polymeric nanomedicines across tumor vasculature. Adv. Drug Deliv. Rev. 2011, 63, 623–639. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Mangala, L.S.; Rodriguez-Aguayo, C.; Kong, X.; Lopez-Berestein, G.; Sood, A.K. RNA interference-based therapy and its delivery systems. Cancer Metastasis Rev. 2018, 37, 107–124. [Google Scholar] [CrossRef] [PubMed]
- Layek, B.; Lipp, L.; Singh, J. Cell penetrating peptide conjugated chitosan for enhanced delivery of nucleic acid. Int. J. Mol. Sci. 2015, 16, 28912–28930. [Google Scholar] [CrossRef] [PubMed]
- Shukla, R.S.; Jain, A.; Zhao, Z.; Cheng, K. Intracellular trafficking and exocytosis of a multi-component siRNA nanocomplex. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 1323–1334. [Google Scholar] [CrossRef] [PubMed]
- Sahay, G.; Querbes, W.; Alabi, C.; Eltoukhy, A.; Sarkar, S.; Zurenko, C.; Karagiannis, E.; Love, K.; Chen, D.; Zoncu, R. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat. Biotechnol. 2013, 31, 653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pezzella, F.; Turley, H.; Kuzu, I.; Tungekar, M.F.; Dunnill, M.S.; Pierce, C.B.; Harris, A.; Gatter, K.C.; Mason, D.Y. bcl-2 protein in non-small-cell lung carcinoma. N. Engl. J. Med. 1993, 329, 690–694. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, Y. Antisense bcl-2 oligodeoxynucleotide enhancing 5-fluorouracil induced apoptosis in human gastric cancer cell line SGC 7901. Tumor 2002, 5, 15. [Google Scholar]
- Joensuu, H.; Pylkkänen, L.; Toikkanen, S. Bcl-2 protein expression and long-term survival in breast cancer. Am. J. Pathol. 1994, 145, 1191. [Google Scholar] [PubMed]
- Sinicrope, F.A.; Hart, J.; Michelassi, F.; Lee, J.J. Prognostic value of bcl-2 oncoprotein expression in stage II colon carcinoma. Clin. Cancer Res. 1995, 1, 1103–1110. [Google Scholar] [PubMed]
- Li, S.-M.; Yao, S.-K.; Yamamura, N.; Nakamura, T. Expression of Bcl-2 and Bax in extrahepatic biliary tract carcinoma and dysplasia. World J. Gastroenterol. 2003, 9, 2579. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.X.; Sato, Y.; Kuwao, S.; Kameya, T. Expression of bcl-2 oncogene protein is prevalent in small cell lung carcinomas. J. Pathol. 1995, 177, 135–138. [Google Scholar] [CrossRef] [PubMed]
- Fu, G.-F.; Lin, X.-H.; Han, Q.-W.; Xu, Y.-F.; Guo, D.; Xu, G.-X.; Hou, Y.-Y. RNA interference remarkably suppresses bcl-2 gene expression in cancer cells in vitro and in vivo. Cancer Biol. Ther. 2005, 4, 822–829. [Google Scholar] [CrossRef] [PubMed]
- Koty, P.P.; Zhang, H.; Levitt, M.L. Antisense bcl-2 treatment increases programmed cell death in non-small cell lung cancer cell lines. Lung Cancer 1999, 23, 115–127. [Google Scholar] [CrossRef]
- Lima, R.T.; Martins, L.M.; Guimaraes, J.E.; Sambade, C.; Vasconcelos, M.H. Specific downregulation of bcl-2 and xIAP by RNAi enhances the effects of chemotherapeutic agents in MCF-7 human breast cancer cells. Cancer Gene ther. 2004, 11, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Weyhenmeyer, B.; Murphy, A.; Prehn, J.; Murphy, B. Targeting the anti-apoptotic Bcl-2 family members for the treatment of cancer. Exp. Oncol. 2012, 34, 192–199. [Google Scholar] [PubMed]
- Garcia-Saez, A. The secrets of the Bcl-2 family. Cell Death Differ. 2012, 19, 1733–1740. [Google Scholar] [CrossRef] [PubMed]
- Sasatomi, E.; Tokunaga, O.; Miyazaki, K. Spontaneous apoptosis in gallbladder carcinoma: Relationships with clinicopathologic factors, expression of E-cadherin, bcl-2 protooncogene, and p53 oncosuppressor gene. Cancer 1996, 78, 2101–2110. [Google Scholar] [CrossRef]
- Mikami, T.; Yanagisawa, N.; Baba, H.; Koike, M.; Okayasu, I. Association of Bcl-2 protein expression with gallbladder carcinoma differentiation and progression and its relation to apoptosis. Cancer 1999, 85, 318–325. [Google Scholar] [CrossRef]
- Geng, Z.-M.; Zhang, M.; Pan, X.-T.; Wang, L. Bcl-2 gene silencing by RNA interference inhibits the growth of the human gallbladder carcinoma cell line GBC-SD in vitro and in vivo. Oncol. Rep. 2013, 30, 793–800. [Google Scholar] [CrossRef] [PubMed]
- Yano, J.; Hirabayashi, K.; Nakagawa, S.-I.; Yamaguchi, T.; Nogawa, M.; Kashimori, I.; Naito, H.; Kitagawa, H.; Ishiyama, K.; Ohgi, T. Antitumor activity of small interfering RNA/cationic liposome complex in mouse models of cancer. Clin. Cancer Res. 2004, 10, 7721–7726. [Google Scholar] [CrossRef] [PubMed]
- Sonoke, S.; Ueda, T.; Fujiwara, K.; Sato, Y.; Takagaki, K.; Hirabayashi, K.; Ohgi, T.; Yano, J. Tumor regression in mice by delivery of Bcl-2 small interfering RNA with pegylated cationic liposomes. Cancer Res. 2008, 68, 8843–8851. [Google Scholar] [CrossRef] [PubMed]
- Grunewald, M.; Avraham, I.; Dor, Y.; Bachar-Lustig, E.; Itin, A.; Yung, S.; Chimenti, S.; Landsman, L.; Abramovitch, R.; Keshet, E. VEGF-induced adult neovascularization: Recruitment, retention, and role of accessory cells. Cell 2006, 124, 175–189. [Google Scholar] [CrossRef] [PubMed]
- Saint-Geniez, M.; D’amore, P.A. Development and pathology of the hyaloid, choroidal and retinal vasculature. Int. J. Dev. Biol. 2004, 48, 1045–1058. [Google Scholar] [CrossRef] [PubMed]
- Stefater, J.A., III; Lewkowich, I.; Rao, S.; Mariggi, G.; Carpenter, A.C.; Burr, A.R.; Fan, J.; Ajima, R.; Molkentin, J.D.; Williams, B.O. Regulation of angiogenesis by a non-canonical Wnt-Flt1 pathway in myeloid cells. Nature 2011, 474, 511–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Folkman, J. Angiogenesis and breast cancer. J. Clin. Oncol. 1994, 12, 441–443. [Google Scholar] [CrossRef] [PubMed]
- Blood, C.H.; Zetter, B.R. Tumor interactions with the vasculature: Angiogenesis and tumor metastasis. Biochim. Biophys. Acta Rev. Cancer 1990, 1032, 89–118. [Google Scholar] [CrossRef]
- Ferrara, N.; Gerber, H.-P.; LeCouter, J. The biology of VEGF and its receptors. Nat. Med. 2003, 9, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Dvorak, H.F.; Detmar, M.; Claffey, K.P.; Nagy, J.A.; van de Water, L.; Senger, D.R. Vascular permeability factor/vascular endothelial growth factor: An important mediator of angiogenesis in malignancy and inflammation. Int. Arch. Allergy Immunol. 1995, 107, 233–235. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, N.; Kerbel, R.S. Angiogenesis as a therapeutic target. Nature 2005, 438, 967–974. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, N. VEGF and the quest for tumour angiogenesis factors. Nat. Rev. Cancer 2002, 2, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Tsai, J.H.; Lee, W.M. Modeling antiangiogenesis gene therapy. Cancer Biol. Ther. 2002, 1, 554–555. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.-P.; Feng, G.-S.; Liang, H.-M.; Zheng, C.-S.; Li, X. Vascular endothelial growth factor antisense oligodeoxynucleotides with lipiodol in arterial embolization of liver cancer in rats. World J. Gastroenterol. 2004, 10, 813. [Google Scholar] [CrossRef] [PubMed]
- Namiecińska, M.; Marciniak, K.; Nowak, J.Z. VEGF jako czynnik angiogenny, neurotroficzny i neuroprotekcyjny* VEGF as an angiogenic, neurotrophic, and neuroprotective factor. Postep. Hig. Med. Dosw. 2005, 59, 573–583. [Google Scholar]
- Takahashi, Y.; Kitadai, Y.; Bucana, C.D.; Cleary, K.R.; Ellis, L.M. Expression of vascular endothelial growth factor and its receptor, KDR, correlates with vascularity, metastasis, and proliferation of human colon cancer. Cancer Res. 1995, 55, 3964–3968. [Google Scholar] [PubMed]
- Toi, M.; Inada, K.; Suzuki, H.; Tominaga, T. Tumor angiogenesis in breast cancer: Its importance as a prognostic indicator and the association with vascular endothelial growth factor expression. Breast Cancer Res. Treat. 1995, 36, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Weidner, N.; Folkman, J.; Pozza, F.; Bevilacqua, P.; Allred, E.N.; Moore, D.H.; Meli, S.; Gasparini, G. Tumor angiogenesis: A new significant and independent prognostic indicator in early-stage breast carcinoma. JNCI J. Natl. Cancer Inst. 1992, 84, 1875–1887. [Google Scholar] [CrossRef] [PubMed]
- Maeda, K.; Chung, Y.; Takatsuka, S.; Ogawa, Y.; Sawada, T.; Yamashita, Y.; Onoda, N.; Kato, Y.; Nitta, A.; Arimoto, Y. Tumor angiogenesis as a predictor of recurrence in gastric carcinoma. J. Clin. Oncol. 1995, 13, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Algire, G.H.; Chalkley, H.W.; Legallais, F.Y.; Park, H.D. Vasculae reactions of normal and malignant tissues in vivo. I. vascular reactions of mice to wounds and to normal and neoplastic transplants. JNCI J. Natl. Cancer Inst. 1945, 6, 73–85. [Google Scholar] [CrossRef]
- Rhee, J.; Hoff, P.M. Angiogenesis inhibitors in the treatment of cancer. Expert Opin. Pharmacother. 2005, 6, 1701–1711. [Google Scholar] [CrossRef] [PubMed]
- Takei, Y.; Kadomatsu, K.; Matsuo, S.; Itoh, H.; Nakazawa, K.; Kubota, S.; Muramatsu, T. Antisense oligodeoxynucleotide targeted to Midkine, a heparin-binding growth factor, suppresses tumorigenicity of mouse rectal carcinoma cells. Cancer Res. 2001, 61, 8486–8491. [Google Scholar] [PubMed]
- Lu, P.Y.; Xie, F.Y.; Woodle, M.C. Modulation of angiogenesis with siRNA inhibitors for novel therapeutics. Trends Mol. Med. 2005, 11, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Fang, X.; Branch, C.; Mazur, W.; French, B.; Roth, J. Generation and identification of recombinant adenovirus by liposome-mediated transfection and PCR analysis. Biotechniques 1993, 15, 868–872. [Google Scholar] [PubMed]
- Jones, N.; Shenk, T. An adenovirus type 5 early gene function regulates expression of other early viral genes. Proc. Natl. Acad. Sci. USA 1979, 76, 3665–3669. [Google Scholar] [CrossRef] [PubMed]
- Im, S.; Kim, J.; Gomez-Manzano, C.; Fueyo, J.; Liu, T.; Cho, M.; Seong, C.; Lee, S.; Hong, Y.; Yung, W. Inhibition of breast cancer growth in vivo by antiangiogenesis gene therapy with adenovirus-mediated antisense-VEGF. Br. J. Cancer 2001, 84, 1252. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, K.; Togawa, H.; Harada, A.; Yasugi, K.; Matsumoto, T.; Katayose, S. Spontaneous formation of polyion complex micelles with narrow distribution from antisense oligonucleotide and cationic block copolymer in physiological saline. Macromolecules 1996, 29, 8556–8557. [Google Scholar] [CrossRef]
- Vinogradov, S.V.; Bronich, T.K.; Kabanov, A.V. Self-Assembly of Polyamine—Poly (ethylene glycol) Copolymers with Phosphorothioate Oligonucleotides. Bioconjugate Chem. 1998, 9, 805–812. [Google Scholar] [CrossRef] [PubMed]
- Schiffelers, R.M.; Ansari, A.; Xu, J.; Zhou, Q.; Tang, Q.; Storm, G.; Molema, G.; Lu, P.Y.; Scaria, P.V.; Woodle, M.C. Cancer siRNA therapy by tumor selective delivery with ligand-targeted sterically stabilized nanoparticle. Nucl. Acids Res. 2004, 32, e149. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Jeong, J.H.; Cho, K.C.; Kim, S.W.; Park, T.G. Target-specific gene silencing by siRNA plasmid DNA complexed with folate-modified poly (ethylenimine). J. Control. Release 2005, 104, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Harada, A.; Kataoka, K. Formation of polyion complex micelles in an aqueous milieu from a pair of oppositely-charged block copolymers with poly (ethylene glycol) segments. Macromolecules 1995, 28, 5294–5299. [Google Scholar] [CrossRef]
