Azure-Based Smart Monitoring System for Anemia-Like Pallor
Abstract
:1. Introduction
2. Related Work
3. System Architecture Overview
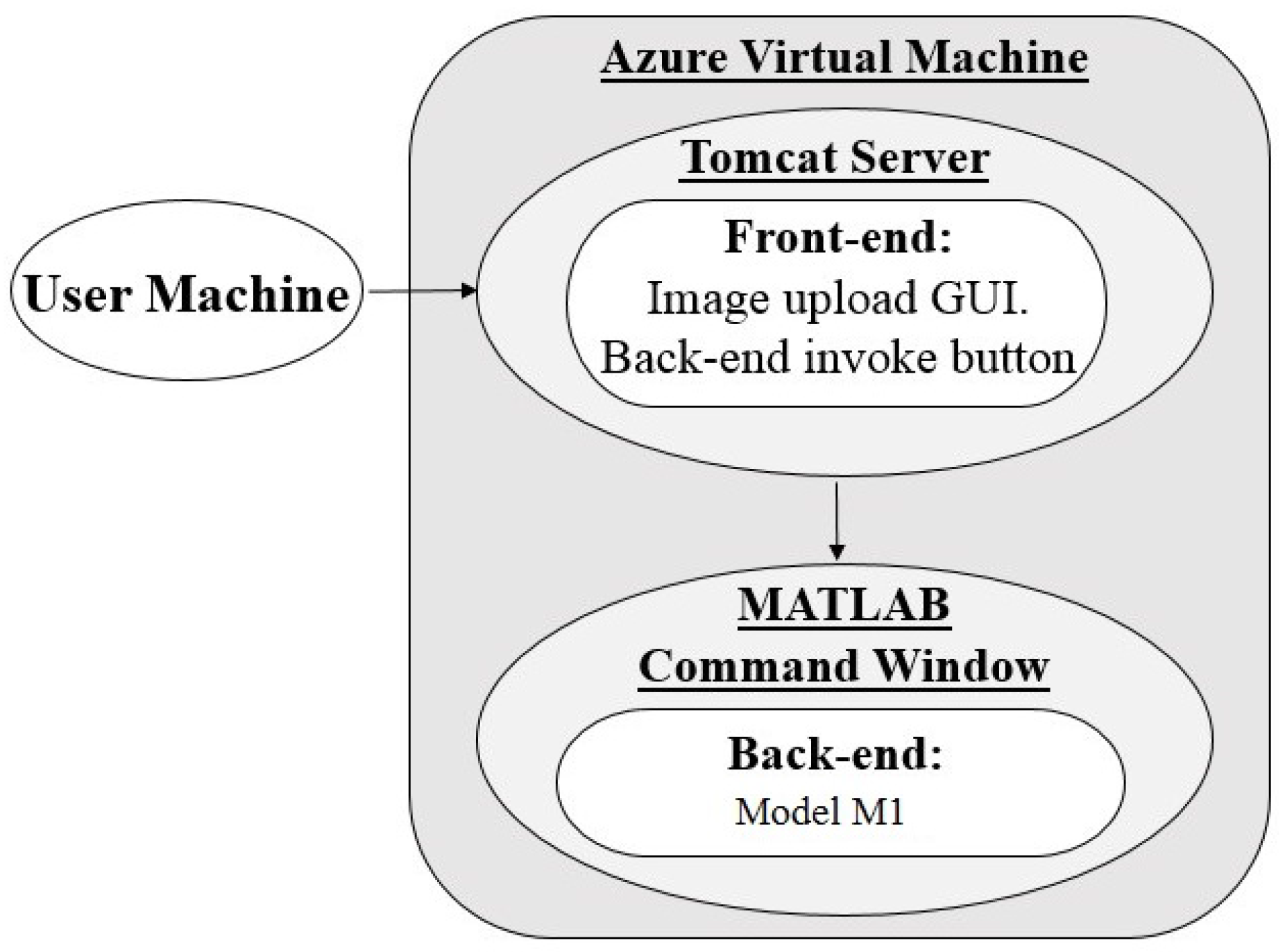
3.1. Architecture 1: Virtual Machine (VM) Hosting With MATLAB Back-End
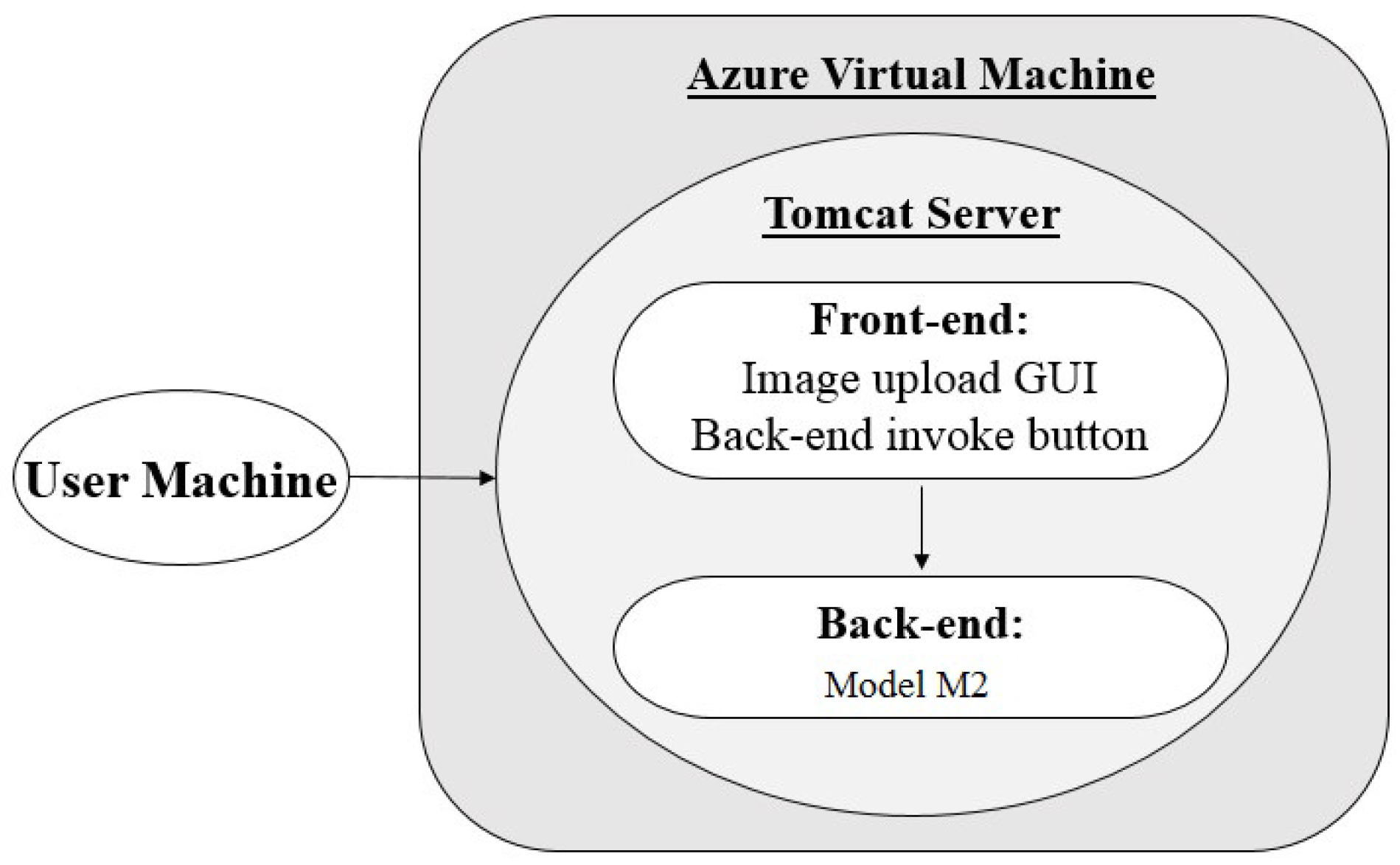
3.2. Architecture 2: Virtual Machine (VM) Hosting with Java Back-end
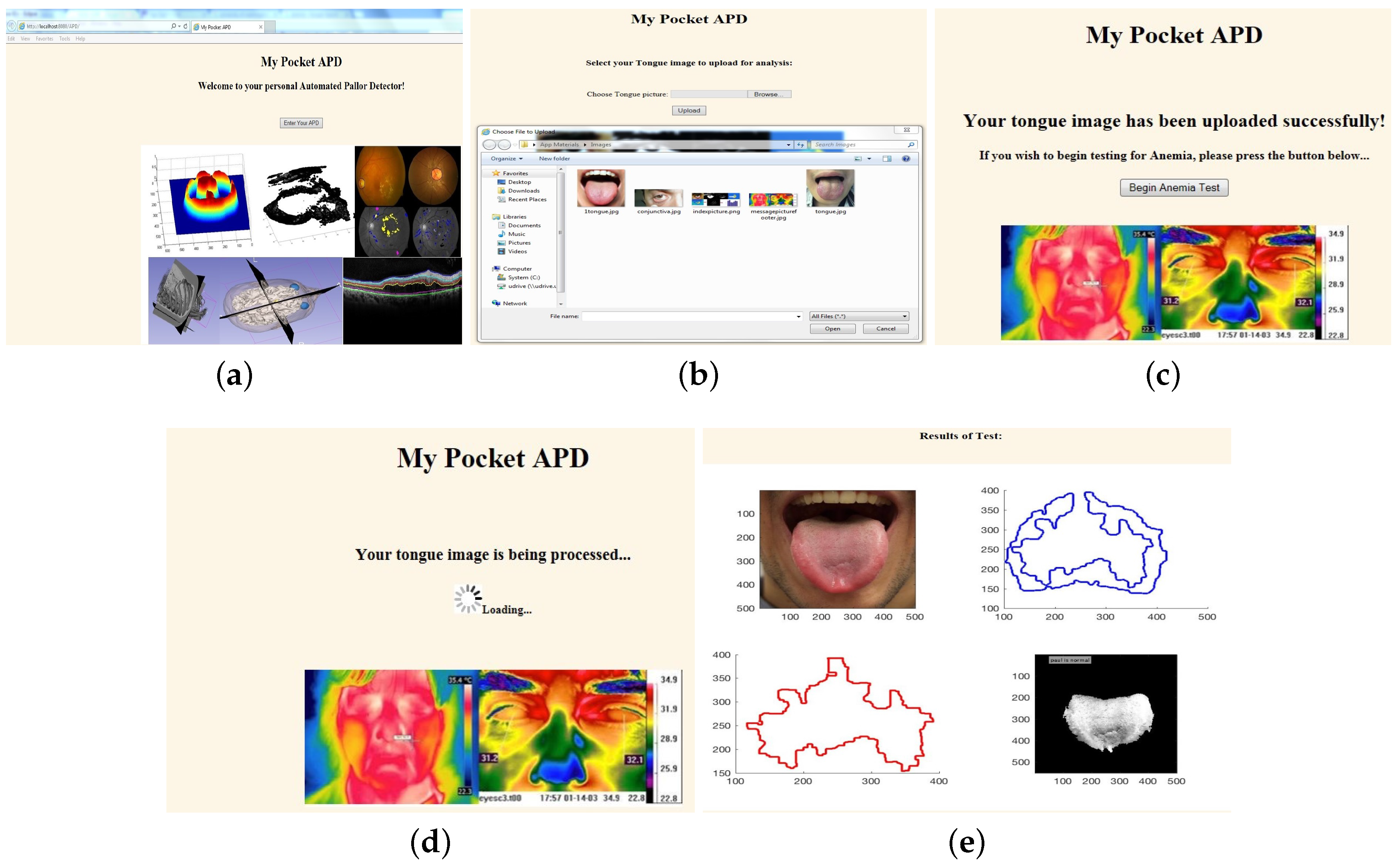
3.3. Application Front-End Design
3.3.1. Integrated Development Environment (IDE): Eclipse
3.3.2. Java Libraries
3.3.3. Web Application Archive (WAR) Deployment
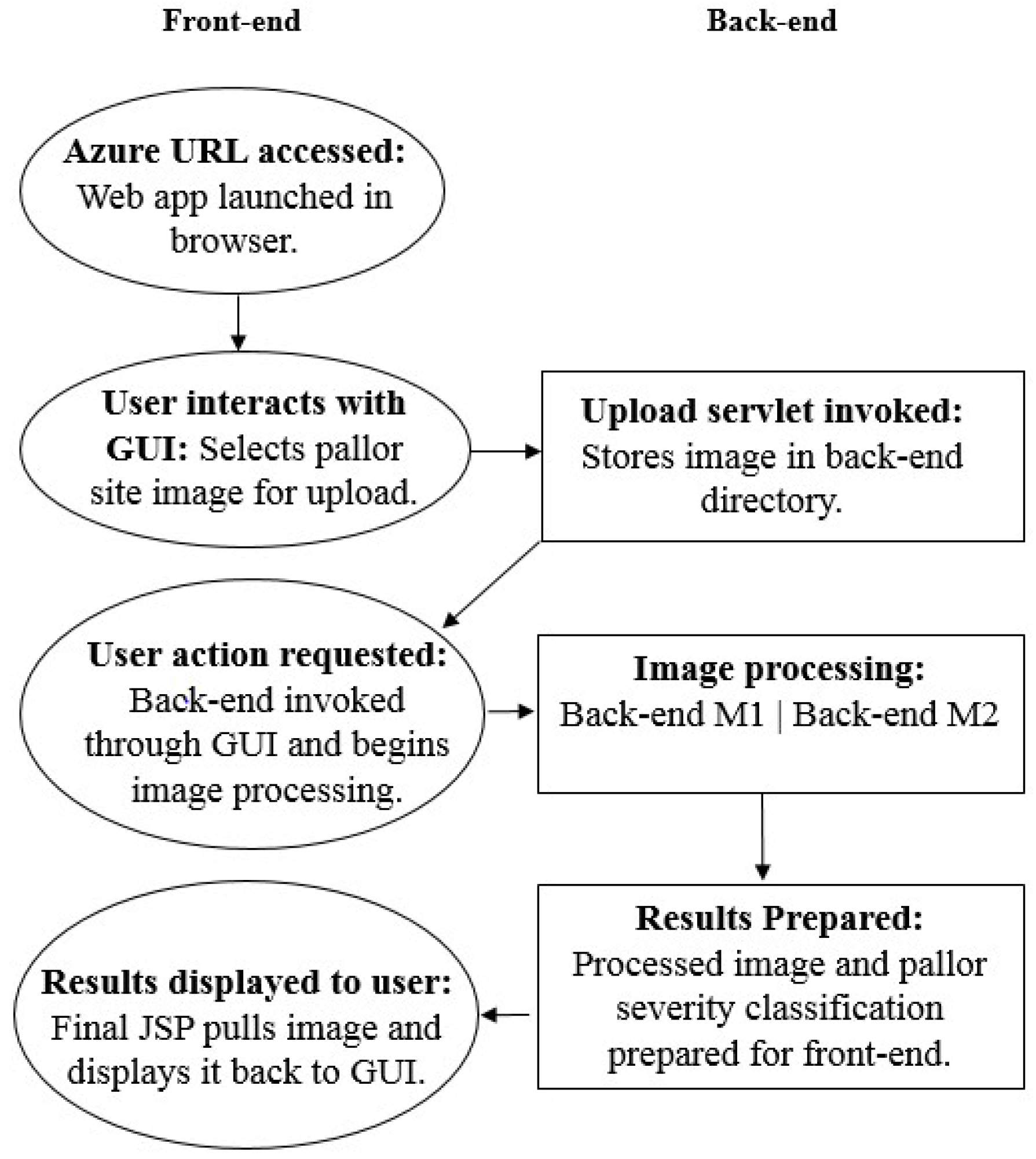
3.3.4. End-to-End Communication
