Integrating Airborne LiDAR and Optical Data to Estimate Forest Aboveground Biomass in Arid and Semi-Arid Regions of China
Abstract
:1. Introduction
2. Materials and Methods
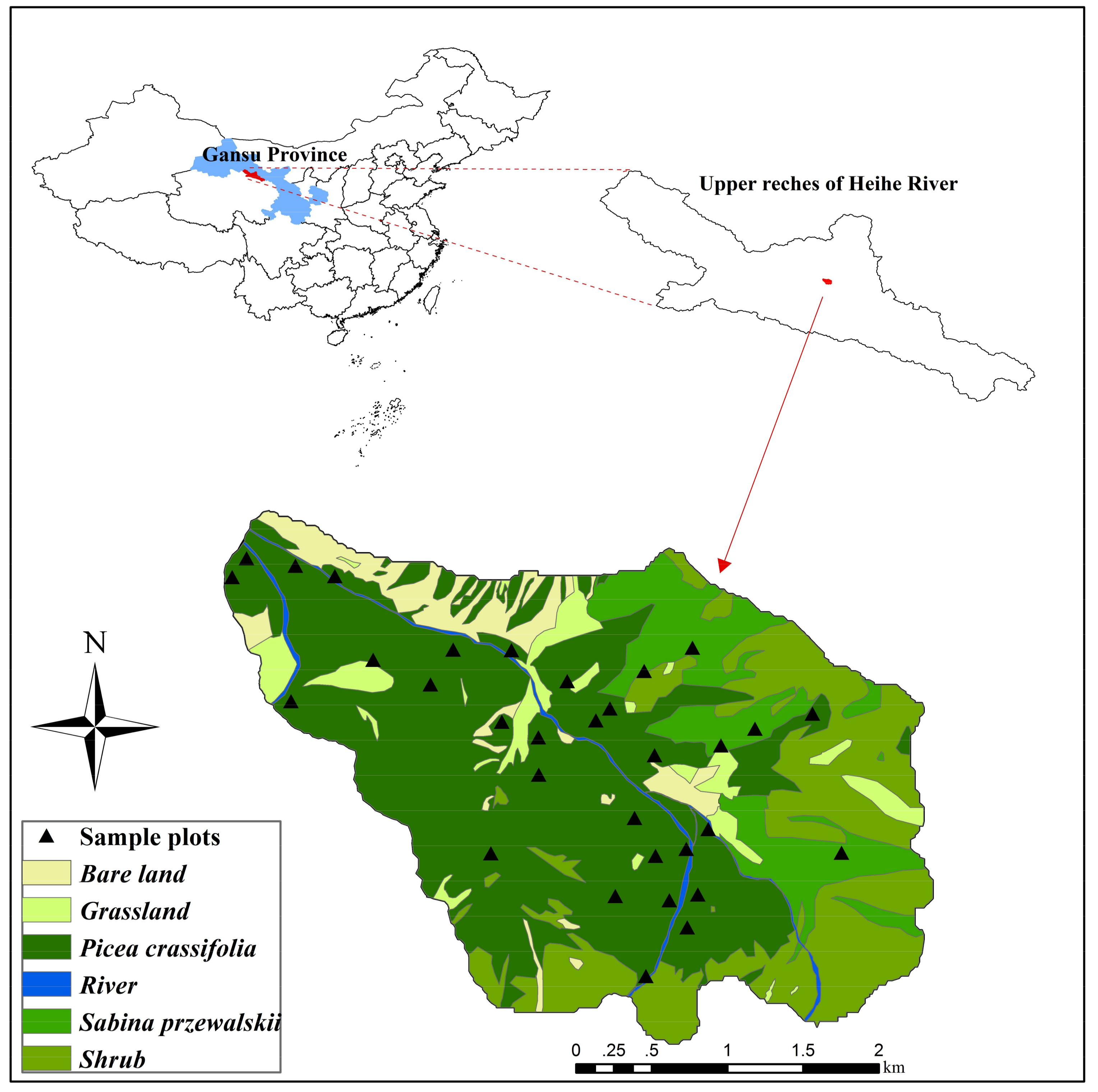
2.1. Study Area
2.2. Field Measurement
2.3. Optical Data
2.4. Airborne LiDAR Data
2.5. Optical Features Extraction for Forest Biomass Estimation
2.5.1. Vegetation Indices
2.5.2. Texture Information
2.6. LiDAR Features Extraction for Forest Biomass Estimation
2.6.1. LiDAR-Derived Variables
2.6.2. Terrain Variables
2.7. Statistical Analysis and Modeling
3. Results
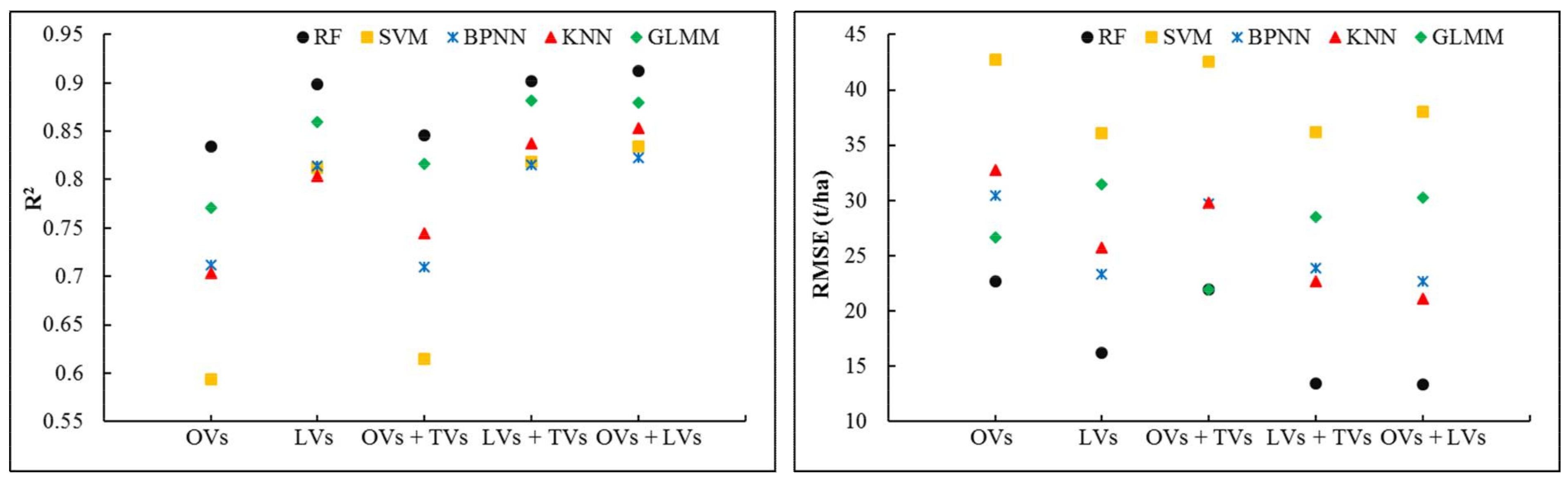
3.1. Comparison of Forest AGB Estimation Using Various Data Types
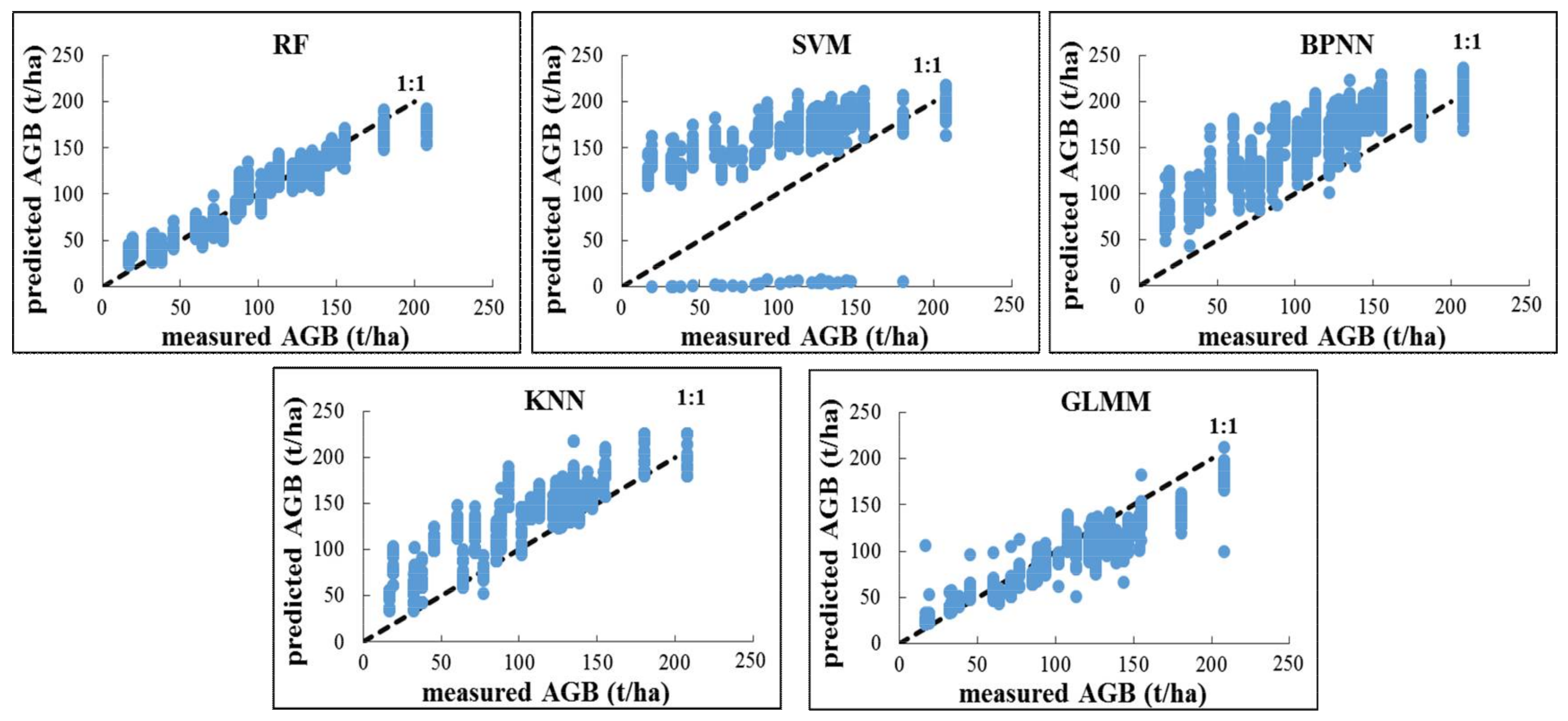
3.2. Optimal Prediction Methods for Forest AGB Estimation
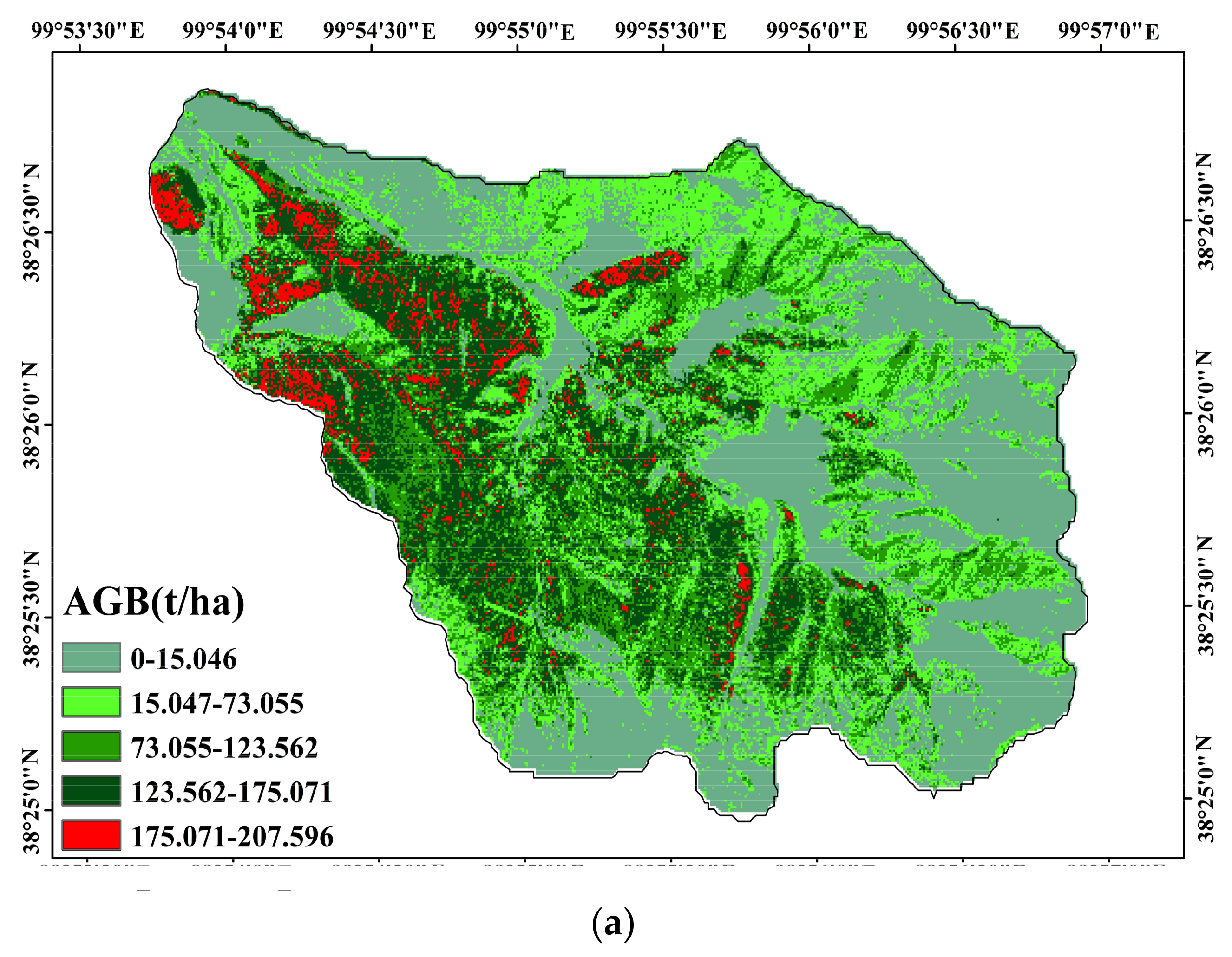
3.3. Mapping the Forest AGB Distribution
4. Discussion
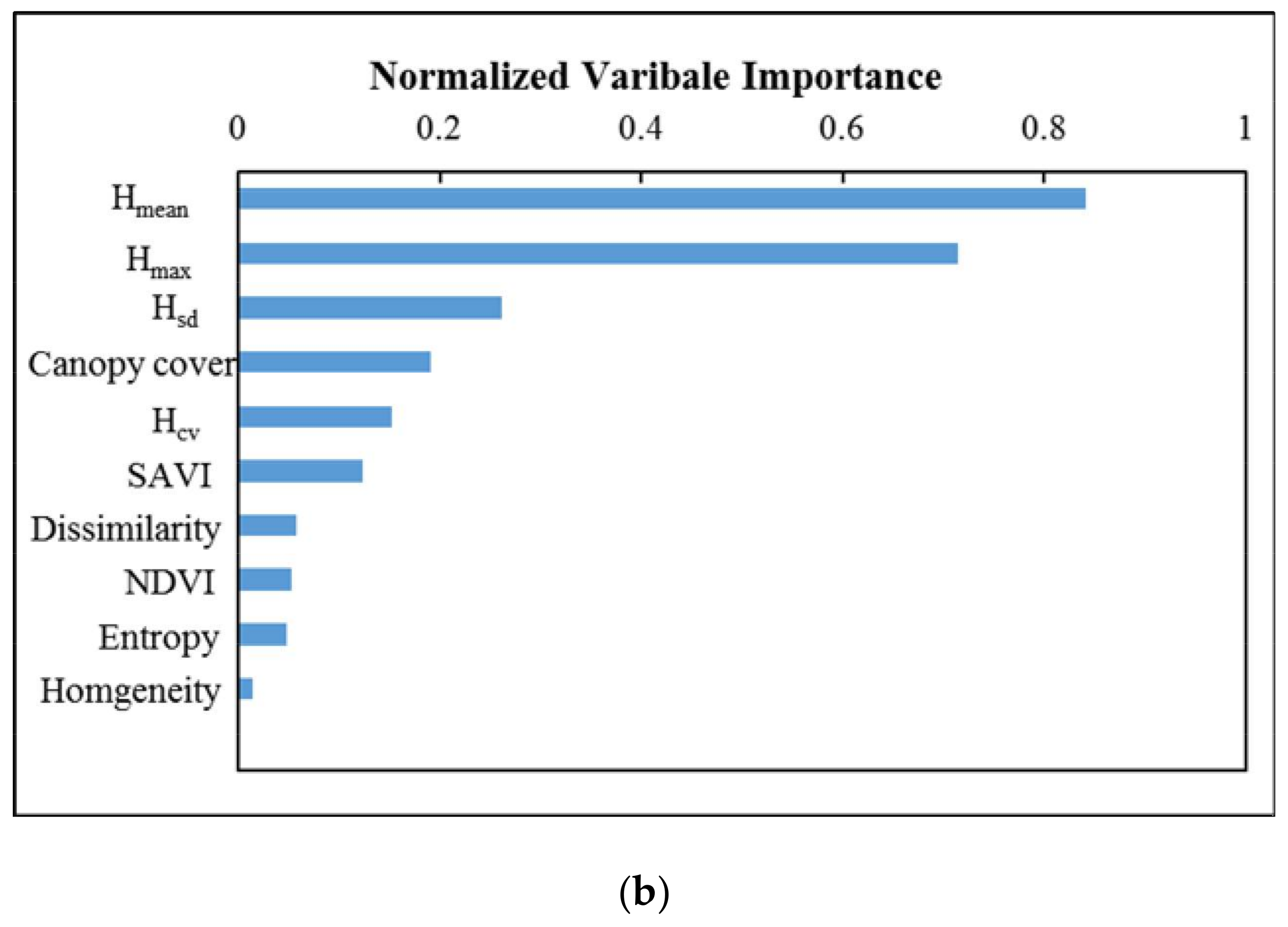
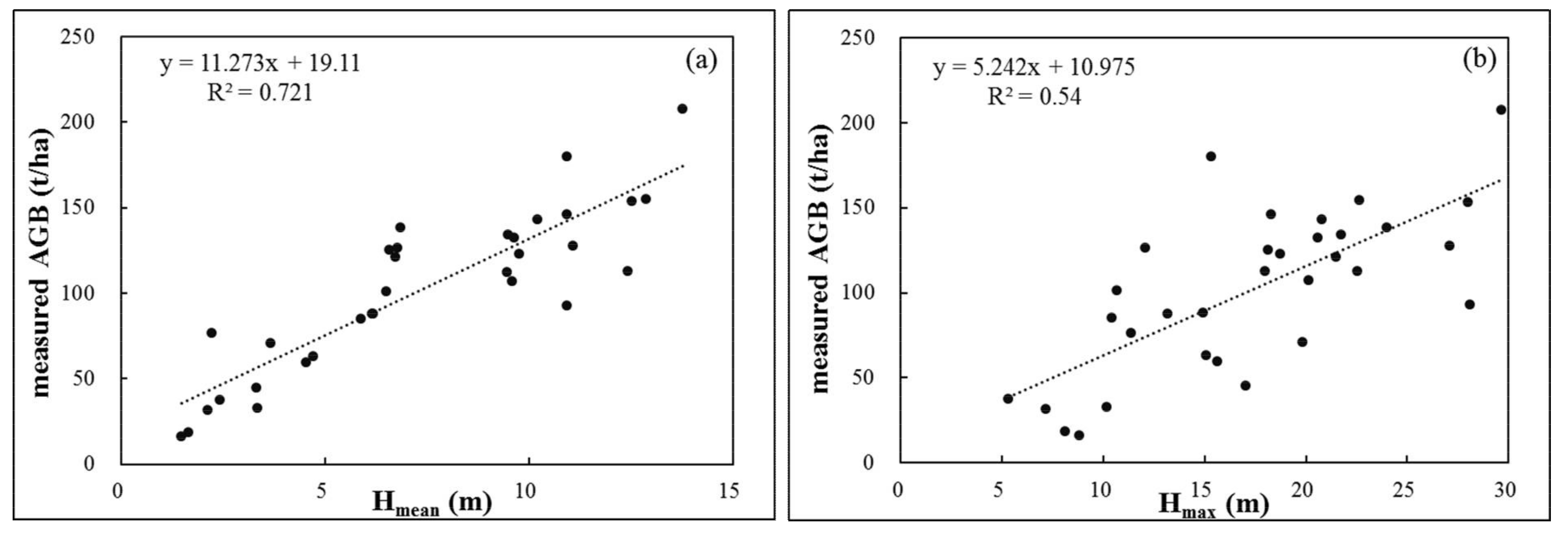
4.1. Importance of Predictive Factors for Model Performance
4.1.1. Single vs. Integrated Data Sources for Estimating Forest AGB
4.1.2. Influence of the Statistical Method on Estimation
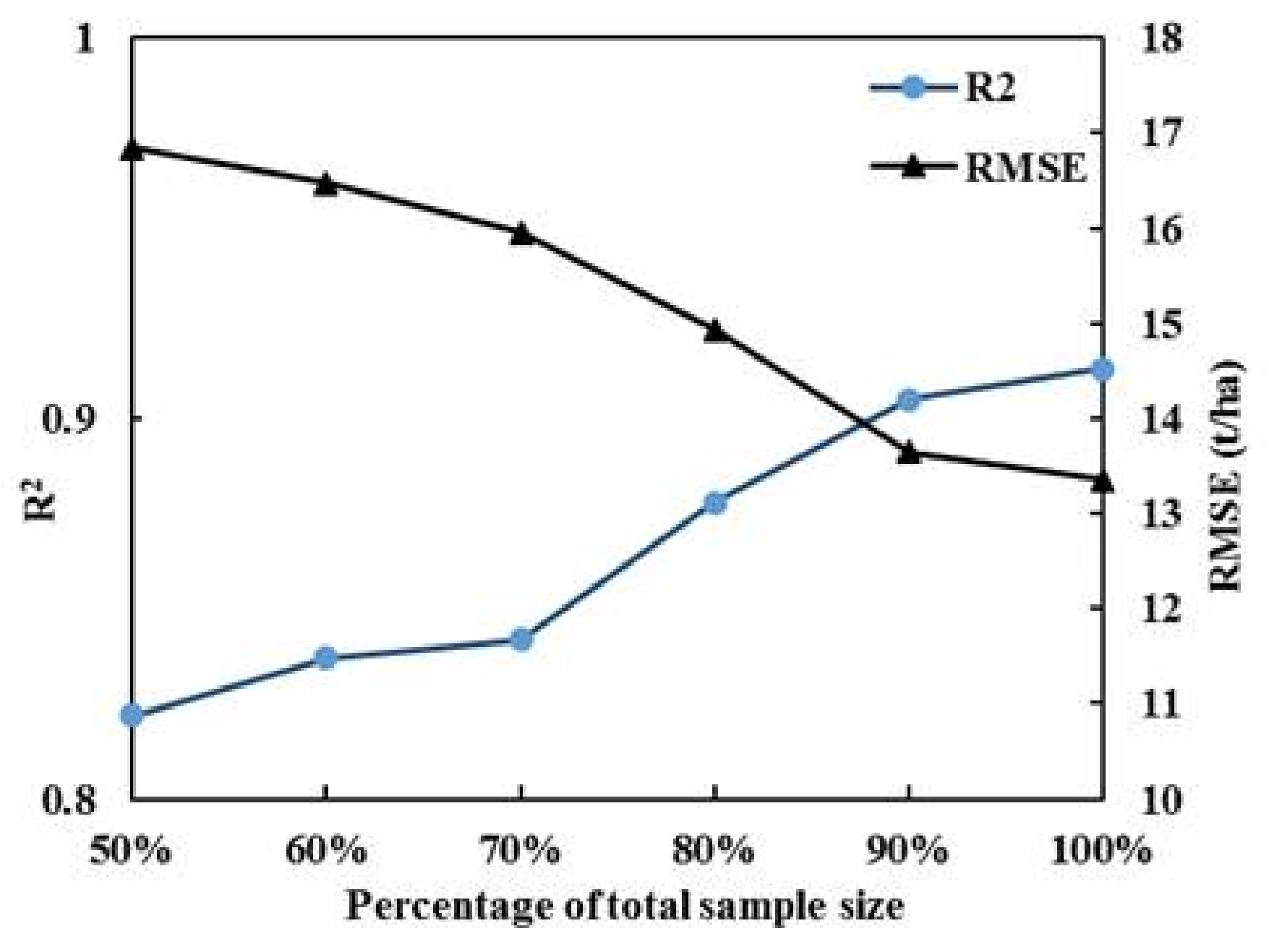
4.2. Impact of Sample Size on Forest AGB Estimation
4.3. Effect of Terrain Variables on the Prediction Model
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Houghton, R.A.; Hall, F.; Goetz, S.J. Importance of biomass in the global carbon cycle. J. Geophys. Res. Biogeosci. 2009, 114, 13. [Google Scholar] [CrossRef]
- Canadell, J.G.; Le Quere, C.; Raupach, M.R.; Field, C.B.; Buitenhuis, E.T.; Ciais, P.; Conway, T.J.; Gillett, N.P.; Houghton, R.A.; Marland, G. Contributions to accelerating atmospheric CO2 growth from economic activity, carbon intensity, and efficiency of natural sinks. Proc. Natl. Acad. Sci. USA 2007, 104, 18866–18870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibbs, H.K.; Brown, S.; Niles, J.O.; Foley, J.A. Monitoring and estimating tropical forest carbon stocks: Making REDD a reality. Environ. Res. Lett. 2007, 2, 13. [Google Scholar] [CrossRef]