- Kim, S.H.; Jeong, J.H.; Lee, S.H.; Kim, S.W.; Park, T.G. Local and systemic delivery of VEGF siRNA using polyelectrolyte complex micelles for effective treatment of cancer. J. Control. Release 2008, 129, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Joukov, V.; Kaipainen, A.; Jeltsch, M.; Pajusola, K.; Olofsson, B.; Kumar, V.; Eriksson, U.; Alitalo, K. Vascular endothelial growth factors VEGF-B and VEGF-C. J. Cell. Physiol. 1997, 173, 211–215. [Google Scholar] [CrossRef]
- Cohen, B.; Addadi, Y.; Sapoznik, S.; Meir, G.; Kalchenko, V.; Harmelin, A.; Ben-Dor, S.; Neeman, M. Transcriptional regulation of vascular endothelial growth factor C by oxidative and thermal stress is mediated by lens epithelium-derived growth factor/p75. Neoplasia 2009, 11, 921. [Google Scholar] [CrossRef] [PubMed]
- Su, J.-L.; Yen, C.; Chen, P.; Chuang, S.; Hong, C.; Kuo, I.; Chen, H.; Hung, M.-C.; Kuo, M. The role of the VEGF-C/VEGFR-3 axis in cancer progression. Br. J. Cancer 2007, 96, 541–545. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, S.; Brown, L.F.; Kodama, S.; Paavonen, K.; Alitalo, K.; Detmar, M. VEGF-C–induced lymphangiogenesis in sentinel lymph nodes promotes tumor metastasis to distant sites. Blood 2007, 109, 1010–1017. [Google Scholar] [CrossRef] [PubMed]
- Jennbacken, K.; Vallbo, C.; Wang, W.; Damber, J.E. Expression of vascular endothelial growth factor C (VEGF-C) and VEGF receptor-3 in human prostate cancer is associated with regional lymph node metastasis. Prostate 2005, 65, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Jenny, B.; Harrison, J.; Baetens, D.; Tille, J.C.; Burkhardt, K.; Mottaz, H.; Kiss, J.Z.; Dietrich, P.Y.; De Tribolet, N.; Pizzolato, G. Expression and localization of VEGF-C and VEGFR-3 in glioblastomas and haemangioblastomas. J. Pathol. 2006, 209, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Huynh, H. AZD6244 (ARRY-142886) enhances the antitumor activity of rapamycin in mouse models of human hepatocellular carcinoma. Cancer 2010, 116, 1315–1325. [Google Scholar] [CrossRef] [PubMed]
- Morelli, M.P.; Brown, A.M.; Pitts, T.M.; Tentler, J.J.; Ciardiello, F.; Ryan, A.; Jürgensmeier, J.M.; Eckhardt, S.G. Targeting vascular endothelial growth factor receptor-1 and-3 with cediranib (AZD2171): Effects on migration and invasion of gastrointestinal cancer cell lines. Mol. Cancer Ther. 2009, 8, 2546–2558. [Google Scholar] [CrossRef] [PubMed]
- Svensson, S.; Jirström, K.; Rydén, L.; Roos, G.; Emdin, S.; Ostrowski, M.C.; Landberg, G. ERK phosphorylation is linked to VEGFR2 expression and Ets-2 phosphorylation in breast cancer and is associated with tamoxifen treatment resistance and small tumours with good prognosis. Oncogene 2005, 24, 4370–4379. [Google Scholar] [CrossRef] [PubMed]
- Brand, S.; Dambacher, J.; Beigel, F.; Olszak, T.; Diebold, J.; Otte, J.-M.; Göke, B.; Eichhorst, S.T. CXCR4 and CXCL12 are inversely expressed in colorectal cancer cells and modulate cancer cell migration, invasion and MMP-9 activation. Exp. Cell Res. 2005, 310, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.-B.; Peek, V.; Zhai, Y.; Paul, D.C.; Lou, Q.; Xia, X.; Eessalu, T.; Kohn, W.; Tang, S. Akt Activation, but not Extracellular Signal–Regulated Kinase Activation, Is Required for SDF-1α/CXCR4–Mediated Migration of Epitheloid Carcinoma Cells. Mol. Cancer Res. 2005, 3, 227–236. [Google Scholar] [PubMed]
- Fournier, E.; Birnbaum, D.; Borg, J.-P. Receptors for factors of the VEGF (vascular endothelial growth family). Bull. Cancer 1997, 84, 397–405. [Google Scholar] [PubMed]
- Deckers, M.M.; Karperien, M.; van der Bent, C.; Yamashita, T.; Papapoulos, S.E.; Löwik, C.W. Expression of vascular endothelial growth factors and their receptors during osteoblast differentiation. Endocrinology 2000, 141, 1667–1674. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.-L.; Zhang, X.; Zhang, J.-Y.; Hou, L.; Tian, R.-H. The mechanisms on apoptosis by inhibiting VEGF expression in human breast cancer cells. Int. Immunopharmacol. 2009, 9, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Kodama, M.; Kitadai, Y.; Tanaka, M.; Kuwai, T.; Tanaka, S.; Oue, N.; Yasui, W.; Chayama, K. Vascular endothelial growth factor C stimulates progression of human gastric cancer via both autocrine and paracrine mechanisms. Clin. Cancer Res. 2008, 14, 7205–7214. [Google Scholar] [CrossRef] [PubMed]
- Jüttner, S.; Wiβmann, C.; Jöns, T.; Vieth, M.; Hertel, J.; Gretschel, S.; Schlag, P.M.; Kemmner, W.; Höcker, M. Vascular endothelial growth factor-D and its receptor VEGFR-3: Two novel independent prognostic markers in gastric adenocarcinoma. J. Clin. Oncol. 2006, 24, 228–240. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.C.; Ma, W.H.; Ge, Y.L.; Xue, M.L.; Zhang, Z.; Zhang, J.Y.; Hou, L.; Mu, R.H. RNAi-mediated gene silencing of vascular endothelial growth factor C suppresses growth and induces apoptosis in mouse breast cancer in vitro and in vivo. Oncol. Lett. 2016, 12, 3896–3904. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Hu, J.; Ma, J.; Feng, K.; Zhang, X.; Yang, S.; Wang, W.; Zhang, J.; Zhang, Y. RNAi-mediated silencing of VEGF-C inhibits non-small cell lung cancer progression by simultaneously down-regulating the CXCR4, CCR7, VEGFR-2 and VEGFR-3-dependent axes-induced ERK, p38 and AKT signalling pathways. Eur. J. Cancer 2011, 47, 2353–2363. [Google Scholar] [CrossRef] [PubMed]
- Ellis, L.M. The role of neuropilins in cancer. Mol. Cancer Ther. 2006, 5, 1099–1107. [Google Scholar] [CrossRef] [PubMed]
- Bielenberg, D.R.; Pettaway, C.A.; Takashima, S.; Klagsbrun, M. Neuropilins in neoplasms: Expression, regulation, and function. Exp. Cell Res. 2006, 312, 584–593. [Google Scholar] [CrossRef] [PubMed]
- Kärpänen, T.; Heckman, C.A.; Keskitalo, S.; Jeltsch, M.; Ollila, H.; Neufeld, G.; Tamagnone, L.; Alitalo, K. Functional interaction of VEGF-C and VEGF-D with neuropilin receptors. FASEB J. 2006, 20, 1462–1472. [Google Scholar] [CrossRef] [PubMed]
- Gray, M.J.; Van Buren, G.; Dallas, N.A.; Xia, L.; Wang, X.; Yang, A.D.; Somcio, R.J.; Lin, Y.G.; Lim, S.; Fan, F. Therapeutic targeting of neuropilin-2 on colorectal carcinoma cells implanted in the murine liver. J. Natl. Cancer Inst. 2008, 100, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, N.; Hillan, K.J.; Gerber, H.-P.; Novotny, W. Discovery and development of bevacizumab, an anti-VEGF antibody for treating cancer. Nat. Rev. Drug Discov. 2004, 3, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Elbashir, S.M.; Harborth, J.; Weber, K.; Tuschl, T. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods 2002, 26, 199–213. [Google Scholar] [CrossRef]
- Paddison, P.; Hannon, G. siRNAs and shRNAs: Skeleton keys to the human genome. Curr. Opin. Mol. Ther. 2003, 5, 217–224. [Google Scholar] [PubMed]
- Carpenter, A.E.; Sabatini, D.M. Systematic genome-wide screens of gene function. Nat. Rev. Genet. 2004, 5, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Ganju, P.; Hall, J. Potential applications of siRNA for pain therapy. Expert Opin. Biol. Ther. 2004, 4, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Woodle, M.; Scaria, P.; Ganesh, S.; Subramanian, K.; Titmas, R.; Cheng, C.; Yang, J.; Pan, Y.; Weng, K.; Gu, C. Sterically stabilized polyplex: Ligand-mediated activity. J. Control. Release 2001, 74, 309–311. [Google Scholar] [CrossRef]
- Langer, R. Drugs on target. Science 2001, 293, 58–59. [Google Scholar] [CrossRef] [PubMed]
- Suh, W.; Han, S.-O.; Yu, L.; Kim, S.W. An angiogenic, endothelial-cell-targeted polymeric gene carrier. Mol. Ther. 2002, 6, 664–672. [Google Scholar] [CrossRef]
- Verbaan, F.; Oussoren, C.; Snel, C.; Crommelin, D.; Hennink, W.; Storm, G. Steric stabilization of poly (2-(dimethylamino) ethyl methacrylate)-based polyplexes mediates prolonged circulation and tumor targeting in mice. J. Gene Med. 2004, 6, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Ogris, M.; Walker, G.; Blessing, T.; Kircheis, R.; Wolschek, M.; Wagner, E. Tumor-targeted gene therapy: Strategies for the preparation of ligand–polyethylene glycol–polyethylenimine/DNA complexes. J. Control. Release 2003, 91, 173–181. [Google Scholar] [CrossRef]
- Wagner, E.; Kircheis, R.; Walker, G.F. Targeted nucleic acid delivery into tumors: New avenues for cancer therapy. Biomed. Pharmacother. 2004, 58, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Hart, S.L.; Knight, A.M.; Harbottle, R.P.; Mistry, A.; Hunger, H.; Cutler, D.F.; Williamson, R.; Coutelle, C. Cell binding and internalization by filamentous phage displaying a cyclic Arg-Gly-Asp-containing peptide. J. Biol. Chem. 1994, 269, 12468–12474. [Google Scholar] [PubMed]
- Janssen, M.L.; Oyen, W.J.; Dijkgraaf, I.; Massuger, L.F.; Frielink, C.; Edwards, D.S.; Rajopadhye, M.; Boonstra, H.; Corstens, F.H.; Boerman, O.C. Tumor targeting with radiolabeled αvβ3 integrin binding peptides in a nude mouse model. Cancer Res. 2002, 62, 6146–6151. [Google Scholar] [PubMed]
- Zitzmann, S.; Ehemann, V.; Schwab, M. Arginine-glycine-aspartic acid (RGD)-peptide binds to both tumor and tumor-endothelial cells in vivo. Cancer Res. 2002, 62, 5139–5143. [Google Scholar] [PubMed]
- Voldborg, B.R.; Damstrup, L.; Spang-Thomsen, M.; Poulsen, H.S. Epidermal growth factor receptor (EGFR) and EGFR mutations, function and possible role in clinical trials. Ann. Oncol. 1997, 8, 1197–1206. [Google Scholar] [CrossRef] [PubMed]
- Slamon, D.J.; Clark, G.M.; Wong, S.G.; Levin, W.J.; Ullrich, A.; McGuire, W.L. Human breast cancer: Correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987, 235, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, E. pH-sensitive nano-crystals of carbonate apatite for smart and cell-specific transgene delivery. Expert Opin. Drug Deliv. 2007, 4, 193–196. [Google Scholar] [CrossRef] [PubMed]
- Tiash, S.; Kamaruzman, N.I.B.; Chowdhury, E.H. Carbonate apatite nanoparticles carry siRNA (s) targeting growth factor receptor genes EGFR1 and ErbB2 to regress mouse breast tumor. Drug Deliv. 2017, 24, 1721–1730. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Fang, F.; Ludewig, G.; Iones, G.; Jones, D. A mutation found in the promoter region of the human survivin gene is correlated to overexpression of survivin in cancer cells. DNA Cell Biol. 2004, 23, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Wheatley, S.P.; McNeish, I.A. Survivin: A protein with dual roles in mitosis and apoptosis. Int. Rev. Cytol. 2005, 247, 35–88. [Google Scholar] [CrossRef]
- Altieri, D.C. Survivin, cancer networks and pathway-directed drug discovery. Nat. Rev. Cancer 2008, 8, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Hunter, A.M.; LaCasse, E.C.; Korneluk, R.G. The inhibitors of apoptosis (IAPs) as cancer targets. Apoptosis 2007, 12, 1543–1568. [Google Scholar] [CrossRef] [PubMed]
- Schimmer, A.D. Inhibitor of apoptosis proteins: Translating basic knowledge into clinical practice. Cancer Res. 2004, 64, 7183–7190. [Google Scholar] [CrossRef] [PubMed]
