Evaluation of the Microbial Viability of Soil Samples from Maize Crops in Freeze-Storage under Different Management Conditions in a Semi-Arid Climate
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Site
2.2. Soil Sampling and Analysis
2.3. DNA Extraction from Cultured Bacteria, PCR Amplification, Sequencing of 16S Ribosomal DNA, and Bacteria Identification
2.4. Biodiversity and Statistical Analysis
3. Results and Discussion
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| CT | conventional tillage |
| DS | direct seeding |
| DSC | direct seeding with a lopsided cover crop |
| OUT | operational taxonomic unit |
| rDNA | Ribosomal DNA |
| SIR | Substrate-Induced Respiration |
| TSA | Triptone Soy Agar |
References
- Verchot, L.V. Cold storage of a tropical soil decreases nitrification potential. Soil Sci. Soc. Am. J. 1999, 63, 1942–1944. [Google Scholar] [CrossRef]
- ISO (International Organization for Standardization). Sampling Part 6: Guidance on collection, handling and storage of soil for assessment of aerobic microbial processes in the laboratory. In Soil Quality; ISO: Geneva, Switzerland, 1993; pp. 10381–10386. [Google Scholar]
- OECD (Organisation for Economic Co-operation and Development). Draft Report of the OECD Workshop on Selection of Soil/Sediments; OECD: Belgirate, Italy, 1995; pp. 29–35. [Google Scholar]
- Stenberg, B.; Johansson, M.; Pell, M.; Sjodahl-Svensson, K.; Stenstrom, J.; Torstensson, L. Microbial biomass and activities in soil as affected by frozen and cold storage. Soil Biol. Biochem. 1998, 30, 393–402. [Google Scholar] [CrossRef]
- Pesaro, M.; Widmer, F.; Nicollier, G.; Zeyer, J. Effects of freeze-thaw stress during soil storage on microbial communities and methidathion degradation. Soil Biol. Biochem. 2003, 35, 1049–1061. [Google Scholar] [CrossRef]
- Christensen, S.; Tiedje, J.M. Brief and vigorous N2O production by soil at spring thaw. Eur. J. Soil Sci. 1990, 41, 1–4. [Google Scholar] [CrossRef]
- Ludwig, B.; Wolf, I.; Teepe, R. Contribution of nitrification and denitrification to the emission of N2O in a freeze-thaw event in an agricultural soil. J. Plant Nutr. 2004, 167, 678–684. [Google Scholar] [CrossRef]
- Sharma, S.; Szele, Z.; Schilling, R.; Munch, J.C.; Schloter, M. Influence of freeze-thaw stress on the structure and function of microbial communities and denitrifying populations in soil. Appl. Environ. Microbiol. 2006, 72, 2148–2154. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, M; Ka, J.O.; Mohn, W.W. Effects of low temperature and freeze-thaw cycles on hydrocarbon biodegradation in Arctic tundra soil. Appl. Environ. Microbiol. 2001, 67, 5107–5112. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.P.E. Handling and storage of soils for pesticide experiments. In Pesticide Effects on Soil Microflora; Somerville, L., Greaves, M.P., Eds.; Taylor and Francis: London, UK, 1987; pp. 45–60. [Google Scholar]