- Saatchi, S.S.; Harris, N.L.; Brown, S.; Lefsky, M.; Mitchard, E.T.A.; Salas, W.; Zutta, B.R.; Buermann, W.; Lewis, S.L.; Hagen, S.; et al. Benchmark map of forest carbon stocks in tropical regions across three continents. Proc. Natl. Acad. Sci. USA 2011, 108, 9899–9904. [Google Scholar] [CrossRef] [PubMed]
- Ene, L.T.; Naesset, E.; Gobakken, T.; Gregoire, T.G.; Stahl, G.; Holm, S. A simulation approach for accuracy assessment of two-phase post-stratified estimation in large-area LiDAR biomass surveys. Remote Sens. Environ. 2013, 133, 210–224. [Google Scholar] [CrossRef]
- Ahmed, R.; Siqueira, P.; Hensley, S. A study of forest biomass estimates from LiDAR in the northern temperate forests of New England. Remote Sens. Environ. 2013, 130, 121–135. [Google Scholar] [CrossRef]
- Ene, L.T.; Naesset, E.; Gobakken, T.; Gregoire, T.G.; Stahl, G.; Nelson, R. Assessing the accuracy of regional LiDAR-based biomass estimation using a simulation approach. Remote Sens. Environ. 2012, 123, 579–592. [Google Scholar] [CrossRef]
- Ioki, K.; Tsuyuki, S.; Hirata, Y.; Phua, M.H.; Wong, W.V.C.; Ling, Z.Y.; Saito, H.; Takao, G. Estimating above-ground biomass of tropical rainforest of different degradation levels in Northern Borneo using airborne LiDAR. For. Ecol. Manag. 2014, 328, 335–341. [Google Scholar] [CrossRef]
- Pflugmacher, D.; Cohen, W.B.; Kennedy, R.E.; Yang, Z.Q. Using Landsat-derived disturbance and recovery history and LiDAR to map forest biomass dynamics. Remote Sens. Environ. 2014, 151, 124–137. [Google Scholar] [CrossRef]
- Lin, Y.; West, G. Reflecting conifer phenology using mobile terrestrial LiDAR: A case study of Pinus sylvestris growing under the Mediterranean climate in Perth, Australia. Ecol. Indic. 2016, 70, 1–9. [Google Scholar] [CrossRef]