- Altieri, D.C. Targeted therapy by disabling crossroad signaling networks: The survivin paradigm. Mol. Cancer Ther. 2006, 5, 478–482. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, S.; Pelus, L.M. Survivin, a cancer target with an emerging role in normal adult tissues. Mol. Cancer Ther. 2006, 5, 1087–1098. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.; Liu, T.; Cotter, M.A.; Florell, S.R.; Robinette, K.; Hanks, A.N.; Grossman, D. Melanocyte expression of survivin promotes development and metastasis of UV-induced melanoma in HGF-transgenic mice. Cancer Res. 2007, 67, 5172–5178. [Google Scholar] [CrossRef] [PubMed]
- Mehrotra, S.; Languino, L.R.; Raskett, C.M.; Mercurio, A.M.; Dohi, T.; Altieri, D.C. IAP regulation of metastasis. Cancer Cell 2010, 17, 53–64. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, J.A.; Liu, T.; Goodson, A.; Grossman, D. Survivin enhances motility of melanoma cells by supporting Akt activation and α5 integrin upregulation. Cancer Res. 2010, 194. [Google Scholar] [CrossRef]
- Kawasaki, H.; Altieri, D.C.; Lu, C.-D.; Toyoda, M.; Tenjo, T.; Tanigawa, N. Inhibition of apoptosis by survivin predicts shorter survival rates in colorectal cancer. Cancer Res. 1998, 58, 5071–5074. [Google Scholar] [PubMed]
- Tanaka, K.; Iwamoto, S.; Gon, G.; Nohara, T.; Iwamoto, M.; Tanigawa, N. Expression of survivin and its relationship to loss of apoptosis in breast carcinomas. Clin. Cancer Res. 2000, 6, 127–134. [Google Scholar] [PubMed]
- Zaffaroni, N.; Pennati, M.; Colella, G.; Perego, P.; Supino, R.; Gatti, L.; Pilotti, S.; Zunino, F.; Daidone, M. Expression of the anti-apoptotic gene survivin correlates with taxol resistance in human ovarian cancer. Cell. Mol. Life Sci. 2002, 59, 1406–1412. [Google Scholar] [CrossRef] [PubMed]
- Tran, J.; Master, Z.; Joanne, L.Y.; Rak, J.; Dumont, D.J.; Kerbel, R.S. A role for survivin in chemoresistance of endothelial cells mediated by VEGF. Proc. Natl. Acad. Sci. USA 2002, 99, 4349–4354. [Google Scholar] [CrossRef] [PubMed]
- Cong, X.L.; Han, Z.C. Survivin and leukemia. Int. J. Hematol. 2004, 80, 232–238. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.; Sun, W.; Kissel, T. Chitosan-based formulations for delivery of DNA and siRNA. Adv. Drug Deliv. Rev. 2010, 62, 12–27. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Jeong, E.J.; Lee, J.; Rhim, T.; Lee, S.K.; Lee, K.Y. Preparation and characterization of nonaarginine-modified chitosan nanoparticles for siRNA delivery. Carbohydr. Polym. 2013, 92, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Sharma, K.; Somavarapu, S.; Colombani, A.; Govind, N.; Taylor, K.M. Nebulised siRNA encapsulated crosslinked chitosan nanoparticles for pulmonary delivery. Int. J. Pharm. 2013, 455, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.W.; Yun, K.-S.; Ban, H.-S.; Choe, W.; Lee, S.K.; Lee, K.Y. Preparation and characterization of chitosan/polyguluronate nanoparticles for siRNA delivery. J. Control. Release 2009, 139, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Guţoaia, A.; Schuster, L.; Margutti, S.; Laufer, S.; Schlosshauer, B.; Krastev, R.; Stoll, D.; Hartmann, H. Fine-tuned PEGylation of chitosan to maintain optimal siRNA-nanoplex bioactivity. Carbohydr. Polym. 2016, 143, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.N.; Xie, H.G.; Yu, W.T.; Liu, X.D.; Xie, W.Y.; Zhu, J.; Ma, X.J. Chitosan-g-MPEG-modified alginate/chitosan hydrogel microcapsules: A quantitative study of the effect of polymer architecture on the resistance to protein adsorption. Langmuir 2010, 26, 17156–17164. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Huang, W.; Jin, M.; Wang, Q.; Fan, B.; Kang, L.; Gao, Z. Chitosan-based nanoparticles for survivin targeted siRNA delivery in breast tumor therapy and preventing its metastasis. Int. J. Nanomed. 2016, 11, 4931. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.; Lins, L.; Divita, G.; Brasseur, R. Realistic modeling approaches of structure–function properties of CPPs in non-covalent complexes. Biochim. Biophys. Acta Biomembr. 2010, 1798, 2217–2222. [Google Scholar] [CrossRef] [PubMed]
- Veiman, K.-L.; Künnapuu, K.; Lehto, T.; Kiisholts, K.; Pärn, K.; Langel, Ü.; Kurrikoff, K. PEG shielded MMP sensitive CPPs for efficient and tumor specific gene delivery in vivo. J. Control. Release 2015, 209, 238–247. [Google Scholar] [CrossRef] [PubMed]
- Bass, J. CPP magnetoresistance of magnetic multilayers: A critical review. J. Magn. Magn. Mater. 2016, 408, 244–320. [Google Scholar] [CrossRef]
- Ronca, F.; Raggi, A. Structure-function relationships in mammalian histidine-proline-rich glycoprotein. Biochimie 2015, 118, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Chou, S.-T.; Hom, K.; Zhang, D.; Leng, Q.; Tricoli, L.J.; Hustedt, J.M.; Lee, A.; Shapiro, M.J.; Seog, J.; Kahn, J.D. Enhanced silencing and stabilization of siRNA polyplexes by histidine-mediated hydrogen bonds. Biomaterials 2014, 35, 846–855. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Guo, Z.; Du, Z.; Fang, R.; Wu, H.; Zeng, X.; Wang, C.; Feng, M.; Pan, S. Serum tolerance and endosomal escape capacity of histidine-modified pDNA-loaded complexes based on polyamidoamine dendrimer derivatives. Biomaterials 2012, 33, 8111–8121. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.R.; Huang, Y.-W.; Winiarz, J.G.; Chiang, H.-J.; Lee, H.-J. Intracellular delivery of quantum dots mediated by a histidine-and arginine-rich HR9 cell-penetrating peptide through the direct membrane translocation mechanism. Biomaterials 2011, 32, 3520–3537. [Google Scholar] [CrossRef] [PubMed]
- Moreira, C.; Oliveira, H.; Pires, L.R.; Simões, S.; Barbosa, M.A.; Pêgo, A.P. Improving chitosan-mediated gene transfer by the introduction of intracellular buffering moieties into the chitosan backbone. Acta Biomater. 2009, 5, 2995–3006. [Google Scholar] [CrossRef] [PubMed]
- Corbet, C.; Ragelle, H.; Pourcelle, V.; Vanvarenberg, K.; Marchand-Brynaert, J.; Préat, V.; Feron, O. Delivery of siRNA targeting tumor metabolism using non-covalent PEGylated chitosan nanoparticles: Identification of an optimal combination of ligand structure, linker and grafting method. J. Control. Release 2016, 223, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Huang, W.; Li, Y.; Liu, S.; Jin, M.; Wang, Y.; Jia, L.; Gao, Z. Anti-tumor effects in mice induced by survivin-targeted siRNA delivered through polysaccharide nanoparticles. Biomaterials 2013, 34, 5689–5699. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Huang, W.; Kang, L.; Jin, M.; Fan, B.; Jin, H.; Wang, Q.-M.; Gao, Z. siRNA-loaded poly (histidine-arginine) 6-modified chitosan nanoparticle with enhanced cell-penetrating and endosomal escape capacities for suppressing breast tumor metastasis. Int. J. Nanomed. 2017, 12, 3221. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.X.; Bos, P.D.; Massagué, J. Metastasis: From dissemination to organ-specific colonization. Nat. Rev. Cancer 2009, 9, 274–284. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Fernando, H.C. Surgical and nonresectional therapies for pulmonary metastasis. Surg. Clin. N. Am. 2010, 90, 1041–1051. [Google Scholar] [CrossRef] [PubMed]
- Soengas, M.S.; Lowe, S.W. Apoptosis and melanoma chemoresistance. Oncogene 2003, 22, 3138–3151. [Google Scholar] [CrossRef] [PubMed]
- Bolcato-Bellemin, A.-L.; Bonnet, M.-E.; Creusat, G.; Erbacher, P.; Behr, J.-P. Sticky overhangs enhance siRNA-mediated gene silencing. Proc. Natl. Acad. Sci. USA 2007, 104, 16050–16055. [Google Scholar] [CrossRef] [PubMed]
- Kedinger, V.; Meulle, A.; Zounib, O.; Bonnet, M.-E.; Gossart, J.-B.; Benoit, E.; Messmer, M.; Shankaranarayanan, P.; Behr, J.-P.; Erbacher, P. Sticky siRNAs targeting survivin and cyclin B1 exert an antitumoral effect on melanoma subcutaneous xenografts and lung metastases. BMC Cancer 2013, 13, 338. [Google Scholar] [CrossRef] [PubMed]
- Parker, B.; Sukumar, S. Distant metastasis in breast cancer: Molecular mechanisms and therapeutic targets. Cancer Biol. Ther. 2003, 2, 13–22. [Google Scholar] [CrossRef]
- Matsumoto, A.; Cabral, H.; Sato, N.; Kataoka, K.; Miyahara, Y. Assessment of Tumor Metastasis by the Direct Determination of Cell-Membrane Sialic Acid Expression. Angew. Chem. Int. Ed. 2010, 49, 5494–5497. [Google Scholar] [CrossRef] [PubMed]
- Büll, C.; Boltje, T.J.; van Dinther, E.A.; Peters, T.; de Graaf, A.M.; Leusen, J.H.; Kreutz, M.; Figdor, C.G.; den Brok, M.H.; Adema, G.J. Targeted delivery of a sialic acid-blocking glycomimetic to cancer cells inhibits metastatic spread. ACS Nano 2015, 9, 733–745. [Google Scholar] [CrossRef] [PubMed]
- Fan, B.; Kang, L.; Chen, L.; Sun, P.; Jin, M.; Wang, Q.; Bae, Y.H.; Huang, W.; Gao, Z. Systemic siRNA delivery with a dual pH-responsive and tumor-targeted nanovector for inhibiting tumor growth and spontaneous metastasis in orthotopic murine model of breast carcinoma. Theranostics 2017, 7, 357. [Google Scholar] [CrossRef] [PubMed]
- Morgan, D.O. Cyclin-dependent kinases: Engines, clocks, and microprocessors. Ann. Rev. Cell Dev. Biol. 1997, 13, 261–291. [Google Scholar] [CrossRef] [PubMed]
- Krek, W.; Nigg, E. Differential phosphorylation of vertebrate p34cdc2 kinase at the G1/S and G2/M transitions of the cell cycle: Identification of major phosphorylation sites. EMBO J. 1991, 10, 305. [Google Scholar] [PubMed]
- Castedo, M.; Perfettini, J.; Roumier, T.; Kroemer, G. Cyclin-dependent kinase-1: Linking apoptosis to cell cycle and mitotic catastrophe. Cell Death Differ. 2002, 9, 1287. [Google Scholar] [CrossRef] [PubMed]
- Jin, P.; Hardy, S.; Morgan, D.O. Nuclear localization of cyclin B1 controls mitotic entry after DNA damage. J. Cell Biol. 1998, 141, 875–885. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Chae, H.-D.; Yun, J.; Jung, M.; Kim, Y.-S.; Kim, S.-H.; Han, M.H.; Shin, D.Y. Constitutive activation of cyclin B1-associated cdc2 kinase overrides p53-mediated G2-M arrest. Cancer Res. 2000, 60, 542–545. [Google Scholar] [PubMed]
- Yin, X.-Y.; Grove, L.; Datta, N.S.; Katula, K.; Long, M.W.; Prochownik, E.V. Inverse regulation of cyclin B1 by c-Myc and p53 and induction of tetraploidy by cyclin B1 overexpression. Cancer Res. 2001, 61, 6487–6493. [Google Scholar] [PubMed]
- Santana, C.; Ortega, E.; García-Carrancá, A. Oncogenic H-ras induces cyclin B1 expression in a p53-independent manner. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2002, 508, 49–58. [Google Scholar] [CrossRef]
- Sarafan-Vasseur, N.; Lamy, A.; Bourguignon, J.; Le Pessot, F.; Hieter, P.; Sesboue, R.; Bastard, C.; Frebourg, T.; Flaman, J.-M. Overexpression of B-type cyclins alters chromosomal segregation. Oncogene 2002, 21, 2051–2057. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Sui, L.; Watanabe, Y.; Sugimoto, K.; Tokuda, M. Clinical relevance of cyclin B1 overexpression in laryngeal squamous cell carcinoma. Cancer Lett. 2002, 177, 13–19. [Google Scholar] [CrossRef]
- Hassan, K.A.; Ang, K.K.; El-Naggar, A.K.; Story, M.D.; Lee, J.I.; Liu, D.; Hong, W.K.; Mao, L. Cyclin B1 overexpression and resistance to radiotherapy in head and neck squamous cell carcinoma. Cancer Res. 2002, 62, 6414–6417. [Google Scholar] [PubMed]