- Trabue, S.L.; Palmquist, D.E.; Lydick, T.M.; Singles, S.K. Effects of soil storage on the microbial community and degradation of metsulfuron-methyl. J. Agric. Food Chem. 2006, 54, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Morley, C.R.; Trofymow, J.A.; Coleman, C.; Cambardella, C. Effects of freeze-thaw stress on bacterial populations in soil microcosms. Microb. Ecol 1983, 9, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Rissanen, A.J.; Kurhela, E.; Aho, T.; Oittinen, T.; Tiirola, M. Storage of environmental samples for guaranteeing nucleic acid yields for molecular microbiological studies. Appl. Microbiol. Biotechnol. 2010, 88, 977–984. [Google Scholar] [CrossRef] [PubMed]
- Tatangelo, V.; Franzetti, A.; Gandolfi, I.; Bestetti, G.; Ambrosini, R. Effect of preservation method on the assessment of bacterial community structure in soil and water samples. FEMS Microbiol. Lett. 2014, 356, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chapman, S.J.; Yao, H. The effect of storage on microbial activity and bacterial community structure of drained and flooded paddy soil. J. Soil Sediments 2015, 15, 880–889. [Google Scholar] [CrossRef]
- Rubin, B.E.; Gibbons, S.M.; Kennedy, S.; Hampton-Marcell, J.; Owens, S.; Gilbert, J.A. Investigating the impact of storage conditions on microbial community composition in soil samples. PLoS ONE 2013, 8, e70460. [Google Scholar] [CrossRef] [PubMed]
- Janssen, P.H.; Yates, P.S.; Grinton, B.E.; Taylor, P.M.; Sait, M. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 2002, 68, 2391–2396. [Google Scholar] [CrossRef] [PubMed]
- Ellis, R.J.; Morgan, P.; Weightman, A.J.; Fry, J.C. Cultivation-dependent and -independent approaches for determining bacterial diversity in heavy-metal-contaminated soil. Appl. Environ. Microbiol. 2003, 69, 3223–3230. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, G.; Sani, R.K. Molecular techniques to assess microbial community structure, function, and dynamics in the environment. In Microbes and Microbial Technology; Ahmad, I., Ahmad, F., Pichtel, J., Eds.; Springer: New York, NY, USA, 2011; pp. 29–57. [Google Scholar]
- Hugenholtz, P.; Goebel, B.M.; Pace, N.R. Impact of culture-Independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 1998, 180, 4765–4774. [Google Scholar] [PubMed]
- Edenborn, S.L.; Sexstone, A.J. DGGE fingerprinting of culturable soil bacterial communities complements culture-independent analyses. Soil Biol. Biochem. 2007, 39, 1570–1579. [Google Scholar] [CrossRef]
- Kisand, V.; Wikner, J. Combining culture-dependent and -independent methodologies for estimation of richness of estuarine bacterioplankton consuming riverine dissolved organic matter. Appl. Environ. Microbiol. 2003, 69, 3607–3616. [Google Scholar] [CrossRef] [PubMed]
- Imfeld, G.; Vuilleumier, S. Measuring the effects of pesticides on bacterial communities in soil: A critical review. Eur. J. Soil Biol. 2012, 49, 22–30. [Google Scholar] [CrossRef]
- Wang, G.C.Y.; Wang, Y. Frequency of formation of chimeric molecules as a consequence of PCR coamplification of 16S rRNA genes from mixed bacterial genomes. Appl. Environ. Microbiol. 1997, 63, 4645–4650. [Google Scholar] [PubMed]