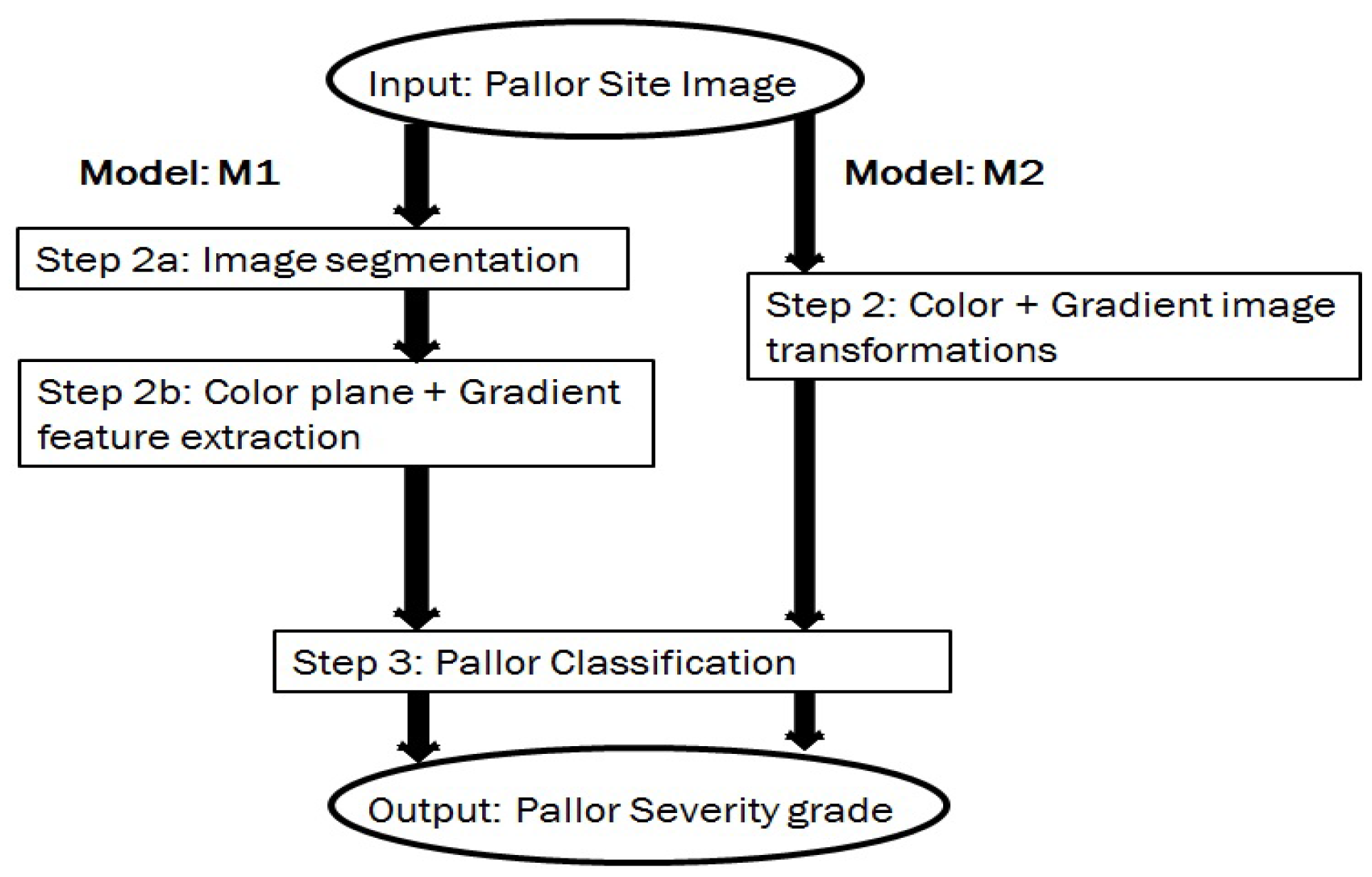
3.4. Back-End Methodology
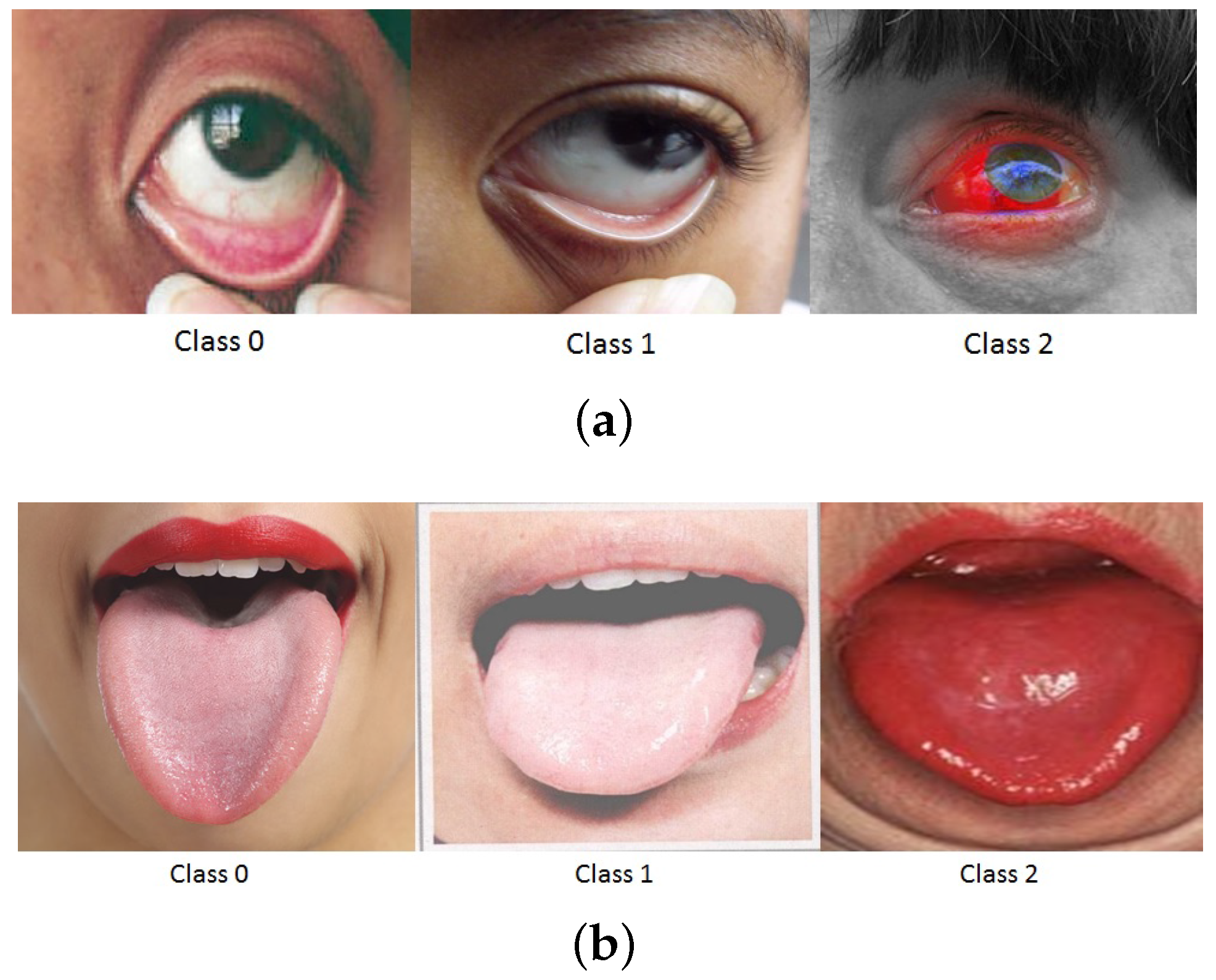
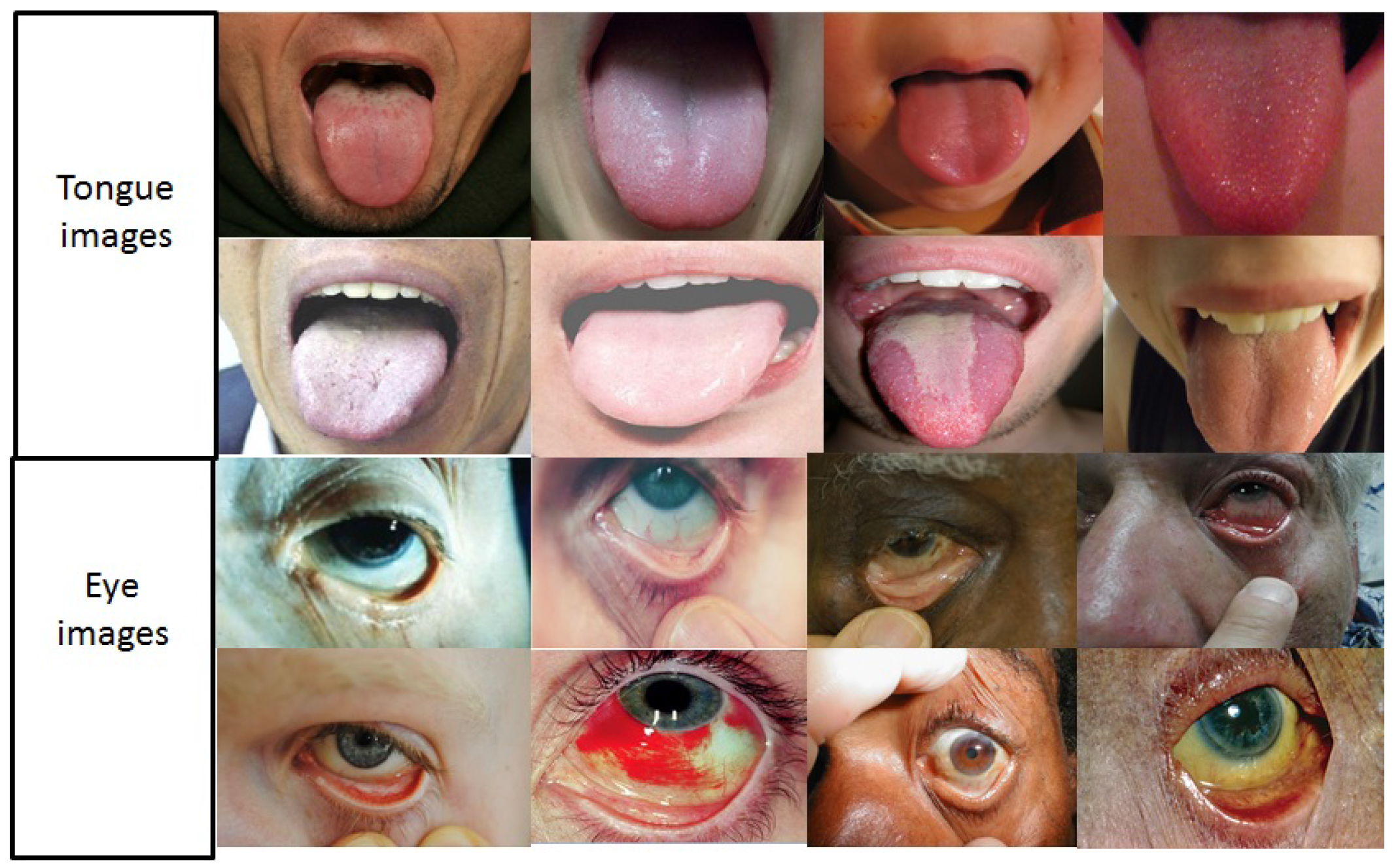
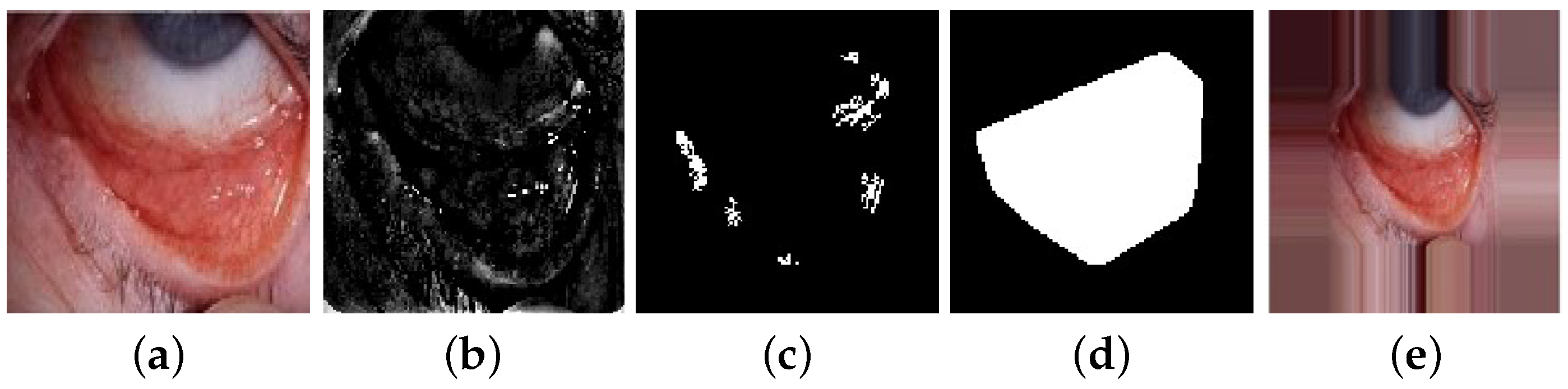
3.4.1. Image Segmentation
3.4.2. Color Planes and Gradient Feature Extraction
3.4.3. Classification
- kNN : The training data set is used to populate the feature space, followed by identification of ‘k’ nearest neighbors to each test sample, and identification of the majority class label [26]. In the training phase, the optimal value of ‘k’ that minimizes validation error is searched in the range [3:25], in