- Takeno, S.; Noguchi, T.; Kikuchi, R.; Uchida, Y.; Yokoyama, S.; Müller, W. Prognostic value of cyclin B1 in patients with esophageal squamous cell carcinoma. Cancer 2002, 94, 2874–2881. [Google Scholar] [CrossRef] [PubMed]
- Goga, A.; Yang, D.; Tward, A.D.; Morgan, D.O.; Bishop, J.M. Inhibition of CDK1 as a potential therapy for tumors over-expressing MYC. Nat. Med. 2007, 13, 820–827. [Google Scholar] [CrossRef] [PubMed]
- Morris, M.C.; Gros, E.; Aldrian-Herrada, G.; Choob, M.; Archdeacon, J.; Heitz, F.; Divita, G. A non-covalent peptide-based carrier for in vivo delivery of DNA mimics. Nucl. Acids Res. 2007, 35, e49. [Google Scholar] [CrossRef] [PubMed]
- Simeoni, F.; Morris, M.C.; Heitz, F.; Divita, G. Insight into the mechanism of the peptide-based gene delivery system MPG: Implications for delivery of siRNA into mammalian cells. Nucl. Acids Res. 2003, 31, 2717–2724. [Google Scholar] [CrossRef] [PubMed]
- Morris, K.V.; Chan, S.W.-L.; Jacobsen, S.E.; Looney, D.J. Small interfering RNA-induced transcriptional gene silencing in human cells. Science 2004, 305, 1289–1292. [Google Scholar] [CrossRef] [PubMed]
- Zeineddine, D.; Papadimou, E.; Chebli, K.; Gineste, M.; Liu, J.; Grey, C.; Thurig, S.; Behfar, A.; Wallace, V.A.; Skerjanc, I.S. Oct-3/4 dose dependently regulates specification of embryonic stem cells toward a cardiac lineage and early heart development. Dev. Cell 2006, 11, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Morris, M.C.; Depollier, J.; Mery, J.; Heitz, F.; Divita, G. A peptide carrier for the delivery of biologically active proteins into mammalian cells. Nat. Biotechnol. 2001, 19, 1173–1176. [Google Scholar] [CrossRef] [PubMed]
- Crombez, L.; Morris, M.C.; Deshayes, S.; Heitz, F.; Divita, G. Peptide-based nanoparticle for ex vivo and in vivo dug delivery. Curr. Pharm. Des. 2008, 14, 3656–3665. [Google Scholar] [CrossRef] [PubMed]
- Zorko, M.; Langel, Ü. Cell-penetrating peptides: Mechanism and kinetics of cargo delivery. Adv. Drug Deliv. Rev. 2005, 57, 529–545. [Google Scholar] [CrossRef] [PubMed]
- Deshayes, S.; Morris, M.; Divita, G.; Heitz, F. Cell-penetrating peptides: Tools for intracellular delivery of therapeutics. Cell. Mol. Life Sci. 2005, 62, 1839–1849. [Google Scholar] [CrossRef] [PubMed]
- Crombez, L.; Morris, M.C.; Dufort, S.; Aldrian-Herrada, G.; Nguyen, Q.; Mc Master, G.; Coll, J.-L.; Heitz, F.; Divita, G. Targeting cyclin B1 through peptide-based delivery of siRNA prevents tumour growth. Nucl. Acids Res. 2009, 37, 4559–4569. [Google Scholar] [CrossRef] [PubMed]
- Aznar, S.; Lacal, J.C. Rho signals to cell growth and apoptosis. Cancer Lett. 2001, 165, 1–10. [Google Scholar] [CrossRef]
- Schmitz, A.A.; Govek, E.-E.; Böttner, B.; Van Aelst, L. Rho GTPases: Signaling, migration, and invasion. Exp. Cell Res. 2000, 261, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Fritz, G.; Just, I.; Kaina, B. Rho GTPases are over-expressed in human tumors. Int. J. Cancer 1999, 81, 682–687. [Google Scholar] [CrossRef]
- Ridley, A.J. Rho GTPases and cell migration. J. Cell Sci. 2001, 114, 2713–2722. [Google Scholar] [PubMed]
- Heasman, S.J.; Ridley, A.J. Mammalian Rho GTPases: New insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 2008, 9, 690–701. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; DiVito, M.M.; Merajver, S.D.; Boyanapalli, M.; Van Golen, K.L. Regulation of pancreatic cancer cell migration and invasion by RhoC GTPase and caveolin-1. Mol. Cancer 2005, 4, 21. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, I.P.; Zohn, I.E.; Der, C.J. Rho GTPase-dependent transformation by G protein-coupled receptors. Oncogene 2001, 20, 1547. [Google Scholar] [CrossRef] [PubMed]
- Gur, S.; Kadowitz, P.J.; Hellstrom, W.J. RhoA/Rho-kinase as a therapeutic target for the male urogenital tract. J. Sex. Med. 2011, 8, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Rathinam, R.; Berrier, A.; Alahari, S.K. Role of Rho GTPases and their regulators in cancer progression. Front. Biosci. 2011, 16, 2561–2571. [Google Scholar] [CrossRef]
- Kwiatkowska, A.; Symons, M. Signaling determinants of glioma cell invasion. In Glioma Signaling; Springer: Berlin, Germany, 2013; pp. 121–141. [Google Scholar]
- Oh, H.K.; Sin, J.-I.; Choi, J.; Park, S.H.; Lee, T.S.; Choi, Y.S. Overexpression of CD73 in epithelial ovarian carcinoma is associated with better prognosis, lower stage, better differentiation and lower regulatory T cell infiltration. J. Gynecol. Oncol. 2012, 23, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Denoyelle, C.; Albanese, P.; Uzan, G.; Hong, L.; Vannier, J.-P.; Soria, J.; Soria, C. Molecular mechanism of the anti-cancer activity of cerivastatin, an inhibitor of HMG-CoA reductase, on aggressive human breast cancer cells. Cell. Signal. 2003, 15, 327–338. [Google Scholar] [CrossRef]
- Van Golen, K.L.; Wu, Z.-F.; Qiao, X.T.; Bao, L.W.; Merajver, S.D. RhoC GTPase, a novel transforming oncogene for human mammary epithelial cells that partially recapitulates the inflammatory breast cancer phenotype. Cancer Res. 2000, 60, 5832–5838. [Google Scholar] [PubMed]
- Ma, L.; Teruya-Feldstein, J.; Weinberg, R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 2007, 449, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Jemal, A.; Siegel, R.; Ward, E.; Murray, T.; Xu, J.; Thun, M.J. Cancer statistics, 2007. CA Cancer J. Clin. 2007, 57, 43–66. [Google Scholar] [CrossRef] [PubMed]
- Jemal, A.; Siegel, R.; Ward, E.; Hao, Y.; Xu, J.; Murray, T.; Thun, M.J. Cancer statistics, 2008. CA Cancer J. Clin. 2008, 58, 71–96. [Google Scholar] [CrossRef] [PubMed]
- Parkin, D.M.; Bray, F.; Ferlay, J.; Pisani, P. Global cancer statistics, 2002. CA Cancer J. Clin. 2005, 55, 74–108. [Google Scholar] [CrossRef] [PubMed]
- Wilke, H.-J.; Van Cutsem, E. Current treatments and future perspectives in colorectal and gastric cancer. Ann. Oncol. 2003, 14, ii49–ii55. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhao, G.; Liu, X.; Sui, A.; Yang, K.; Yao, R.; Wang, Z.; Shi, Q. Silencing of RhoA and RhoC expression by RNA interference suppresses human colorectal carcinoma growth in vivo. J. Exp. Clin. Cancer Res. 2010, 29, 123. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, J.-R.; Pottier, M.; Vekris, A.; Opolon, P.; Maksimenko, A.; Malvy, C. Comparison of antisense oligonucleotides and siRNAs in cell culture and in vivo. Biochem. Biophys. Res. Commun. 2002, 296, 1000–1004. [Google Scholar] [CrossRef]
- Pillé, J.-Y.; Denoyelle, C.; Varet, J.; Bertrand, J.-R.; Soria, J.; Opolon, P.; Lu, H.; Pritchard, L.-L.; Vannier, J.-P.; Malvy, C. Anti-RhoA and anti-RhoC siRNAs inhibit the proliferation and invasiveness of MDA-MB-231 breast cancer cells in vitro and in vivo. Mol. Ther. 2005, 11, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Pillé, J.-Y.; Li, H.; Blot, E.; Bertrand, J.-R.; Pritchard, L.-L.; Opolon, P.; Maksimenko, A.; Lu, H.; Vannier, J.-P.; Soria, J. Intravenous delivery of anti-RhoA small interfering RNA loaded in nanoparticles of chitosan in mice: Safety and efficacy in xenografted aggressive breast cancer. Hum. Gene Ther. 2006, 17, 1019–1026. [Google Scholar] [CrossRef] [PubMed]
- Suwa, H.; Ohshio, G.; Imamura, T.; Watanabe, G.; Arii, S.; Imamura, M.; Narumiya, S.; Hiai, H.; Fukumoto, M. Overexpression of the RhoC gene correlates with progression of ductal adenocarcinoma of the pancreas. Br. J. Cancer 1998, 77, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Zhu, M.; Lv, G.; Zhang, Q.; Wang, G. The role of RhoC in the proliferation and apoptosis of hepatocellular carcinoma cells. Med. Oncol. 2012, 29, 1802–1809. [Google Scholar] [CrossRef] [PubMed]
- Robertson, F.M.; Bondy, M.; Yang, W.; Yamauchi, H.; Wiggins, S.; Kamrudin, S.; Krishnamurthy, S.; Le-Petross, H.; Bidaut, L.; Player, A.N. Inflammatory breast cancer: The disease, the biology, the treatment. CA Cancer J. Clin. 2010, 60, 351–375. [Google Scholar] [CrossRef] [PubMed]
- Anderson, W.F.; Schairer, C.; Chen, B.E.; Hance, K.W.; Levine, P.H. Epidemiology of inflammatory breast cancer (IBC) 1. Breast Dis. 2006, 22, 9–23. [Google Scholar] [CrossRef]
- Van Golen, K.L.; Bao, L.W.; Pan, Q.; Miller, F.R.; Wu, Z.F.; Merajver, S.D. Mitogen activated protein kinase pathway is involved in RhoC GTPase induced motility, invasion and angiogenesis in inflammatory breast cancer. Clin. Exp. Metastasis 2002, 19, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.-D.; Shen, H.-B.; Zhu, L.; Lu, J.-Q.; Zhang, L.; Luo, Z.-Y.; Wu, Y.-Q. Anti-rhoc sirnas inhibit the proliferation and invasiveness of breast cancer cells via modulating the Kai1, MMP9, and CXCR4 expression. OncoTargets Ther. 2017, 10, 1827. [Google Scholar] [CrossRef] [PubMed]
- Kraus, C.; Liehr, T.; Hülsken, J.; Behrens, J.; Birchmeier, W.; Grzeschik, K.-H.; Ballhausen, W.G. Localization of the human β-catenin gene (CTNNB1) to 3p21: A region implicated in tumor development. Genomics 1994, 23, 272–274. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, B.T.; Tamai, K.; He, X. Wnt/β-catenin signaling: Components, mechanisms, and diseases. Dev. Cell 2009, 17, 9–26. [Google Scholar] [CrossRef] [PubMed]
- Peifer, M.; Rauskolb, C.; Williams, M.; Riggleman, B.; Wieschaus, E. The segment polarity gene armadillo interacts with the wingless signaling pathway in both embryonic and adult pattern formation. Development 1991, 111, 1029–1043. [Google Scholar] [PubMed]
- Noordermeer, J.; Klingensmith, J.; Perrimonl, N. Dishevelled and armadillo act in the Wingless signalling pathway in Drosophila. Nature 1994, 367, 80–83. [Google Scholar] [CrossRef] [PubMed]
- Peifer, M.; Berg, S.; Reynolds, A.B. A repeating amino acid motif shared by proteins with diverse cellular roles. Cell 1994, 76, 789–791. [Google Scholar] [CrossRef]
- Bilić, J.; Huang, Y.-L.; Davidson, G.; Zimmermann, T.; Cruciat, C.-M.; Bienz, M.; Niehrs, C. Wnt induces LRP6 signalosomes and promotes dishevelled-dependent LRP6 phosphorylation. Science 2007, 316, 1619–1622. [Google Scholar] [CrossRef] [PubMed]
- Schwarz-Romond, T.; Fiedler, M.; Shibata, N.; Butler, P.J.G.; Kikuchi, A.; Higuchi, Y.; Bienz, M. The DIX domain of Dishevelled confers Wnt signaling by dynamic polymerization. Nat. Struct. Mol. Biol. 2007, 14, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Behrens, J.; von Kries, J.P.; Kühl, M.; Bruhn, L.; Wedlich, D.; Grosschedl, R.; Birchmeier, W. Functional interaction of β-catenin with the transcription factor LEF-1. Nature 1996, 382, 638–642. [Google Scholar] [CrossRef] [PubMed]
- Molenaar, M.; van de Wetering, M.; Oosterwegel, M.; Peterson-Maduro, J.; Godsave, S.; Korinek, V.; Roose, J.; Destrée, O.; Clevers, H. XTcf-3 transcription factor mediates β-catenin-induced axis formation in Xenopus embryos. Cell 1996, 86, 391–399. [Google Scholar] [CrossRef]
- Grigoryan, T.; Wend, P.; Klaus, A.; Birchmeier, W. Deciphering the function of canonical Wnt signals in development and disease: Conditional loss-and gain-of-function mutations of β-catenin in mice. Genes Dev. 2008, 22, 2308–2341. [Google Scholar] [CrossRef] [PubMed]