- Chroni, C.; Kyriacou, A.; Manios, T.; Lasaridi, K.E. Investigation of the microbial community structure and activity as indicators of compost stability and composting process evolution. Bioresour. Technol. 2009, 100, 3745–3750. [Google Scholar] [CrossRef] [PubMed]
- Pham, V.H.; Kim, J. Cultivation of unculturable soil bacteria. Trends Biotechnol. 2012, 30, 475–484. [Google Scholar] [CrossRef] [PubMed]
- Truu, M.; Juhanson, J.; Truu, J. Microbial biomass, activity and community composition in constructed wetlands. Sci. Total Environ. 2009, 407, 3958–3971. [Google Scholar] [CrossRef] [PubMed]
- Fakruddin, M.; Mannan, K. Methods for analyzing diversity of microbial communities in natural environments. Ceylon J. Sci. 2013, 42, 19–33. [Google Scholar] [CrossRef]
- Da Silva, P.C.G.; Foloni, J.S.S.; Fabris, L.B.; Tiritan, C.S. Fitomassa e relação C/N em consórcios de sorgo e milho com espécies de cobertura. Pesqui. Agropecu. Bras. 2009, 44, 1504–1512. (In Portuguese) [Google Scholar] [CrossRef]
- Dignam, B.E.; O’Callaghan, M.; Condron, L.M.; Raaijmakers, J.M.; Kowalchuk, G.A.; Wakelin, S.A. Challenges and opportunities in harnessing soil disease suppressiveness for sustainable pasture production. Soil Biol. Biochem. 2016, 95, 100–111. [Google Scholar] [CrossRef]
- Bahadur, I.; Maurya, B.R.; Kumar, A.; Meena, V.S.; Raghuwanshi, R. Towards the soil sustainability and potassium-solubilizing microorganisms. In Potassium Solubilizing Microorganisms for Sustainable Agriculture; Springer: New Delhi, India, 2016; pp. 255–266. [Google Scholar]
- Kumari, S.; Varma, A.; Tuteja, N.; Choudhary, D.K. Bacterial ACC-deaminase: An Eco-friendly Strategy to Cope Abiotic Stresses for Sustainable Agriculture. In Plant-Microbe Interaction: An Approach to Sustainable Agriculture; Springer: Singapore, 2016; pp. 165–185. [Google Scholar]
- Ciancio, A.; Corné, M.J.P.; Jesús, M.-B. Editorial: Harnessing useful rhizosphere microorganisms for pathogen and pest biocontrol. Front. Microbiol. 2016, 7, 1620. [Google Scholar] [CrossRef] [PubMed]
- Embarcadero-Jiménez, S.; Flor, N.R.-O.; Wang, E.N. Bacterial communities estimated by pyrosequencing in the soils of chinampa, a traditional sustainable agro-ecosystem in Mexico. J. Soils Sediments 2016, 16, 1001–1011. [Google Scholar] [CrossRef]
- Bender, S.F.; Cameron, W.; Van der Marcel, G.A.H. An underground revolution: biodiversity and soil ecological engineering for agricultural sustainability. Trends Ecol. Evol. 2016, 31, 440–452. [Google Scholar] [CrossRef] [PubMed]
- Keesstra, S.D.; Quinton, J.N.; van der Putten, W.H.; Bardgett, R.D.; Fresco, L.O. The significance of soils and soil science towards realization of the United Nations Sustainable Development Goals. Soil 2016, 2, 111. [Google Scholar] [CrossRef]
- Wolff, F.; Timo, K. The UN Convention on biological diversity and soils: Status and future options. In International Yearbook of Soil Law and Policy 2016; Springer: Cham, Switzerland, 2017; pp. 129–148. [Google Scholar]
- UNESCO (The United Nations Educational, Scientific and Cultural Organization). Map of the World Distribution of Arid Regions; MAB Technical Notes 7; UNESCO: Paris, France, 1977. [Google Scholar]