- Huete, A.; Justice, C.; Liu, H. Development of vegetation and soil indexes for MODIS-EOS. Remote Sens. Environ. 1994, 49, 224–234. [Google Scholar] [CrossRef]
- Blackard, J.A.; Finco, M.V.; Helmer, E.H.; Holden, G.R.; Hoppus, M.L.; Jacobs, D.M.; Lister, A.J.; Moisen, G.G.; Nelson, M.D.; Riemann, R.; et al. Mapping us forest biomass using nationwide forest inventory data and moderate resolution information. Remote Sens. Environ. 2008, 112, 1658–1677. [Google Scholar] [CrossRef]
- Foody, G.M.; Boyd, D.S.; Cutler, M.E.J. Predictive relations of tropical forest biomass from Landsat TM data and their transferability between regions. Remote Sens. Environ. 2003, 85, 463–474. [Google Scholar] [CrossRef]
- Pu, R.L.; Cheng, J. Mapping forest leaf area index using reflectance and textural information derived from worldview-2 imagery in a mixed natural forest area in Florida, US. Int. J. Appl. Earth Obs. Geoinf. 2015, 42, 11–23. [Google Scholar] [CrossRef]
- Avitabile, V.; Baccini, A.; Friedl, M.A.; Schmullius, C. Capabilities and limitations of Landsat and land cover data for aboveground woody biomass estimation of Uganda. Remote Sens. Environ. 2012, 117, 366–380. [Google Scholar] [CrossRef]
- Gao, M.L.; Zhao, W.J.; Gong, Z.N.; Gong, H.L.; Chen, Z.; Tang, X.M. Topographic correction of ZY-3 satellite images and its effects on estimation of shrub leaf biomass in mountainous areas. Remote Sens. 2014, 6, 2745–2764. [Google Scholar] [CrossRef]
- Chi, H.; Sun, G.Q.; Huang, J.L.; Guo, Z.F.; Ni, W.J.; Fu, A.M. National forest aboveground biomass mapping from ICESat/GLAS data and MODIS imagery in China. Remote Sens. 2015, 7, 5534–5564. [Google Scholar] [CrossRef]
- Kumar, L.; Mutanga, O. Remote sensing of above-ground biomass. Remote Sens. 2017, 9, 935. [Google Scholar] [CrossRef]
- Ni, W.; Sun, G.; Ranson, K.J.; Pang, Y.; Zhang, Z.; Yao, W. Extraction of ground surface elevation from ZY-3 winter stereo imagery over deciduous forested areas. Remote Sens. Environ. 2015, 159, 194–202. [Google Scholar] [CrossRef]