- Hajra, K.M.; Fearon, E.R. Cadherin and catenin alterations in human cancer. Genes Chromosom. Cancer 2002, 34, 255–268. [Google Scholar] [CrossRef] [PubMed]
- Ilyas, M.; Tomlinson, I.; Rowan, A.; Pignatelli, M.; Bodmer, W. β-Catenin mutations in cell lines established from human colorectal cancers. Proc. Natl. Acad. Sci. USA 1997, 94, 10330–10334. [Google Scholar] [CrossRef] [PubMed]
- Morin, P.J.; Sparks, A.B.; Korinek, V.; Barker, N.; Clevers, H.; Vogelstein, B.; Kinzler, K.W. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science 1997, 275, 1787–1790. [Google Scholar] [CrossRef] [PubMed]
- Polakis, P. Wnt signaling and cancer. Genes Dev. 2000, 14, 1837–1851. [Google Scholar] [CrossRef] [PubMed]
- Sparks, A.B.; Morin, P.J.; Vogelstein, B.; Kinzler, K.W. Mutational analysis of the APC/β-catenin/Tcf pathway in colorectal cancer. Cancer Res. 1998, 58, 1130–1134. [Google Scholar] [PubMed]
- Su, L.-K.; Vogelstein, B.; Kinzler, K.W. Association of the APC tumor suppressor protein with catenins. Science 1993, 262, 1734–1738. [Google Scholar] [CrossRef] [PubMed]
- Korinek, V.; Barker, N.; Morin, P.J.; Van Wichen, D.; De Weger, R.; Kinzler, K.W.; Vogelstein, B.; Clevers, H. Constitutive transcriptional activation by a β-catenin-Tcf complex in APC−/− colon carcinoma. Science 1997, 275, 1784–1787. [Google Scholar] [CrossRef] [PubMed]
- Bass, B.L. Double-stranded RNA as a template for gene silencing. Cell 2000, 101, 235–238. [Google Scholar] [CrossRef]
- Verma, U.N.; Surabhi, R.M.; Schmaltieg, A.; Becerra, C.; Gaynor, R.B. Small interfering RNAs directed against β-catenin inhibit the in vitro and in vivo growth of colon cancer cells. Clin. Cancer Res. 2003, 9, 1291–1300. [Google Scholar] [PubMed]
- Barker, N.; Ridgway, R.A.; van Es, J.H.; van de Wetering, M.; Begthel, H.; van den Born, M.; Danenberg, E.; Clarke, A.R.; Sansom, O.J.; Clevers, H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 2009, 457, 608–611. [Google Scholar] [CrossRef] [PubMed]
- Barker, N.; Van Es, J.H.; Kuipers, J.; Kujala, P.; Van Den Born, M.; Cozijnsen, M.; Haegebarth, A.; Korving, J.; Begthel, H.; Peters, P.J. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007, 449, 1003–1007. [Google Scholar] [CrossRef] [PubMed]
- Rishi, A.K.; Zhang, L.; Yu, Y.; Jiang, Y.; Nautiyal, J.; Wali, A.; Fontana, J.A.; Levi, E.; Majumdar, A.P. Cell cycle-and apoptosis-regulatory protein-1 is involved in apoptosis signaling by epidermal growth factor receptor. J. Biol. Chem. 2006, 281, 13188–13198. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Yang, C.K.; Heo, K.; Roeder, R.G.; An, W.; Stallcup, M.R. CCAR1, a key regulator of mediator complex recruitment to nuclear receptor transcription complexes. Mol. Cell 2008, 31, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Ou, C.-Y.; Kim, J.H.; Yang, C.K.; Stallcup, M.R. Requirement of cell cycle and apoptosis regulator 1 for target gene activation by Wnt and β-catenin and for anchorage-independent growth of human colon carcinoma cells. J. Biol. Chem. 2009, 284, 20629–20637. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.-S.; Wei, K.-L.; Lu, C.-K.; Chen, Y.-H.; Cheng, Y.-T.; Tung, S.-Y.; Wu, C.-S.; Chiang, M.-K. Inhibition of CCAR1, a Coactivator of β-Catenin, Suppresses the Proliferation and Migration of Gastric Cancer Cells. Int. J. Mol. Sci. 2017, 18, 460. [Google Scholar] [CrossRef] [PubMed]
- Amit, I.; Wides, R.; Yarden, Y. Evolvable signaling networks of receptor tyrosine kinases: Relevance of robustness to malignancy and to cancer therapy. Mol. Syst. Biol. 2007, 3, 151. [Google Scholar] [CrossRef] [PubMed]
- Zelinski, D.P.; Zantek, N.D.; Stewart, J.C.; Irizarry, A.R.; Kinch, M.S. EphA2 overexpression causes tumorigenesis of mammary epithelial cells. Cancer Res. 2001, 61, 2301–2306. [Google Scholar] [PubMed]
- Kullander, K.; Klein, R. Mechanisms and functions of Eph and ephrin signalling. Nat. Rev. Mol. Cell Biol. 2002, 3, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Hafner, C.; Schmitz, G.; Meyer, S.; Bataille, F.; Hau, P.; Langmann, T.; Dietmaier, W.; Landthaler, M.; Vogt, T. Differential gene expression of Eph receptors and ephrins in benign human tissues and cancers. Clin. Chem. 2004, 50, 490–499. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-F.; Fokas, E.; Bieker, M.; Rose, F.; Rexin, P.; Zhu, Y.; Pagenstecher, A.; Engenhart-Cabillic, R.; An, H.-X. Increased expression of EphA2 correlates with adverse outcome in primary and recurrent glioblastoma multiforme patients. Oncol. Rep. 2008, 19, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Brannan, J.M.; Dong, W.; Prudkin, L.; Behrens, C.; Lotan, R.; Bekele, B.N.; Wistuba, I.; Johnson, F.M. Expression of the receptor tyrosine kinase EphA2 is increased in smokers and predicts poor survival in non–small cell lung cancer. Clin. Cancer Res. 2009, 15, 4423–4430. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.D.; Lee, M.J.; Yu, G.R.; Kim, I.H.; Yu, H.C.; Song, E.Y.; Kim, D.G. EFNA1 ligand and its receptor EphA2: Potential biomarkers for hepatocellular carcinoma. Int. J. Cancer 2010, 126, 940–949. [Google Scholar] [CrossRef] [PubMed]
- Baeten, C.I.; Hillen, F.; Pauwels, P.; de Bruine, A.P.; Baeten, C.G. Prognostic role of vasculogenic mimicry in colorectal cancer. Dis. Colon Rectum 2009, 52, 2028–2035. [Google Scholar] [CrossRef] [PubMed]
- Merritt, W.M.; Kamat, A.A.; Hwang, J.-Y.; Bottsford-Miller, J.; Lu, C.; Lin, Y.G.; Coffey, D.; Spannuth, W.A.; Nugent, E.; Han, L.Y. Clinical and biological impact of EphA2 overexpression and angiogenesis in endometrial cancer. Cancer Biol. Ther. 2010, 10, 1306–1314. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.-J.; Ge, J.; Chen, Z.-K.; Wu, S.-B.; Shen, H.; Yang, P.; Hu, B.; Zhang, G.-W.; Chen, Z.-H. Over-expression of EphA2 and EphrinA-1 in human gastric adenocarcinoma and its prognostic value for postoperative patients. Dig. Dis. Sci. 2009, 54, 2410. [Google Scholar] [CrossRef] [PubMed]
- Hirai, H.; Maru, Y.; Hagiwara, K.; Nishida, J.; Takaku, F. A novel putative tyrosine kinase receptor encoded by the eph gene. Science 1987, 238, 1717–1721. [Google Scholar] [CrossRef] [PubMed]
- Thaker, P.H.; Deavers, M.; Celestino, J.; Thornton, A.; Fletcher, M.S.; Landen, C.N.; Kinch, M.S.; Kiener, P.A.; Sood, A.K. EphA2 expression is associated with aggressive features in ovarian carcinoma. Clin. Cancer Res. 2004, 10, 5145–5150. [Google Scholar] [CrossRef] [PubMed]
- Zeng, G.; Hu, Z.; Kinch, M.S.; Pan, C.-X.; Flockhart, D.A.; Kao, C.; Gardner, T.A.; Zhang, S.; Li, L.; Baldridge, L.A. High-level expression of EphA2 receptor tyrosine kinase in prostatic intraepithelial neoplasia. Am. J. Pathol. 2003, 163, 2271–2276. [Google Scholar] [CrossRef]
- Wykosky, J.; Gibo, D.M.; Stanton, C.; Debinski, W. Interleukin-13 receptor α2, EphA2, and Fos-related antigen 1 as molecular denominators of high-grade astrocytomas and specific targets for combinatorial therapy. Clin. Cancer Res. 2008, 14, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Lazar, D.F.; Saltiel, A.R.; Dixit, V.M. Activation of the Eck receptor protein tyrosine kinase stimulates phosphatidylinositol 3-kinase activity. J. Biol. Chem. 1994, 269, 30154–30157. [Google Scholar] [PubMed]
- Landen, C.N.; Chavez-Reyes, A.; Bucana, C.; Schmandt, R.; Deavers, M.T.; Lopez-Berestein, G.; Sood, A.K. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 2005, 65, 6910–6918. [Google Scholar] [CrossRef] [PubMed]
- Ayaki, M.; Komatsu, K.; Mukai, M.; Murata, K.; Kameyama, M.; Ishiguro, S.; Miyoshi, J.; Tatsuta, M.; Nakamura, H. Reduced expression of focal adhesion kinase in liver metastases compared with matched primary human colorectal adenocarcinomas. Clin. Cancer Res. 2001, 70, 3106–3112. [Google Scholar]
- Cance, W.G.; Harris, J.E.; Iacocca, M.V.; Roche, E.; Yang, X.; Chang, J.; Simkins, S.; Xu, L. Immunohistochemical analyses of focal adhesion kinase expression in benign and malignant human breast and colon tissues: correlation with preinvasive and invasive phenotypes. Clin. Cancer Res. 2000, 6, 2417–2423. [Google Scholar] [PubMed]
- Owens, L.V.; Xu, L.; Craven, R.J.; Dent, G.A.; Weiner, T.M.; Kornberg, L.; Liu, E.T.; Cance, W.G. Overexpression of the focal adhesion kinase (p125FAK) in invasive human tumors. Cancer Res. 1995, 55, 2752–2755. [Google Scholar] [PubMed]
- Owens, L.V.; Xu, L.; Dent, G.A.; Yang, X.; Sturge, G.C.; Craven, R.J.; Cance, W.G. Focal adhesion kinase as a marker of invasive potential in differentiated human thyroid cancer. Ann. Surg. Oncol. 1996, 3, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, L.; Hauck, W.; Aprikian, A.G.; Begin, L.R.; Chapdelaine, A.; Chevalier, S. Focal adhesion kinase (pp125FAK) expression, activation and association with paxillin and p50CSK in human metastatic prostate carcinoma. Int. J. Cancer 1996, 68, 164–171. [Google Scholar] [CrossRef]
- Sood, A.K.; Coffin, J.E.; Schneider, G.B.; Fletcher, M.S.; DeYoung, B.R.; Gruman, L.M.; Gershenson, D.M.; Schaller, M.D.; Hendrix, M.J. Biological significance of focal adhesion kinase in ovarian cancer: role in migration and invasion. Am. J. Pathol. 2004, 165, 1087–1095. [Google Scholar] [CrossRef]
- Ishizawar, R.; Parsons, S.J. c-Src and cooperating partners in human cancer. Cancer Cell 2004, 6, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, M.M.; Lu, C.; Lee, J.-W.; Stone, R.L.; Mitra, R.; Mangala, L.S.; Lu, Y.; Baggerly, K.A.; Danes, C.G.; Nick, A.M. Dual targeting of EphA2 and FAK in ovarian carcinoma. Cancer Biol. Ther. 2009, 8, 1027–1034. [Google Scholar] [CrossRef] [PubMed]
- Sulman, E.P.; Tang, X.X.; Allen, C.; Biegel, J.A.; Pleasure, D.E.; Brodeur, G.M.; Ikegaki, N. ECK, a HumanEPH-Related Gene, Maps to 1p36. 1, a Common Region of Alteration in Human Cancers. Genomics 1997, 40, 371–374. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Chen, Z.; Wu, S.; Guo, J.; Ge, J.; Yang, P.; Huang, J. Silencing of EphA2 inhibits invasion of human gastric cancer SGC-7901 cells in vitro and in vivo. Neoplasma 2012, 59, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Higashiyama, M.; Doi, O.; Kodama, K.; Yokouchi, H.; Kasugai, T.; Ishiguro, S.; Takami, K.; Nakayama, T.; Nishisho, I. MDM2 gene amplification and expression in non-small-cell lung cancer: Immunohistochemical expression of its protein is a favourable prognostic marker in patients without p53 protein accumulation. Br. J. Cancer 1997, 75, 1302–1308. [Google Scholar] [CrossRef] [PubMed]
- Graves, B.; Thompson, T.; Xia, M.; Janson, C.; Lukacs, C.; Deo, D.; Di Lello, P.; Fry, D.; Garvie, C.; Huang, K.-S. Activation of the p53 pathway by small-molecule-induced MDM2 and MDMX dimerization. Proc. Natl. Acad. Sci. USA 2012, 109, 11788–11793. [Google Scholar] [CrossRef] [PubMed]
- Koster, R.; Timmer-Bosscha, H.; Bischoff, R.; Gietema, J.A.; de Jong, S. Disruption of the MDM2–p53 interaction strongly potentiates p53-dependent apoptosis in cisplatin-resistant human testicular carcinoma cells via the Fas/FasL pathway. Cell Death Dis. 2011, 2, e148. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Wagner, E. Bioresponsive polymers for nonviral gene delivery. Curr. Opin. Mol. Ther. 2009, 11, 165–178. [Google Scholar] [PubMed]