- Muñoz, A.; López-Piñeiro, A.; Ramírez, M. Soil quality attributes as influenced by conservation management regimes in semi-arid regions, south-western Spain. Soil Tillage Res. 2007, 95, 255–265. [Google Scholar] [CrossRef]
- Ron, Z.; Kohler, R.E.; Davis, B.D. Polysomes extracted from Escherichia coli by freeze-thaw-lysozyme lysis. Science 1966, 153, 1119–1120. [Google Scholar] [CrossRef] [PubMed]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Goodfellow, E.S.M., Stackebrant, E., Eds.; John Wiley & Sons Ltd.: London, UK, 1991; pp. 115–175. [Google Scholar]
- González, J.M.; Portillo, M.C.; Saiz-Jimenez, C. Multiple displacement amplification as a pre-polymerase chain reaction (pre-PCR) to process difficult to amplify samples and low copy number sequences from natural environments. Environ. Microbiol. 2005, 7, 1024–1028. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Camargo, F.A.O.; Okeke, B.C.; Bento, F.M.; Frankenberger, W.T. Diversity of chromium-resistant bacteria isolated from soils contaminated with dichromate. Appl. Soil Ecol. 2005, 29, 193–202. [Google Scholar] [CrossRef]
- Shen, T.; Urrutia Benet, G.; Brul, S.; Knorr, D. Influence of high-pressure–low-temperature treatment on the inactivation of Bacillus subtilis cells. Innov. Food Sci. Emerg. Technol. 2005, 6, 271–278. [Google Scholar] [CrossRef]
- Shen, T.; Bos, A.P.; Brul, S. Assessing freeze–thaw and high pressure low temperature induced damage to Bacillus subtilis cells with flow cytometry. Innov. Food Sci. Emerg. Technol. 2009, 10, 9–15. [Google Scholar] [CrossRef]
- Seki, K.; Horikawa, D.D. Diversity of Tardigrada. In Biodiversity in Agricultural Production Systems; Benckiser, G., Schnell, S., Eds.; CRC/Taylor and Francis: Boca Raton, FL, USA; London, UK, 2007; pp. 237–247. [Google Scholar]
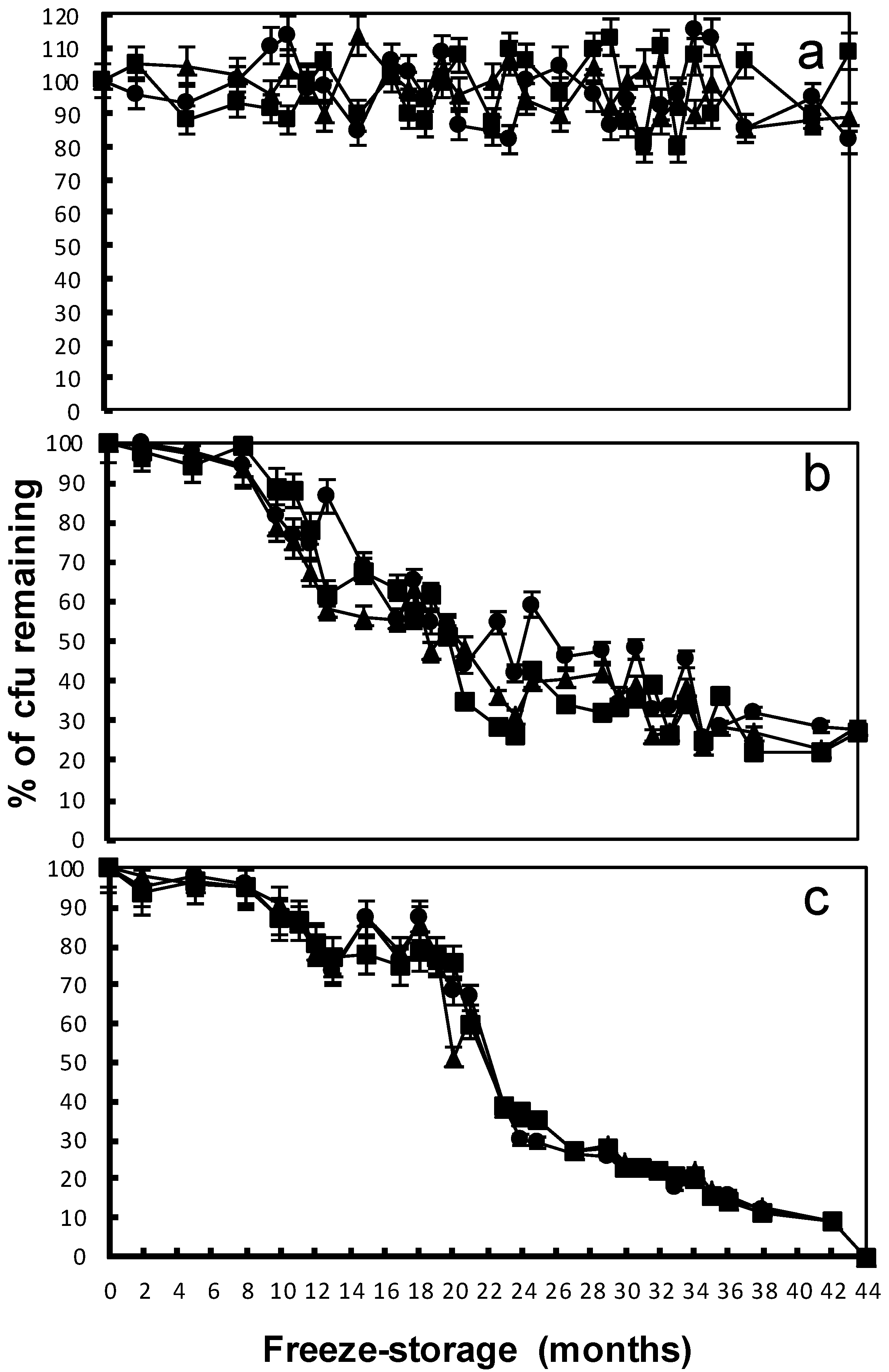
 ); after eight months of freeze-storage (
); after eight months of freeze-storage (  ); and after 44 months of freeze-storage (
); and after 44 months of freeze-storage (  ). Columns in the same treatment with the same lower case letter were not significantly different at the p < 0.05 level. Columns for the same freeze-storage time in different soils with the same capital letter were not significantly different at the p < 0.05 level.
). Columns in the same treatment with the same lower case letter were not significantly different at the p < 0.05 level. Columns for the same freeze-storage time in different soils with the same capital letter were not significantly different at the p < 0.05 level.
 ); after eight months of freeze-storage (
); after eight months of freeze-storage (  ); and after 44 months of freeze-storage (
); and after 44 months of freeze-storage (  ). Columns in the same treatment with the same lower case letter were not significantly different at the p < 0.05 level. Columns for the same freeze-storage time in different soils with the same capital letter were not significantly different at the p < 0.05 level.