- Sun, G.; Ni, W.; Zhang, Z.; Xiong, C. Forest abovegroundbiomass mapping using spaceborne stereo imagery acquired by Chinese ZY-3. In Proceedings of the AGU Fall Meeting, San Francisco, CA, USA, 14–18 December 2015; Volume 12, p. 2089. [Google Scholar]
- Santi, E.; Paloscia, S.; Pettinato, S.; Fontanelli, G.; Mura, M.; Zolli, C.; Maselli, F.; Chiesi, M.; Bottai, L.; Chirici, G. The potential of multifrequency SAR images for estimating forest biomass in Mediterranean areas. Remote Sens. Environ. 2017, 200, 63–73. [Google Scholar] [CrossRef]
- Zhou, T.; Li, Z.; Pan, J. Multi-feature classification of multi-sensor satellite imagery based on dual-polarimetric Sentinel-1a, Landsat-8 oli, and hyperion images for urban land-cover classification. Sensors 2018, 18, 373. [Google Scholar] [CrossRef] [PubMed]
- Laurin, G.V.; Chen, Q.; Lindsell, J.A.; Coomes, D.A.; Del Frate, F.; Guerriero, L.; Pirotti, F.; Valentini, R. Above ground biomass estimation in an African tropical forest with LiDAR and hyperspectral data. ISPRS-J. Photogramm. Remote Sens. 2014, 89, 49–58. [Google Scholar] [CrossRef]
- He, Q.S.; Chen, E.X.; An, R.; Li, Y. Above-ground biomass and biomass components estimation using LiDAR data in a coniferous forest. Forests 2013, 4, 984–1002. [Google Scholar] [CrossRef]
- Singh, K.K.; Chen, G.; Vogler, J.B.; Meentemeyer, R.K. When big data are too much: Effects of LiDAR returns and point density on estimation of forest biomass. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2016, 9, 3210–3218. [Google Scholar] [CrossRef]
- Lefsky, M.A.; Cohen, W.B.; Parker, G.G.; Harding, D.J. LiDAR remote sensing for ecosystem studies. Bioscience 2002, 52, 19–30. [Google Scholar] [CrossRef]
- Zolkos, S.G.; Goetz, S.J.; Dubayah, R. A meta-analysis of terrestrial aboveground biomass estimation using LiDAR remote sensing. Remote Sens. Environ. 2013, 128, 289–298. [Google Scholar] [CrossRef]