- Yu, H.; Zou, Y.; Jiang, L.; Yin, Q.; He, X.; Chen, L.; Zhang, Z.; Gu, W.; Li, Y. Induction of apoptosis in non-small cell lung cancer by downregulation of MDM2 using pH-responsive PMPC-b-PDPA/siRNA complex nanoparticles. Biomaterials 2013, 34, 2738–2747. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-D.; Chono, S.; Huang, L. Efficient oncogene silencing and metastasis inhibition via systemic delivery of siRNA. Mol. Ther. 2008, 16, 942–946. [Google Scholar] [PubMed]
- Pollak, M. Insulin and insulin-like growth factor signalling in neoplasia. Nat. Rev. Cancer 2008, 8, 915–928. [Google Scholar] [CrossRef] [PubMed]
- Beech, D.J.; Parekh, N.; Pang, Y. Insulin-like growth factor-I receptor antagonism results in increased cytotoxicity of breast cancer cells to doxorubicin and taxol. Oncol. Rep. 2001, 8, 325–329. [Google Scholar] [CrossRef] [PubMed]
- Cantley, L.C.; Auger, K.R.; Carpenter, C.; Duckworth, B.; Graziani, A.; Kapeller, R.; Soltoff, S. Oncogenes and signal transduction. Cell 1991, 64, 281–302. [Google Scholar] [CrossRef]
- Chernicky, C.L.; Yi, L.; Tan, H.; Gan, S.U.; Ilan, J. Treatment of human breast cancer cells with antisense RNA to the type I insulin-like growth factor receptor inhibits cell growth, suppresses tumorigenesis, alters the metastatic potential, and prolongs survival in vivo. Cancer Gene Ther. 2000, 7, 384. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Tewari, M.; Cui, S.; Rubin, R. Activation of the insulin-like growth factor-I receptor inhibits tumor necrosis factor-induced cell death. J. Cell. Physiol. 1996, 168, 499–509. [Google Scholar] [CrossRef]
- Baserga, R. The IGF-I receptor in cancer research. Exp. Cell Res. 1999, 253, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.; Yee, D. Insulin-like growth factors and breast cancer. Biomed. Pharmacother. 1995, 49, 415–421. [Google Scholar] [CrossRef]
- Iravani, S.; Zhang, H.Q.; Yuan, Z.Q.; Cheng, J.Q.; Karl, R.C.; Jove, R.; Coppola, D. Modification of insulin-like growth factor 1 receptor, c-Src, and Bcl-X L protein expression during the progression of barrett’s neoplasia. Hum. Pathol. 2003, 34, 975–982. [Google Scholar] [CrossRef]
- LeRoith, D.; Baserga, R.; Helman, L.; Roberts, C.T., Jr. Insulin-like growth factors and cancer. Ann. Intern. Med. 1995, 122, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Jemal, A.; Siegel, R.; Xu, J.; Ward, E. Cancer statistics, 2010. CA Cancer J. Clin. 2010, 60, 277–300. [Google Scholar] [CrossRef] [PubMed]
- Gridelli, C. Targeted therapy developments in the treatment of non-small cell lung cancer: A promising but long and winding road. Curr. Opin. Oncol. 2008, 20, 145–147. [Google Scholar] [CrossRef] [PubMed]
- Hansen, H.H. Treatment of advanced non-small cell lung cancer: Should include short courses of radiation, with palliation as the aim. BMJ Br. Med. J. 2002, 325, 452. [Google Scholar] [CrossRef]
- Singh, P. Insulin-like growth factor system in growth, development and carcinogenesis. J. Clin. Ligand Assay 2000, 23, 214–232. [Google Scholar]
- Dong, A.-Q.; Kong, M.-J.; Ma, Z.-Y.; Qian, J.-F.; Xu, X.-H. Down-regulation of IGF-IR using small, interfering, hairpin RNA (siRNA) inhibits growth of human lung cancer cell line A549 in vitro and in nude mice. Cell Biol. Int. 2007, 31, 500–507. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Ding, C.; Kong, M.; Dong, A.; Qian, J.; Jiang, D.; Shen, Z. Tumor-targeting magnetic lipoplex delivery of short hairpin RNA suppresses IGF-1R overexpression of lung adenocarcinoma A549 cells in vitro and in vivo. Biochem. Biophys. Res. Commun. 2011, 410, 537–542. [Google Scholar] [CrossRef] [PubMed]
- Deveraux, Q.L.; Stennicke, H.R.; Salvesen, G.S.; Reed, J.C. Endogenous inhibitors of caspases. J. Clin. Immunol. 1999, 19, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, A.; Yang, A.Y.-P.; Srivastava, M. Regulators of IAP function: Coming to grips with the grim reaper. Curr. Opin. Cell Biol. 2003, 15, 717–724. [Google Scholar] [CrossRef] [PubMed]
- Yagihashi, A.; Asanuma, K.; Tsuji, N.; Torigoe, T.; Sato, N.; Hirata, K.; Watanabe, N. Detection of anti-livin antibody in gastrointestinal cancer patients. Clin. Chem. 2003, 49, 1206–1208. [Google Scholar] [CrossRef] [PubMed]
- Ambrosini, G.; Adida, C.; Altieri, D.C. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat. Med. 1997, 3, 917–921. [Google Scholar] [CrossRef] [PubMed]
- Richter, B.W.; Duckett, C.S. The IAP proteins: Caspase inhibitors and beyond. Sci. Signal. 2000, 2000, pe1. [Google Scholar] [CrossRef] [PubMed]
- Ashhab, Y.; Alian, A.; Polliack, A.; Panet, A.; Yehuda, D.B. Two splicing variants of a new inhibitor of apoptosis gene with different biological properties and tissue distribution pattern. FEBS Lett. 2001, 495, 56–60. [Google Scholar] [CrossRef]
- Ka, H.; Hunt, J.S. Temporal and spatial patterns of expression of inhibitors of apoptosis in human placentas. Am. J. Pathol. 2003, 163, 413–422. [Google Scholar] [CrossRef]
- Lv, J.; Chen, Z. Resent research about Livin in cancer. Chin. J. Cancer Prev. Treat. 2006, 13, 1347–1350. [Google Scholar]
- Yagihashi, A.; Ohmura, T.; Asanuma, K.; Kobayashi, D.; Tsuji, N.; Torigoe, T.; Sato, N.; Hirata, K.; Watanabe, N. Detection of autoantibodies to survivin and livin in sera from patients with breast cancer. Clin. Chim. Acta 2005, 362, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Hariu, H.; Hirohashi, Y.; Torigoe, T.; Asanuma, H.; Hariu, M.; Tamura, Y.; Aketa, K.; Nabeta, C.; Nakanishi, K.; Kamiguchi, K. Aberrant expression and potency as a cancer immunotherapy target of inhibitor of apoptosis protein family, Livin/ML-IAP in lung cancer. Clin. Cancer Res. 2005, 11, 1000–1009. [Google Scholar] [PubMed]
- Jemal, A.; Bray, F.; Center, M.M.; Ferlay, J.; Ward, E.; Forman, D. Global cancer statistics. CA Cancer J. Clin. 2011, 61, 69–90. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2015. CA Cancer J. Clin. 2015, 65, 5–29. [Google Scholar] [CrossRef] [PubMed]
- Bar-Eli, M. Role of AP-2 in tumor growth and metastasis of human melanoma. Cancer Metastasis Rev. 1999, 18, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Bar-Eli, M. Gene regulation in melanoma progression by the AP-2 transcription factor. Pigment. Cell Melanoma Res. 2001, 14, 78–85. [Google Scholar] [CrossRef]
- Soutschek, J.; Akinc, A.; Bramlage, B.; Charisse, K.; Constien, R.; Donoghue, M.; Elbashir, S.; Geick, A.; Hadwiger, P.; Harborth, J. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 2004, 432, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Dykxhoorn, D.M.; Lieberman, J. The silent revolution: RNA interference as basic biology, research tool, and therapeutic. Annu. Rev. Med. 2005, 56, 401–423. [Google Scholar] [CrossRef] [PubMed]
- Ryther, R.; Flynt, A.; Phillips, J.; Patton, J. siRNA therapeutics: Big potential from small RNAs. Gene Ther. 2005, 12, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, Y.; Wang, W.; Guan, B.; Xun, M.; Zhang, H.; Wang, Z.; Zhao, Y. Single-chain antibody–delivered Livin siRNA inhibits human malignant melanoma growth in vitro and in vivo. Tumor Biol. 2017, 39. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Lin, F.; Wang, X.; Gao, P.; Dong, K.; Zou, A.; Cheng, S.; Wei, S.; Zhang, H. Silencing Livin gene expression to inhibit proliferation and enhance chemosensitivity in tumor cells. Cancer Gene Ther. 2008, 15, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Call, K.M.; Glaser, T.; Ito, C.Y.; Buckler, A.J.; Pelletier, J.; Haber, D.A.; Rose, E.A.; Kral, A.; Yeger, H.; Lewis, W.H. Isolation and characterization of a zinc finger polypeptide gene at the human chromosome 11 Wilms’ tumor locus. Cell 1990, 60, 509–520. [Google Scholar] [CrossRef]
- Gessler, M.; Poustka, A.; Cavenee, W.; Neve, R.L.; Orkin, S.H.; Bruns, G.A. Homozygous deletion in Wilms tumours of a zinc-finger gene identified by chromosome jumping. Nature 1990, 343, 774. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, H. Wilms’ tumor gene WT1: Its oncogenic function and clinical application. Int. J. Hematol. 2001, 73, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Coppes, M.J.; Campbell, C.E.; Williams, B. The role of WT1 in Wilms tumorigenesis. FASEB J. 1993, 7, 886–895. [Google Scholar] [CrossRef] [PubMed]
- Rauscher, F. The WT1 Wilms tumor gene product: A developmentally regulated transcription factor in the kidney that functions as a tumor suppressor. FASEB J. 1993, 7, 896–903. [Google Scholar] [CrossRef] [PubMed]
- Haber, D.; Park, S.; Maheswaran, S.; Englert, C.; Re, G.; Hazen-Martin, D.; Sens, D.; Garvin, A. WT1-mediated growth suppression of Wilms’ tumor cells expressing a WT1 splicing variant. Science 1993, 262, 2057–2059. [Google Scholar] [CrossRef] [PubMed]
- Algar, E.M.; Kenney, M.T.; Simms, L.A.; Smith, S.I.; Kida, Y.; Smith, P.J. Homozygous intragenic deletion in the WT1 gene in a sporadic Wilms’ tumour associated with high levels of expression of a truncated transcript. Hum. Mutat. 1995, 5, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Little, M.; Wells, C. A clinical overview of WT1 gene mutations. Hum. Mutat. 1997, 9, 209–225. [Google Scholar] [CrossRef]
- Sugiyama, H. Cancer immunotherapy targeting Wilms’ tumor gene WT1 product. Expert Rev. Vaccines 2005, 4, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Miwa, H.; Beran, M.; Saunders, G. Expression of the Wilms’ tumor gene (WT1) in human leukemias. Leukemia 1992, 6, 405–409. [Google Scholar] [PubMed]
- Moore, A.W.; McInnes, L.; Kreidberg, J.; Hastie, N.D.; Schedl, A. YAC complementation shows a requirement for Wt1 in the development of epicardium, adrenal gland and throughout nephrogenesis. Development 1999, 126, 1845–1857. [Google Scholar] [PubMed]
- Inoue, K.; Sugiyama, H.; Ogawa, H.; Nakagawa, M.; Yamagami, T.; Miwa, H.; Kita, K.; Hiraoka, A.; Masaoka, T.; Nasu, K. WT1 as a new prognostic factor and a new marker for the detection of minimal residual disease in acute leukemia. Blood 1994, 84, 3071–3079. [Google Scholar] [PubMed]
- Oji, Y.; Miyoshi, S.; Maeda, H.; Hayashi, S.; Tamaki, H.; Nakatsuka, S.I.; Yao, M.; Takahashi, E.; Nakano, Y.; Hirabayashi, H. Overexpression of the Wilms’ tumor gene WT1 in de novo lung cancers. Int. J. Cancer 2002, 100, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Loeb, D.M.; Evron, E.; Patel, C.B.; Sharma, P.M.; Niranjan, B.; Buluwela, L.; Weitzman, S.A.; Korz, D.; Sukumar, S. Wilms’ tumor suppressor gene (WT1) is expressed in primary breast tumors despite tumor-specific promoter methylation. Cancer Res. 2001, 61, 921–925. [Google Scholar] [PubMed]
- Miyoshi, Y.; Ando, A.; Egawa, C.; Taguchi, T.; Tamaki, Y.; Tamaki, H.; Sugiyama, H.; Noguchi, S. High expression of Wilms’ tumor suppressor gene predicts poor prognosis in breast cancer patients. Clin. Cancer Res. 2002, 8, 1167–1171. [Google Scholar] [PubMed]