). Columns in the same treatment with the same lower case letter were not significantly different at the p < 0.05 level. Columns for the same freeze-storage time in different soils with the same capital letter were not significantly different at the p < 0.05 level.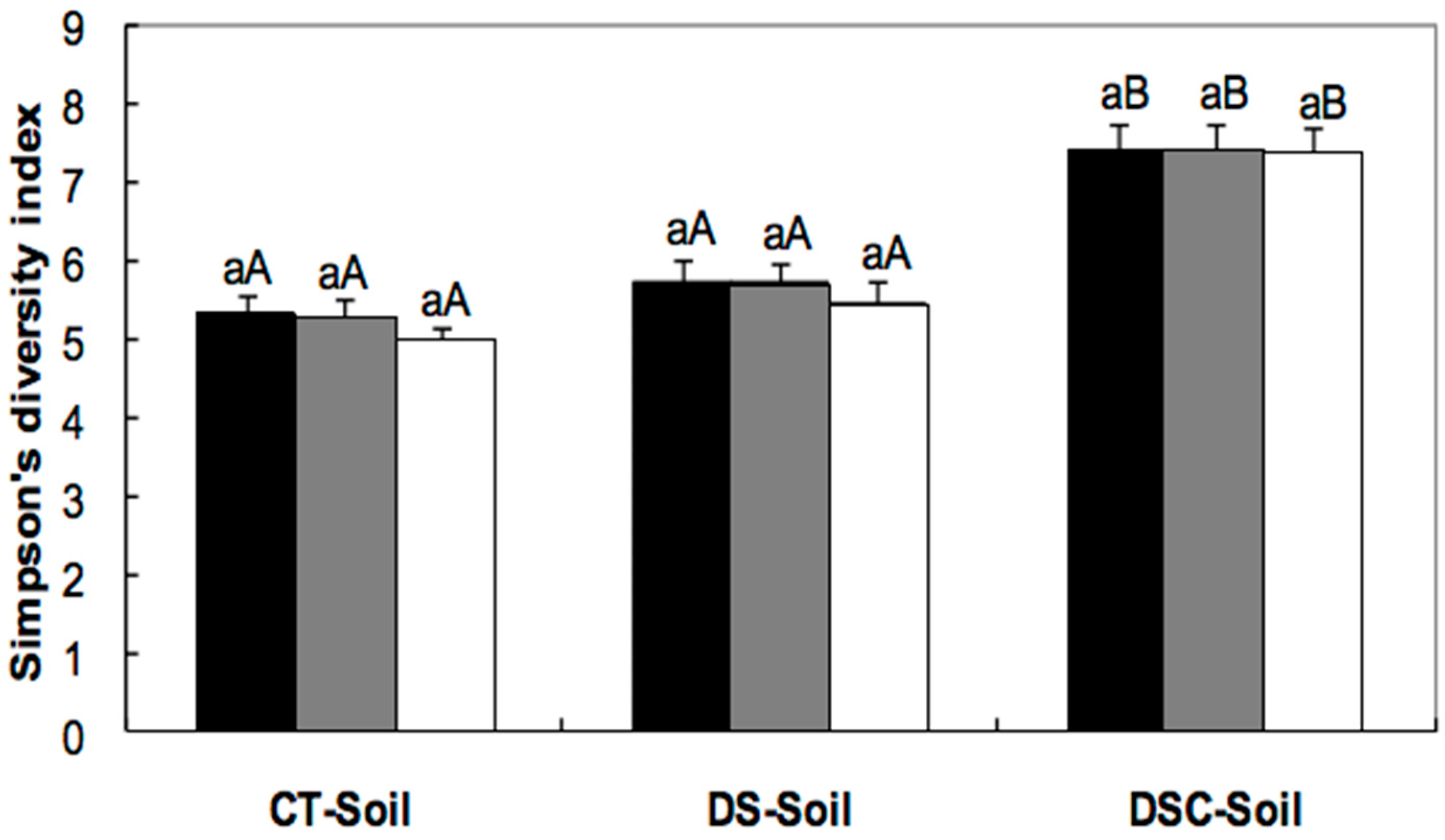
| Type | Colony Morphology | Spores | Cell Shape | Gram | Microorganism (OTU) † | ||||
|---|---|---|---|---|---|---|---|---|---|
| Form | Margin | Texture | Opacity | Color | |||||
| T1 | Circular | Entire | Shiny | Opaque | White | No | Cocci-rods | + | Arthrobacter dextranolyticus |
| T2 | Circular | Entire | Shiny | Opaque | Yellow | No | Cocci-rods | + | Arthrobacter nicotinovorans |
| T3 | Rhizoid | Curled | Wrinkled | Opaque | Beige | Yes | Rods | + | Bacillus mycoides |
| T4 | Irregular | Entire | Granular | Opaque | White | Yes | Rods | + | Bacillus megaterium |
| T5 | Irregular | Entire | Mucoid | Opaque | Beige | Yes | Rods | + | Bacillus simplex |
| T6 | Irregular | Lobate | Mucoid | Translucent | Beige | Yes | Rods | + | Bacillus subtilis |
| T7 | Irregular | Lobate | Granular | Translucent | Beige | Yes | Rods | + | Bacillus thuringiensis |
| T8 | Circular | Entire | Rough | Opaque | Beige | Yes | Rods | + | Bacillus weihenstephanensis |
| T9 | Irregular | Entire | Smooth | Opaque | Beige | No | Rods | - | Chryseobacterium indologenes |
| T10 | Circular | Curled | Wrinkled | Opaque | Violet | No | Rods | - | Janthinobacterium lividum |
| T11 | Circular | Entire | Smooth | Opaque | White | Yes | Rods | + | Paenibacillus polymyxa |
| T12 | Circular | Entire | Shiny | Translucent | White | No | Rods | - | Pantoea agglomerans |
| T13 | Irregular | Entire | Smooth | Transparent | n.a. | No | Rods | - | Pseudomonas cedrella |
| T14 | Circular | Entire | Shiny | Opaque | White | No | Rods | - | Pseudomonas filiscindens |
| T15 | Irregular | Entire | Rough | Opaque | Beige | No | Rods | - | Pseudomonas fluorecens |
| T16 | Circular | Entire | Shiny | Opaque | Brown | No | Rods | - | Pseudomonas jessenii |