steps of 2. For model , corresponding to each image I, 54 region-based features are extracted, which represent the feature space, followed by determination of pallor class label for each test image. Due to low computational complexity of this classifier, the final models are deployed for pallor classification using the kNN classifier.
- CNN: This category of classifiers, with high computational complexity, is motivated by the prior works in [5,6]. Here, each input image is subjected to several hidden layers of feature learning to generate an output vector of probability scores, for each image to belong to an output class label. For this analysis, we implement CNN architecture with the following 7 hidden layers: convolutional (C)-subsampling (S)-activation (A)-convolutional (C)-subsampling (S)-activation (A)-neural network (NN). Each C-layer convolves the input image with a set of kernels/filters, the S-layer performs pixel pooling, A-layer performs pixel scaling in the range [−1, 1] and the final NN-layer implements classification using 200 hidden neurons. Dropout was performed with probability of 0.5. Kernels for the low-dimensional model were selected as [3 × 3 × 3], while for the model they were [7 × 7 × 10], per convolutional layer. Pooling was performed with [2 × 2] and stride 2. The kernels/filters per hidden layer were randomly initialized, followed by error back-propagation from the training samples, finally resulting in trained activated feature maps (AFMs). For each test image, the CNN output corresponds to the output class with maximum probability score assigned by the NN-layer. Due to lack of data samples for CNN parameter tuning, the trained AFMs from the hidden layers are analyzed for qualitative feature learning, and to assess the significance of color planes and ROIs towards pallor classification.