- Oji, Y.; Suzuki, T.; Nakano, Y.; Maruno, M.; Nakatsuka, S.i.; Jomgeow, T.; Abeno, S.; Tatsumi, N.; Yokota, A.; Aoyagi, S. Overexpression of the Wilms’ tumor gene WT1 in primary astrocytic tumors. Cancer Sci. 2004, 95, 822–827. [Google Scholar] [CrossRef] [PubMed]
- Bowman, T.; Garcia, R.; Turkson, J.; Jove, R. STATs in oncogenesis. Oncogene 2000, 19, 2474. [Google Scholar] [CrossRef] [PubMed]
- Bromberg, J. Stat proteins and oncogenesis. J. Clin. Investig. 2002, 109, 1139. [Google Scholar] [CrossRef] [PubMed]
- Darnell, J.E. Validating Stat3 in cancer therapy. Nat. Med. 2005, 11, 595–596. [Google Scholar] [CrossRef] [PubMed]
- Sinibaldi, D.; Wharton, W.; Turkson, J.; Bowman, T.; Pledger, W.J.; Jove, R. Induction of p21 WAF1/CIP1 and cyclin D1 expression by the Src oncoprotein in mouse fibroblasts: Role of activated STAT3 signaling. Oncogene 2000, 19, 5419–5427. [Google Scholar] [CrossRef] [PubMed]
- Catlett-Falcone, R.; Dalton, W.S.; Jove, R. STAT proteins as novel targets for cancer therapy. Curr. Opin. Oncol. 1999, 11, 490. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.; Hassan, R. Immunotherapies for non-small-cell lung cancer and mesothelioma. Lancet Oncol. 2012, 13, e301–e310. [Google Scholar] [CrossRef]
- Xu, C.; Wu, C.; Xia, Y.; Zhong, Z.; Liu, X.; Xu, J.; Cui, F.; Chen, B.; Røe, O.D.; Li, A. WT1 promotes cell proliferation in non-small cell lung cancer cell lines through up-regulating cyclin D1 and p-pRb in vitro and in vivo. PLoS ONE 2013, 8, e68837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derycke, A.S.; De Witte, P.A. Transferrin-mediated targeting of hypericin embedded in sterically stabilized PEG-liposomes. Int. J. Oncol. 2002, 20, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Stavridis, J.; Deliconstantinos, G.; Psallidopoulos, M.; Armenakas, N.; Hadjiminas, D.; Hadjiminas, J. Construction of transferrin-coated liposomes for in vivo transport of exogenous DNA to bone marrow erythroblasts in rabbits. Exp. Cell Res. 1986, 164, 568–572. [Google Scholar] [CrossRef]
- Voinea, M.; Dragomir, E.; Manduteanu, I.; Simionescu, M. Binding and uptake of transferrin-bound liposomes targeted to transferrin receptors of endothelial cells. Vasc. Pharmacol. 2002, 39, 13–20. [Google Scholar] [CrossRef]
- Kobayashi, T.; Ishida, T.; Okada, Y.; Ise, S.; Harashima, H.; Kiwada, H. Effect of transferrin receptor-targeted liposomal doxorubicin in P-glycoprotein-mediated drug resistant tumor cells. Int. J. Pharm. 2007, 329, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Saavedra-Alonso, S.; Zapata-Benavides, P.; Chavez-Escamilla, A.K.; Manilla-Muñoz, E.; Zamora-Avila, D.E.; Franco-Molina, M.A.; Rodriguez-Padilla, C. WT1 shRNA delivery using transferrin-conjugated PEG liposomes in an in vivo model of melanoma. Exp. Ther. Med. 2016, 12, 3778–3784. [Google Scholar] [CrossRef] [PubMed]
- Hinitt, C.; Wood, J.; Lee, S.; Williams, A.; Howarth, J.; Glover, C.; Uney, J.; Hague, A. BAG-1 enhances cell–cell adhesion, reduces proliferation and induces chaperone-independent suppression of hepatocyte growth factor-induced epidermal keratinocyte migration. Exp. Cell Res. 2010, 316, 2042–2060. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Lu, S.; Gu, L.; Gao, Y.; Wang, T.; Zhao, J.; Rao, J.; Chen, J.; Hao, X.; Tang, S.-C. Modulation of BAG-1 expression alters the sensitivity of breast cancer cells to tamoxifen. Cell. Physiol. Biochem. 2014, 33, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Ozfiliz, P.; Arisan, E.D.; Coker-Gurkan, A.; Obakan, P.; Eralp, T.N.; Dinler-Doganay, G.; Palavan-Unsal, N. Bag-1L is a stress-withstand molecule prevents the downregulation of Mcl-1 and c-Raf under control of heat shock proteins in cisplatin treated HeLa cervix cancer cells. Asian Pac. J. Cancer Prev. 2014, 15, 4475–4482. [Google Scholar] [CrossRef] [PubMed]
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed]
- El-Shami, K.; Oeffinger, K.C.; Erb, N.L.; Willis, A.; Bretsch, J.K.; Pratt-Chapman, M.L.; Cannady, R.S.; Wong, S.L.; Rose, J.; Barbour, A.L. American Cancer Society colorectal cancer survivorship care guidelines. CA Cancer J. Clin. 2015, 65, 427–455. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Meng, Q.; Tian, A. Expressions of the anti-apoptotic genes Bag-1 and Bcl-2 in colon cancer and their relationship. Am. J. Surg. 2010, 200, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Zhou, G.; Ling, J.; Tian, A.; Sun, N. Silencing Bag-1 gene via magnetic gold nanoparticle-delivered siRNA plasmid for colorectal cancer therapy in vivo and in vitro. Tumor Biol. 2016, 37, 10365–10374. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; McGarry, T.J.; Bernal, T.; Kirschner, M.W. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science 1999, 285, 418–422. [Google Scholar] [CrossRef] [PubMed]
- Jallepalli, P.V.; Waizenegger, I.C.; Bunz, F.; Langer, S.; Speicher, M.R.; Peters, J.-M.; Kinzler, K.W.; Vogelstein, B.; Lengauer, C. Securin is required for chromosomal stability in human cells. Cell 2001, 105, 445–457. [Google Scholar] [CrossRef]
- Yu, R.; Lu, W.; Chen, J.; McCabe, C.J.; Melmed, S. Overexpressed pituitary tumor-transforming gene causes aneuploidy in live human cells. Endocrinology 2003, 144, 4991–4998. [Google Scholar] [CrossRef] [PubMed]
- Christopoulou, L.; Moore, J.D.; Tyler-Smith, C. Over-expression of wild-type Securin leads to aneuploidy in human cells. Cancer Lett. 2003, 202, 213–218. [Google Scholar] [CrossRef]
- Weaver, B.A.; Cleveland, D.W. Decoding the links between mitosis, cancer, and chemotherapy: The mitotic checkpoint, adaptation, and cell death. Cancer Cell 2005, 8, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Horwitz, G.A.; Heaney, A.P.; Nakashima, M.; Prezant, T.R.; Bronstein, M.D.; Melmed, S. Pituitary tumor transforming gene (PTTG) expression in pituitary adenomas. J. Clin. Endocrinol. Metab. 1999, 84, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Heaney, A.P.; Singson, R.; McCabe, C.J.; Nelson, V.; Nakashima, M.; Melmed, S. Expression of pituitary-tumour transforming gene in colorectal tumours. Lancet 2000, 355, 716–719. [Google Scholar] [CrossRef]
- Puri, R.; Tousson, A.; Chen, L.; Kakar, S.S. Molecular cloning of pituitary tumor transforming gene 1 from ovarian tumors and its expression in tumors. Cancer Lett. 2001, 163, 131–139. [Google Scholar] [CrossRef]
- Solbach, C.; Roller, M.; Fellbaum, C.; Nicoletti, M.; Kaufmann, M. PTTG mRNA expression in primary breast cancer: A prognostic marker for lymph node invasion and tumor recurrence. Breast 2004, 13, 80–81. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, A.; Ramos-Morales, F.; Romero, F.; Rios, R.M.; Dreyfus, F.; Tortolero, M.; Pintor-Toro, J.A. hpttg, a human homologue of rat pttg, is overexpressed in hematopoietic neoplasms. Evidence for a transcriptional activation function of hPTTG. Oncogene 1998, 17, 2187. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Melmed, S. Pituitary tumor transforming gene: An update. In Molecular Pathology of the Pituitary; Karger Publishers: Basel, Switzerland, 2004; Volume 32, pp. 175–185. [Google Scholar]
- Thorgeirsson, S.S.; Grisham, J.W. Molecular pathogenesis of human hepatocellular carcinoma. Nat. Genet. 2002, 31, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Bruix, J.; Llovet, J.M. Prognostic prediction and treatment strategy in hepatocellular carcinoma. Hepatology 2002, 35, 519–524. [Google Scholar] [CrossRef] [PubMed]
- Bemal, J.; Luna, R.; Espina, A. Human securin interacts with p53 and modulates p53–mediated transcriptional activity and apoptosis. Nat. Genet. 2002, 32, 306–311. [Google Scholar]
- Jung, C.R.; Yoo, J.; Jang, Y.J.; Kim, S.; Chu, I.S.; Yeom, Y.I.; Choi, J.Y.; Im, D.S. Adenovirus-mediated transfer of siRNA against PTTG1 inhibits liver cancer cell growth in vitro and in vivo. Hepatology 2006, 43, 1042–1052. [Google Scholar] [CrossRef] [PubMed]
- Frazier, W.A.; Gao, A.-G.; Dimitry, J.; Chung, J.; Brown, E.J.; Lindberg, F.P.; Linder, M.E. The thrombospondin receptor integrin-associated protein (CD47) functionally couples to heterotrimeric Gi. J. Biol. Chem. 1999, 274, 8554–8560. [Google Scholar] [CrossRef] [PubMed]
- Oldenborg, P.-A.; Zheleznyak, A.; Fang, Y.-F.; Lagenaur, C.F.; Gresham, H.D.; Lindberg, F.P. Role of CD47 as a marker of self on red blood cells. Science 2000, 288, 2051–2054. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, S.; Jamieson, C.H.; Pang, W.W.; Park, C.Y.; Chao, M.P.; Majeti, R.; Traver, D.; van Rooijen, N.; Weissman, I.L. CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell 2009, 138, 271–285. [Google Scholar] [CrossRef] [PubMed]
- Majeti, R.; Chao, M.P.; Alizadeh, A.A.; Pang, W.W.; Jaiswal, S.; Gibbs, K.D.; van Rooijen, N.; Weissman, I.L. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell 2009, 138, 286–299. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.S.; Espinosa, I.; Chao, M.; Wong, D.; Ailles, L.; Diehn, M.; Gill, H.; Presti, J.; Chang, H.Y.; van de Rijn, M. Identification, molecular characterization, clinical prognosis, and therapeutic targeting of human bladder tumor-initiating cells. Proc. Natl. Acad. Sci. USA 2009, 106, 14016–14021. [Google Scholar] [CrossRef] [PubMed]
- Chao, M.P.; Alizadeh, A.A.; Tang, C.; Myklebust, J.H.; Varghese, B.; Gill, S.; Jan, M.; Cha, A.C.; Chan, C.K.; Tan, B.T. Anti-CD47 antibody synergizes with rituximab to promote phagocytosis and eradicate non-Hodgkin lymphoma. Cell 2010, 142, 699–713. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, Z.; Guo, S.; Zhang, L.; Sharma, A.; Robertson, G.P.; Huang, L. Intravenous delivery of siRNA targeting CD47 effectively inhibits melanoma tumor growth and lung metastasis. Mol. Ther. 2013, 21, 1919–1929. [Google Scholar] [CrossRef] [PubMed]






| Drug Formulation | Target Gene | NPs | Treatment | Diseases | Phase | Status | Identifier Trial Number (https://clinicaltrails.gov) |
|---|---|---|---|---|---|---|---|
| DCR-MYC | MYC | Lipid | siRNAs | Hepatocellular carcinoma | 1/2 | Ongoing, not recruiting 2014-present | NCT02314052 |
| DCR-MYC | MYC | Lipid | siRNAs | Solid tumors, multiple myeloma, non-Hodgkin lymphoma, or pancreatic neuroendocrine tumors | 1 | Ongoing, not recruiting 2014-present | NCT02110563 |
| ALN-VSP02 | KSP and VEGF | Lipid | siRNAs | Solid tumors | 1 | Completed | NCT00882180 |
| Atu 027 | PKN3 | Lipid Nanoparticles | siRNAs | Advanced cancers | 1 | Completed | NCT00938574 |
| TKM-080301 | PLK1 | Lipid | siRNAs | Primary and secondary liver cancer | 1 | completed | NCT01437007 |
| PLK1 | Lipid | siRNAs | Neuroendocrine tumors | 1/2 | completed | NCT01262235 | |
| PLK1 | Lipid | siRNAs | Advanced hepatocellular carcinoma | 1/2 | completed | NCT02191878 | |
| siRNA-EphA2-DOPC | EphA2 | Lipid | siRNAs | Advanced solid tumors | 1 | Recruiting | NCT01591356 |
| siG12D-LODER | KRAS | LODER polymer | siRNAs | Ductal adenocarcinoma or pancreatic cancer | 1 | completed | NCT01188785 |
| siG12D-LODER | KRAS | LODER polymer | siRNAs | pancreatic cancer | 2 | Not yet recruiting | NCT01676259 |
| SNS01-T | eIF5A | polyethyleneimine | siRNAs plasmids | Multiple myeloma | 1/2 | unknown | NCT01435720 |