| T17 | Circular | Entire | Shiny | Translucent | Brown | No | Rods | - | Pseudomonas mediterranea |
| T18 | Circular | Entire | Shiny | Opaque | White | No | Rods | - | Pseudomonas mosselii |
| T19 | Circular | Entire | Shiny | Opaque | Beige | No | Rods | - | Pseudomonas synxantha |
| T20 | Circular | Entire | Dull | Opaque | White | No | Cocci | + | Staphylococcus epidermidis |
| T21 | Irregular | Undulated | Wrinkled | Opaque | Beige | Yes | Large rods | + | Streptomyces scabrisporus |
| T22 | Circular | Entire | Dusty | Opaque | Beige | Yes | Large rods | + | Streptomyces flavovirens |
| T23 | Circular | Entire | Dusty | Opaque | Grey | Yes | Large rods | + | Streptomyces xanthophaeus |
| T24 | Circular | Entire | Dusty | Opaque | White | Yes | Large rods | + | Streptomyces ciscaucasicus |
| T25 | Circular | Entire | Dusty | Opaque | Brown | Yes | Large rods | + | Streptomyces griseoaurantiacus |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramírez, M.; Muñoz, A.; López-Piñeiro, A.; Albarrán, Á.; Peña, D.; Nunes, J.M.R.; Gama, J.; Loures, L. Evaluation of the Microbial Viability of Soil Samples from Maize Crops in Freeze-Storage under Different Management Conditions in a Semi-Arid Climate. Sustainability 2017, 9, 690. https://doi.org/10.3390/su9050690
Ramírez M, Muñoz A, López-Piñeiro A, Albarrán Á, Peña D, Nunes JMR, Gama J, Loures L. Evaluation of the Microbial Viability of Soil Samples from Maize Crops in Freeze-Storage under Different Management Conditions in a Semi-Arid Climate. Sustainability. 2017; 9(5):690. https://doi.org/10.3390/su9050690
Chicago/Turabian StyleRamírez, Manuel, Ana Muñoz, Antonio López-Piñeiro, Ángel Albarrán, David Peña, José Manuel Rato Nunes, José Gama, and Luis Loures. 2017. "Evaluation of the Microbial Viability of Soil Samples from Maize Crops in Freeze-Storage under Different Management Conditions in a Semi-Arid Climate" Sustainability 9, no. 5: 690. https://doi.org/10.3390/su9050690