- Su, Y.J.; Guo, Q.H.; Xue, B.L.; Hu, T.Y.; Alvarez, O.; Tao, S.L.; Fang, J.Y. Spatial distribution of forest aboveground biomass in China: Estimation through combination of spaceborne LiDAR, optical imagery, and forest inventory data. Remote Sens. Environ. 2016, 173, 187–199. [Google Scholar] [CrossRef]
- Chi, H.; Sun, G.Q.; Huang, J.L.; Li, R.D.; Ren, X.Y.; Ni, W.J.; Fu, A.M. Estimation of forest aboveground biomass in Changbai mountain region using ICESat/GLAS and Landsat/TM data. Remote Sens. 2017, 9, 707. [Google Scholar] [CrossRef]
- Luo, S.Z.; Wang, C.; Xi, X.H.; Pan, F.F.; Peng, D.L.; Zou, J.; Nie, S.; Qin, H.M. Fusion of airborne LiDAR data and hyperspectral imagery for aboveground and belowground forest biomass estimation. Ecol. Indic. 2017, 73, 378–387. [Google Scholar] [CrossRef]
- Brovkina, O.; Novotny, J.; Cienciala, E.; Zemek, F.; Russ, R. Mapping forest aboveground biomass using airborne hyperspectral and LiDAR data in the mountainous conditions of central Europe. Ecol. Eng. 2017, 100, 219–230. [Google Scholar] [CrossRef]
- Swatantran, A.; Dubayah, R.; Roberts, D.; Hofton, M.; Blair, J.B. Mapping biomass and stress in the Sierra Nevada using LiDAR and hyperspectral data fusion. Remote Sens. Environ. 2011, 115, 2917–2930. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Hartig, F.; Latifi, H.; Berger, C.; Hernandez, J.; Corvalan, P.; Koch, B. Importance of sample size, data type and prediction method for remote sensing-based estimations of aboveground forest biomass. Remote Sens. Environ. 2014, 154, 102–114. [Google Scholar] [CrossRef]
- Liu, K.; Wang, J.; Zeng, W.; Song, J. Comparison and evaluation of three methods for estimating forest above ground biomass using TM and GLAS data. Remote Sens. 2017, 9, 341. [Google Scholar] [CrossRef]
- Shao, Z.; Zhang, L. Estimating forest aboveground biomass by combining optical and SAR data: A case study in Genhe, Inner Mongolia, China. Sensors 2016, 16, 834. [Google Scholar] [CrossRef] [PubMed]