4. Experiments and Results
4.1. Processing Speed Analysis
4.1.1. Front-End Upload Time
4.1.2. Back-end Model Processing Time
4.1.3. Back-end Model Processing Time
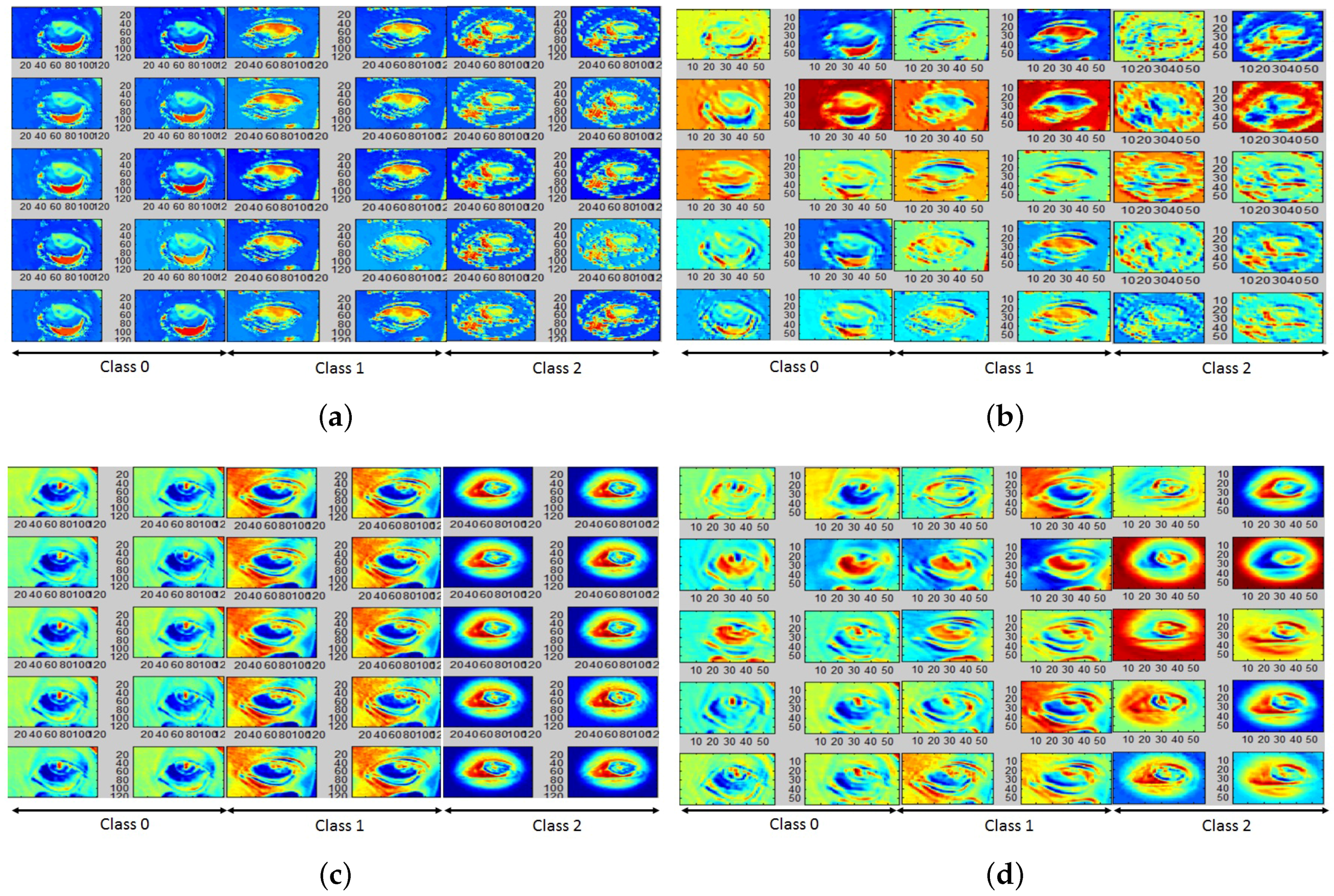
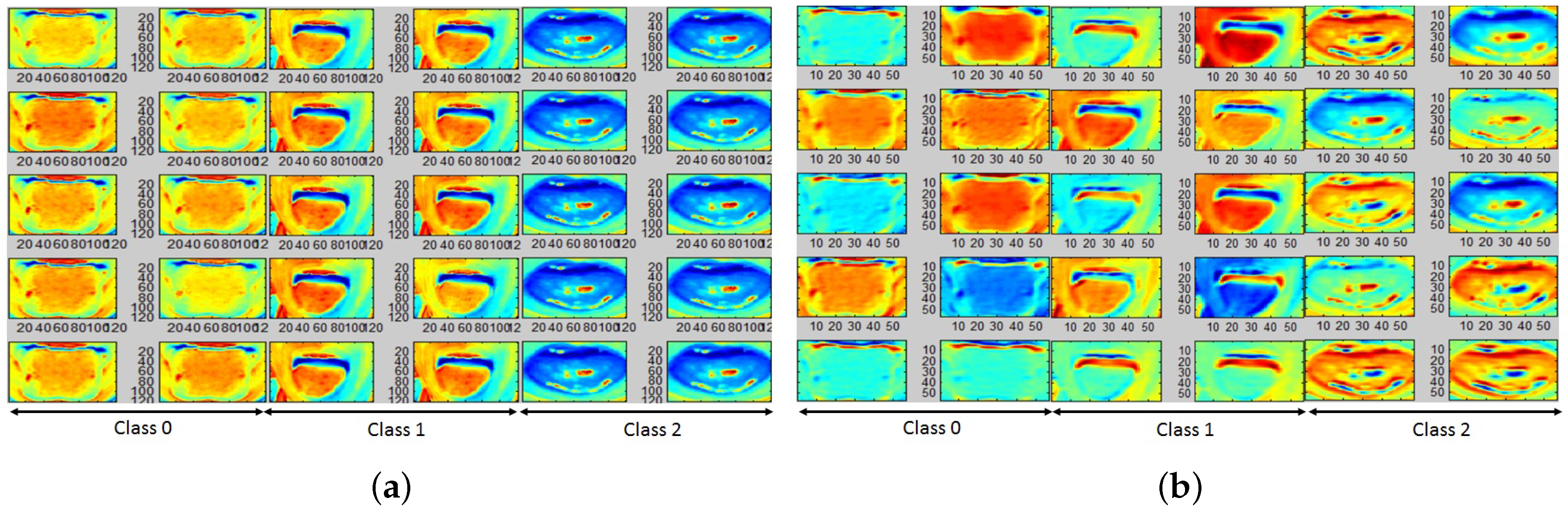
4.2. Color and Gradient-Based Feature Learning
4.3. Classification Performance Analysis
5. Conclusions and Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lewis, G.G.; Phillips, S.T. Quantitative Point-of-Care (POC) Assays Using Measurements of Time as the Readout: A New Type of Readout for mHealth. Method. Mol. Biol. 2015, 1256, 213–229. [Google Scholar]
- Centers for Medicare and Medicaid Services. National Health Expenditures 2014 Highlights. Available online: https://ccf.georgetown.edu/wp-content/uploads/2017/03/highlights.pdf (accessed on 15 May 2017).