| Target Genes | Role of Genes | Delivery Vehicle | Treatment | Preclinical Studies Application | Preclinical Studies Outcome | Refs. |
|---|---|---|---|---|---|---|
| Bcl-2 | Inhibits apoptosis pathways and promote cellular growth and survival in breast, lung, liver and gastric cancer | Liposome-protamine | siRNA | Balb/c mouse model inoculated with H22 liver tumor cells | 66.5% reduction of tumor growth by suppressing Bcl-2 gene expression | 65 |
| Bcl-2 | pSilencerTM-EGFP sh515 | shRNA | Balb/C mouse model inoculated with GBC-SD, gallbladder carcinoma cells | 50% reduction of tumor volume and decreased tumor growth rate | 72 | |
| Bcl-2 | Cationic liposome, LIC-101 | siRNA | Balb/C nu++ mouse model inoculated with PC-3 prostate cancer cells | 63% reduction of tumor volume | 73 | |
| Bcl-2 | PEG-LIC complex | siRNA | Balb/C mouse model inoculated with PC-3 human prostate cancer cells | Increased siRNA uptake, 65% tumor reduction without any systemic toxicity | 74 | |
| VEGF | Stimulates angiogenesis and vascular permeability | Adenoviral vector (Ad5CMV) | siRNA | Athymic female mouse model inoculated with MDA251-MB, human breast cancer cells | Reduced 80% of tumor through anti-angiogenesis mechanism | 97 |
| VEGF | Polyelectrolyte complex (PEG/PEI PEC micelles) | siRNA | Female nude mice (nu/nu) model inoculated with PC-3 human prostate cancer cells | Intratumoral injection caused 79% tumor inhibition; Intravenous administration reduced 86% of tumor volume | 103 | |
| VEGF-C | Promotes lymphogenesis, tumorigenesis and initiates metastasis | Hifectin-mediated transfection | siRNA | Balb/C mouse model inoculated with 4T1 cells, mouse breast cancer cells | Reduced 28% of tumor volume. | 120 |
| VEGF-C | Lentivirus vector (Lv) | siRNA | Balb/C mouse model inoculated with A549, human NSCLC cells | 64% tumor inhibition and 48% reduction of tumor volume by decreasing VEGF-C expression | 121 | |
| NRP-2 | Binds with VEGF and regulates vascularization and lymphogenesis of various tumors | DOPC (neutral lipid 1,2-dioleoyl-sn-glycero-3-phosphatidyl choline) | siRNA | Male athymic nude mouse model inoculated with HTC-116, human colorectal carcinoma cell lines | Reduced 91.3% of tumor volume via increasing anti-angiogenic mechanism | 125 |
| VEGF R2 | Regulates angiogenesis and tumor growth | RGD (Ar3-Gly-Asp peptides)-PEG-PEI nanoplexes | siRNA | Female nude mouse model inoculated with N2A, mouse neuroblastoma cells | Enabled tissue-specific delivery and inhibited more than 90% of tumor volume | 100 |
| EGFR 1 & ERBB2 | Activate downstream signaling pathways and play key role in cell division and proliferation | Carbonate apatite Nano-particle | siRNA | Female Balb/C mouse model inoculated with 4T1 cells, mouse breast cancer cells | 61% reduction of tumor volume without any toxicity | 143 |
| Survivin | Suppresses apoptosis by inhibiting both intrinsic and extrinsic pathways of apoptosis, as well as improves chemo-resistance to various chemotherapeutics and increases tumor recurrence rate | PEGylated chitosan (PEG-CS) | siRNA | Female Balb/C mouse model inoculated with 4T1 cells, mouse breast cancer cells | Increased biological stability and targeted gene delivery, reduced 55% of tumor volume | 165 |
| Survivin | Chiosan-6-poly arginine and histidine (H6R6-CS) | siRNA | Female Balb/C mouse model inoculated with 4T1 cells, mouse breast cancer cells | Improved cellular uptake and endosomal escape with 63% tumor inhibition | 176 | |
| Survivin | Cationic linear polyethyleneimine (PEI) | Sticky siRNA (ssiRNA) | NMRI nude female mouse model inoculated with B16-F10 cells, murine melanoma cell lines | Reduced 50% of tumor volume through silencing of Survivin gene | 181 | |
| Survivin | PCPP (PEG-CPB-PEI) nano-particle | siRNA | Balb/C mouse model inoculated with 4T1 cells, mouse breast cancer cells | Increased tumor accumulation and improved cellular uptake with 66% reduction of tumor volume | 185 | |
| Cyclin-B1 | As a mitosis promoting factor triggers uncontrolled cell proliferation and hampers the stability of chromosomes | MPG-8 (Primary amphipathic peptide carrier)-cholesterol (MPG-8/chol) | siRNA | Swiss nude mouse model inoculated with PC-3 cells, human prostate cancer cells | 90% tumor size inhibition for maximum dose, 60–80% reduction of Cyclin B1 expression and extended survival rate | 206 |
| Cyclin-B1 | Cationic linear polyethyleneimine (PEI) | Sticky siRNA (ssiRNA) | NMRI nude female mouse model inoculated with B16-F10 cells, Murine melanoma cell lines | Reduced 44% of tumor volume via down regulating Cyclin B1 expression | 181 | |
| RhoA & RhoC | Triggers signal transduction and drives a series of pathologies of cancer including cell motility, proliferation, apoptosis inhibition, cell cycle progression, invasion, metastasis and inflammation | Adenoviral vector | shRNA | Male Balb/C mouse model inoculated with HTC-116, human colorectal carcinoma cell lines | Slowed tumor growth (2.38 fold) and reduced 37% of tumor volume | 225 |
| RhoA & RhoC | Cytofectin-mediated transfection | siRNA | Athymic female mouse model inoculated with MDA251-MB, human breast cancer cells | Reduced tumor volume 85% (anti-RhoA) and 53% (anti-RhoC), lowered angiogenesis index | 227 | |
| RhoA | Chitosan-PIHCA (polyisohexylcyanoacrylate) | siRNA | Athymic female mouse model inoculated with MDA251-MB, human breast cancer cells | At higher dose the tumor were completely removed | 228 | |
| RhoC | Lipofectamine-mediated transfection | siRNA | Balb/C-nu mouse model inoculated with SUM149, human IBC cells | Reduced tumor volume by 35%, increased survival rate to 85%, up-regulated metastasis suppressor gene KAll | 234 | |
| β-Catenin | Regulates cell-cell adhesion and gene transcription, ultimately controlling cellular proliferation | Oligofectamine-mediated transfection | siRNA | Female nude/nu mouse model inoculated with HTC-116, human colorectal carcinoma cell lines | Three-fold smaller in size of tumor in comparison to control with extended survival rate | 253 |
| β-Catenin | Lentivirus vector | shRNA | Male athymic nude mouse model inoculated with AGS cells, human gastric cancer cells | 75% reduction of tumor volume by inhibiting CCAR1 gene expression | 259 | |
| EphA2 | Enhances cell-extracellular matrix (ECM) adhesion, anchorage-dependent growth and metastasis | DOPC (neutral lipid 1,2-dioleoyl-sn-glycero-3-phosphatidyl choline) | siRNA | Female athymic nude (Ncr-nu) mouse model inoculated with SkOV3ip1 cells, ovarian cancer cell lines | Reduced 35–50% of tumor size | 275 |
| EphA2 and FAK | DOPC (neutral lipid 1,2-dioleoyl-sn-glycero-3-phosphatidyl choline) | siRNA | Female athymic nude (Ncr-nu) mouse model inoculated with SkOV3ip1 cells, ovarian cancer cell lines | Reduced 62–82% of tumor metastasis and slowed down tumor growth rate | 283 | |
| EphA2 | Liposome | siRNA | Balb/C mouse model inoculated with SGC 7901, human gastric adenocarcinoma cells | 43.1% inhibition of tumor growth, with reduction in expression of metastatic gene MMP-9 | 285 | |
| MDM-2 | Inhibits the regulation of p53 tumor suppressor gene | PMPC-b-PDPA (di-block copolymer of poly (methacryloyloxy ethyl phosphorylcholine)-b-poly (diisopropanolamine ethyl methacrylate) | siRNA | Athymic mouse model inoculated with H2009 cells, NSCLC cells | 67% reduction of tumor growth via down regulation of MDM-2 gene expression without any systemic toxicity | 290 |
| MDM-2, c-myc and VEGF | cationic lipid-PEG | siRNA | Female C57B216 mouse model inoculated with B16-F10 cells, murine melanoma cell lines | 20–30% reduction of tumor load with extended survival rate | 291 | |
| IGF-1R | Promotes cellular metabolism, differentiation, apoptosis, chemo resistance and angiogenesis as well as protecting cells from UV irradiation, cytokine and gamma radiation-induced apoptosis | Plasmids-PEI | siRNA | Male nude mouse model inoculated with A549 cells, human lung adenocarcinoma cell lines | 60% of reduction of tumor volume, Increasing apoptotic cells | 305 |
| IGF-1R | Magnetic lipoplexes | shRNA | Male Balb/C AnNcrj-nu mouse model inoculated with A549 cells (NSCLC cells line with overexpression of IGF-1R) | Improved site specificity and cellular uptake, and reduced 85.1 ± 3% of IGF-1R gene expression | 306 | |
| Livin | Thwarts both extrinsic and intrinsic apoptosis pathways by interacting with specific cysteine proteases or caspases, while playing a significant role in tumor progression and chemo resistance development | Single chain antibody | siRNA | Nude mouse model inoculated with LiBr cells, malignant melanoma cell lines. | Reduces approximately 64% of tumor size. | 324 |
| Livin | Plasmid vector | siRNA | Balb/C nu/nu mouse model inoculated with SPCA-1 cells, human lung cancer cell lines. | 73% reduction of mean tumor size, with increased apoptotic fraction and improved survival rate | 325 | |
| WT1 | The key drivers that control cell proliferation and apoptosis via regulation of the expression of proliferative genes | Plasmid vector | shRNA | Balb/C nu/nu mouse model inoculated with A549, H1299 and H1650 cells | 69–76% reduction of tumor volume without any systemic toxicity | 348 |
| WT1 | Liposome-PEG | shRNA | Female C57BL/6 mouse model inoculated with B16F10 cell, murine melanoma cell lines | Reduced 34% of tumor weight and extended survival rate (62.5%) | 353 | |
| Bag-1 | Regulates Bcl-2 gene expression and mimics the anti-apoptotic activities via bridging between the growth factor and anti-apoptotic mechanisms | Magnetic gold nanoparticles | siRNA | Balb/C nude mouse model inoculated with LoVo cell, human colon cancer cell lines | 69% of tumor inhibition without toxicity | 360 |
| PTTG1 | Plays a vital role in several cellular processes like mitosis, DNA repair, apoptosis and gene regulation and causes aneuploidy. | Adenoviral vector | siRNA | Balb/C nude mouse model inoculated with SH-J1 cells, hepatoma cell lines | Significant tumor inhibition efficacy (84%) | 375 |
| CD-47 | Causes the tumor cells escape from immunosurveillance with the result of tumor progression | Liposome-protamine-hyaluronic acid | siRNA | C57B2/6 mouse model inoculated with B16F10 cell, murine melanoma cell lines | Above 90% tumor inhibition without toxicity | 382 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karim, M.E.; Tha, K.K.; Othman, I.; Borhan Uddin, M.; Chowdhury, E.H. Therapeutic Potency of Nanoformulations of siRNAs and shRNAs in Animal Models of Cancers. Pharmaceutics 2018, 10, 65. https://doi.org/10.3390/pharmaceutics10020065
Karim ME, Tha KK, Othman I, Borhan Uddin M, Chowdhury EH. Therapeutic Potency of Nanoformulations of siRNAs and shRNAs in Animal Models of Cancers. Pharmaceutics. 2018; 10(2):65. https://doi.org/10.3390/pharmaceutics10020065
Chicago/Turabian StyleKarim, Md. Emranul, Kyi Kyi Tha, Iekhsan Othman, Mohammad Borhan Uddin, and Ezharul Hoque Chowdhury. 2018. "Therapeutic Potency of Nanoformulations of siRNAs and shRNAs in Animal Models of Cancers" Pharmaceutics 10, no. 2: 65. https://doi.org/10.3390/pharmaceutics10020065