- De Toledo, J.J.; Magnusson, W.E.; Castilho, C.V.; Nascimento, H.E.M. Tree mode of death in central Amazonia: Effects of soil and topography on tree mortality associated with storm disturbances. For. Ecol. Manag. 2012, 263, 253–261. [Google Scholar] [CrossRef]
- Ferry, B.; Morneau, F.; Bontemps, J.-D.; Blanc, L.; Freycon, V. Higher treefall rates on slopes and waterlogged soils result in lower stand biomass and productivity in a tropical rain forest. J. Ecol. 2010, 98, 106–116. [Google Scholar] [CrossRef]
- Singh, K.K.; Bianchetti, R.A.; Chen, G.; Meentemeyer, R.K. Assessing effect of dominant land-cover types and pattern on urban forest biomass estimated using LiDAR metrics. Urban Ecosyst. 2017, 20, 265–275. [Google Scholar] [CrossRef]
- Viana, H.; Aranha, J.; Lopes, D.; Cohen, W.B. Estimation of crown biomass of Pinus pinaster stands and shrubland above-ground biomass using forest inventory data, remotely sensed imagery and spatial prediction models. Ecol. Model. 2012, 226, 22–35. [Google Scholar] [CrossRef]
- Lee, I.S.; Ge, L.L. The performance of RTK-GPS for surveying under challenging environmental conditions. Earth Planets Space 2006, 58, 515–522. [Google Scholar] [CrossRef]
- Wang, J.; Ju, K.; Fu, H.; Chang, X. Study on biomass of water conservation forest on North Slope of Qilian Mountains. J. Fujian Coll. For. 1998, 18, 319–323. [Google Scholar]
- Xiao, Q.; Wen, J. HIWATER: Airborne LiDAR Raw Data in Tianlaochi Catchment; Heihe Plan Science Data Center: Heihe, China, 2014. [Google Scholar]
- Qi, J.; Chehbouni, A.; Huete, A.R.; Kerr, Y.H.; Sorooshian, S. A modified soil adjusted vegetation index. Remote Sens. Environ. 1994, 48, 119–126. [Google Scholar] [CrossRef]
- Rondeaux, G.; Steven, M.; Baret, F. Optimization of soil-adjusted vegetation indices. Remote Sens. Environ. 1996, 55, 95–107. [Google Scholar] [CrossRef]
- Mutanga, O.; Skidmore, A.K. Narrow band vegetation indices overcome the saturation problem in biomass estimation. Int. J. Remote Sens. 2004, 25, 3999–4014. [Google Scholar] [CrossRef]