- Diogenes, Y. Azure Data Security and Encryption Best Practices. Available online: https://docs.microsoft.com/en-us/azure/security/azure-security-data-encryption-best-practices (accessed on 28 April 2017).
- Roychowdhury, S.; Sun, D.; Bihis, M.; Ren, J.; Hage, P.; Rahman, H.H. Computer aided detection of anemia-like pallor. In Proceedings of the 2017 IEEE EMBS International Conference on Biomedical Health Informatics (BHI), Orlando, FL, USA, 16–19 February 2017; pp. 461–464. [Google Scholar]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Roychowdhury, S.; Ren, J. Non-deep CNN for multi-modal image classification and feature learning: An Azure-based model. Proceesings of the 2016 IEEE International Conference on Big Data (IEEE BigData), Washington, DC, USA, 5–8 December 2016. [Google Scholar]
- Roychowdhury, S.; Koozekanani, D.; Parhi, K. Dream: Diabetic retinopathy analysis using machine learning. IEEE J. Biomed. Health Inf. 2014, 18, 1717–1728. [Google Scholar] [CrossRef] [PubMed]
- Morales, S.; Naranjo, V.; Navea, A.; Alcaniz, M. Computer-Aided Diagnosis Software for Hypertensive Risk Determination Through Fundus Image Processing. IEEE J. Biomed. Health Inf. 2014, 18, 1757–1763. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-de Gea, J.M.; García-Mateos, G.; Fernández-Alemán, J.L.; Hernández-Hernández, J.L. A Computer-Aided Detection System for Digital Chest Radiographs. J. Healthc. Eng. 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- McLean, E.; Cogswell, M.; Egli, I.; Wojdyla, D.; De Benoist, B. Worldwide prevalence of anaemia, WHO vitamin and mineral nutrition information system, 1993–2005. Public Health Nutr. 2009, 12, 444–454. [Google Scholar] [CrossRef] [PubMed]
- Silva, R.M.d.; Machado, C.A. Clinical evaluation of the paleness: Agreement between observers and comparison with hemoglobin levels. Rev. Bras. Hematol. Hemoter. 2010, 32, 444–448. [Google Scholar] [CrossRef]
- Stevens, G.A.; Finucane, M.M.; De-Regil, L.M.; Paciorek, C.J.; Flaxman, S.R.; Branca, F.; Peña-Rosas, J.P.; Bhutta, Z.A.; Ezzati, M.; Group, N.I.M.S.; et al. Global, regional, and national trends in haemoglobin concentration and prevalence of total and severe anaemia in children and pregnant and non-pregnant women for 1995–2011: A systematic analysis of population-representative data. Lancet Glob. Health 2013, 1, e16–e25. [Google Scholar] [CrossRef]
- Benseñor, I.M.; Calich, A.L.G.; Brunoni, A.R.; Espírito-Santo, F.F.d.; Mancini, R.L.; Drager, L.F.; Lotufo, P.A. Accuracy of anemia diagnosis by physical examination. Sao Paulo Med. J. 2007, 125, 170–173. [Google Scholar] [CrossRef] [PubMed]
- Yalçin, S.S.; Ünal, S.; Gümrük, F.; Yurdakök, K. The validity of pallor as a clinical sign of anemia in cases with beta-thalassemia. Turk. J. Pediatr. 2007, 49, 408–412. [Google Scholar] [PubMed]
- Roychowdhury, S.; Emmons, M. A Survey of the Trends in Facial and Expression Recognition Databases and Methods. arXiv, 2015; arXiv:1511.02407. [Google Scholar]
- Shaik, K.B.; Ganesan, P.; Kalist, V.; Sathish, B.; Jenitha, J.M.M. Comparative study of skin color detection and segmentation in HSV and YCbCr color space. Procedia Comput. Sci. 2015, 57, 41–48. [Google Scholar] [CrossRef]
- Pujol, F.A.; Pujol, M.; Jimeno-Morenilla, A.; Pujol, M.J. Face detection based on skin color segmentation using fuzzy entropy. Entropy 2017, 19, 26. [Google Scholar] [CrossRef]
- Minaee, S.; Abdolrashidi, A.; Wang, Y. Iris recognition using scattering transform and textural features. In Proceedings of the 2015 IEEE Signal Processing and Signal Processing Education Workshop (SP/SPE), Salt Lake City, UT, USA, 9–12 Auguest 2015; pp. 37–42. [Google Scholar]
- Roychowdhury, S.; Koozekanani, D.D.; Parhi, K.K. Blood vessel segmentation of fundus images by major vessel extraction and subimage classification. IEEE J. Biomed. Health Inf. 2015, 19, 1118–1128. [Google Scholar]
- Roychowdhury, S.; Koozekanani, D.D.; Parhi, K.K. Automated detection of neovascularization for proliferative diabetic retinopathy screening. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 17–20 August 2016; pp. 1300–1303. [Google Scholar]
- Ginige, A.; Murugesan, S. Web engineering: A methodology for developing scalable, maintainable Web applications. Cut. IT J. 2001, 14, 24–35. [Google Scholar]
- Jadeja, Y.; Modi, K. Cloud computing-concepts, architecture and challenges. In Proceedings of the IEEE 2012 International Conference on Computing, Electronics and Electrical Technologies (ICCEET), Nagercoil, India, 21–22 March 2012; pp. 877–880. [Google Scholar]
- Roychowdhury, S.; Bihis, M. AG-MIC: Azure-Based Generalized Flow for Medical Image Classification. IEEE Access 2016, 4, 5243–5257. [Google Scholar] [CrossRef]
- Chieu, T.C.; Mohindra, A.; Karve, A.A.; Segal, A. Dynamic scaling of web applications in a virtualized cloud computing environment. In Proceedings of the IEEE International Conference on E-Business Engineering (ICEBE’09), Hangzhou, China, 9–11 September 2009; pp. 281–286. [Google Scholar]
- Kalantri, A.; Karambelkar, M.; Joshi, R.; Kalantri, S.; Jajoo, U. Accuracy and reliability of pallor for detecting anaemia: A hospital-based diagnostic accuracy study. PLoS ONE 2010, 5, e8545. [Google Scholar] [CrossRef] [PubMed]
- Cherkassky, V.; Mullier, F. Learning from Data; John Wiley and Sons: New York, NY, USA, 1998. [Google Scholar]
- Gonzalez, R.C.; Woods, R.E. Digital Image Processing, 2nd ed.; Addison-Wesley Longman Publishing Co., Inc.: Boston, MA, USA, 1992. [Google Scholar]
- Veksler, O. Machine Learning in Computer Vision. Available online: http://www.csd.uwo.ca/courses/CS9840a/Lecture2knn.pdf (accessed on 24 May 2017).