- Liu, D.; Pu, H.B.; Sun, D.W.; Wang, L.; Zeng, X.A. Combination of spectra and texture data of hyperspectral imaging for prediction of pH in salted meat. Food Chem. 2014, 160, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Wulder, M.A.; LeDrew, E.F.; Franklin, S.E.; Lavigne, M.B. Aerial image texture information in the estimation of northern deciduous and mixed wood forest leaf area index (LAI). Remote Sens. Environ. 1998, 64, 64–76. [Google Scholar] [CrossRef]
- Pan, J.P.; Gong, J.Y.; Lu, J.; Ye, H.Z.; Chen, X.L.; Yang, J.L. Classification based on texture feature of wavelet transform. In Instruments, Science, and Methods for Geospace and Planetary Remote Sensing; Nardell, C.A., Lucey, P.G., Yee, J.H., Garvin, J.B., Eds.; SPIE: Bellingham, WA, USA, 2004; Volume 5660, pp. 208–217. [Google Scholar]
- Frazer, G.W.; Magnussen, S.; Wulder, M.A.; Niemann, K.O. Simulated impact of sample plot size and co-registration error on the accuracy and uncertainty of LiDAR-derived estimates of forest stand biomass. Remote Sens. Environ. 2011, 115, 636–649. [Google Scholar] [CrossRef]
- Dubayah, R.O.; Sheldon, S.L.; Clark, D.B.; Hofton, M.A.; Blair, J.B.; Hurtt, G.C.; Chazdon, R.L. Estimation of tropical forest height and biomass dynamics using LiDAR remote sensing at La Selva, Costa Rica. J. Geophys. Res. Biogeosci. 2010, 115. [Google Scholar] [CrossRef]
- Nie, S.; Wang, C.; Zeng, H.C.; Xi, X.H.; Li, G.C. Above-ground biomass estimation using airborne discrete-return and full-waveform LiDAR data in a coniferous forest. Ecol. Indic. 2017, 78, 221–228. [Google Scholar] [CrossRef]
- Lang, M.W.; McCarty, G.W. LiDAR intensity for improved detection of inundation below the forest canopy. Wetlands 2009, 29, 1166–1178. [Google Scholar] [CrossRef]