- The University of Utah. Spatial Filtering. Available online: http://www.coe.utah.edu/~cs4640/slides/Lecture5.pdf (accessed on 30 April 2017).

| Feature | # Features | Meaning |
|---|---|---|
| Pallor Site: | Eye | |
| Color Planes in | (6 × 3) = 18 | Max, mean, variance of pixels in red, green, blue, hue, saturation, intensity color planes. |
| Gradient intensity in | ||
| for | 5 | Max, min, mean, variance and intensity of pixels. |
| Frangi-filtered intensity in for | 4 | Max, mean, variance and intensity of pixels. |
| Color Planes in | (6 × 3) = 18 | Max, mean, variance of pixels in red, green, blue, hue, saturation, intensity color planes. |
| Gradient intensity in | ||
| for | 5 | Max, min, mean, variance and intensity of pixels. |
| Frangi-filtered intensity in for | 4 | Max, mean, variance and intensity of pixels. |
| Pallor Site: | Tongue | |
| Color Planes in | (6 × 3) = 18 | Max, mean, variance of pixels in red, green, blue, hue, saturation, intensity color planes. |
| Gradient intensity in | ||
| for | 5 | Max, min, mean, variance and intensity of pixels. |
| Frangi-filtered intensity in | ||
| for | 4 | Max, mean, variance and intensity of pixels. |
| Color Planes in | (6 × 3) = 18 | Max, mean, variance of pixels in red, green, blue, hue, saturation, intensity color planes. |
| Gradient intensity in | ||
| for | 5 | Max, min, mean, variance and intensity of pixels. |
| Frangi-filtered intensity in for | 4 | Max, mean, variance and intensity of pixels. |
| Time | Eye, | Eye, | Tongue, | Tongue, |
|---|---|---|---|---|
| (s) | 8.72 (7.6 × ) | 1.26 (0.33) | 31.49 (0.44) | 2.71 (0.57) |
| Time Complexity | O(6 × 62,500n) + O() + O() | O() | O(10 × 62,500n) + O() + O() | O() |
| t (s) | 8.74 (7.6 × ) | 1.30 (0.33) | 31.51 (0.04) | 3.94 (0.52) |
| 1.002 (5.9 × ) | 1.02 (0.003) | 1.00 (1.7 × ) | 1.08 (0.0014) | |
| 0.002 (5.9 × ) | 0.017 (0.003) | 0.006 (1.7 × ) | 0.01 (0.0014) |
| Model | , kNN | , kNN | , CNN | , CNN | ||||
|---|---|---|---|---|---|---|---|---|
| Task | 0/1,2 | 1/2 | 0/1,2 | 1/2 | 0/1,2 | 1/2 | 1/0,2 | 0/2 |
| PR | 0.85 | 0.57 | 0.87 | 0.61 | 0.74 | 0.50 | 0.51 | 0.94 |
| RE | 0.99 | 0.84 | 0.97 | 0.80 | 1.00 | 0.80 | 0.03 | 0.19 |
| Acc | 0.86 | 0.67 | 0.87 | 0.53 | 0.74 | 0.5 | 0.41 | 0.56 |
| AUC | 0.75 | 0.675 | 0.5 | 0.5 | 0.68 | 0.4 | 0.57 | 0.57 |
| Model | , kNN | , kNN | , CNN | , CNN | ||||
|---|---|---|---|---|---|---|---|---|
| Task | 0/1,2 | 1/2 | 0/1,2 | 1/2 | 1/0,2 | 0/2 | 2/0,1 | 0/1 |
| PR | 0.982 | 0.51 | 0.69 | 0.9 | 0.95 | 0.66 | 0.71 | 0.68 |
| RE | 1.00 | 0.53 | 0.9 | 0.98 | 1.00 | 1.00 | 0.55 | 0.69 |
| Acc | 0.982 | 0.61 | 0.65 | 0.88 | 0.95 | 0.66 | 0.54 | 0.57 |
| AUC | 0.83 | 0.574 | 0.57 | 0.51 | 0.83 | 0.5 | 0.65 | 0.5 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roychowdhury, S.; Hage, P.; Vasquez, J. Azure-Based Smart Monitoring System for Anemia-Like Pallor. Future Internet 2017, 9, 39. https://doi.org/10.3390/fi9030039
Roychowdhury S, Hage P, Vasquez J. Azure-Based Smart Monitoring System for Anemia-Like Pallor. Future Internet. 2017; 9(3):39. https://doi.org/10.3390/fi9030039
Chicago/Turabian StyleRoychowdhury, Sohini, Paul Hage, and Joseph Vasquez. 2017. "Azure-Based Smart Monitoring System for Anemia-Like Pallor" Future Internet 9, no. 3: 39. https://doi.org/10.3390/fi9030039