- Ni-Meister, W.; Lee, S.Y.; Strahler, A.H.; Woodcock, C.E.; Schaaf, C.; Yao, T.A.; Ranson, K.J.; Sun, G.Q.; Blair, J.B. Assessing general relationships between aboveground biomass and vegetation structure parameters for improved carbon estimate from LiDAR remote sensing. J. Geophys. Res. Biogeosci. 2010, 115, 12. [Google Scholar] [CrossRef]
- Korhonen, L.; Korpela, I.; Heiskanen, J.; Maltamo, M. Airborne discrete-return LiDAR data in the estimation of vertical canopy cover, angular canopy closure and leaf area index. Remote Sens. Environ. 2011, 115, 1065–1080. [Google Scholar] [CrossRef]
- Zhang, G.; Ganguly, S.; Nemani, R.R.; White, M.A.; Milesi, C.; Hashimoto, H.; Wang, W.L.; Saatchi, S.; Yu, Y.F.; Myneni, R.B. Estimation of forest aboveground biomass in California using canopy height and leaf area index estimated from satellite data. Remote Sens. Environ. 2014, 151, 44–56. [Google Scholar] [CrossRef]
- Jensen, J.L.R.; Mathews, A.J. Assessment of image-based point cloud products to generate a bare earth surface and estimate canopy heights in a woodland ecosystem. Remote Sens. 2016, 8, 50. [Google Scholar] [CrossRef]
- Glenn, N.F.; Streutker, D.R.; Chadwick, D.J.; Thackray, G.D.; Dorsch, S.J. Analysis of LiDAR-derived topographic information for characterizing and differentiating landslide morphology and activity. Geomorphology 2006, 73, 131–148. [Google Scholar] [CrossRef]
- O’Loughlin, E.J.; Chin, Y.P. Quantification and characterization of dissolved organic carbon and iron in sedimentary porewater from green bay, WI, USA. Biogeochemistry 2004, 71, 371–386. [Google Scholar] [CrossRef]
- Karlson, M.; Ostwald, M.; Reese, H.; Sanou, J.; Tankoano, B.; Mattsson, E. Mapping tree canopy cover and aboveground biomass in Sudano-Sahelian woodlands using Landsat 8 and random forest. Remote Sens. 2015, 7, 10017–10041. [Google Scholar] [CrossRef]
- Mountrakis, G.; Im, J.; Ogole, C. Support vector machines in remote sensing: A review. ISPRS-J. Photogramm. Remote Sens. 2011, 66, 247–259. [Google Scholar] [CrossRef]
- Wang, L.; Zeng, Y.; Chen, T. Back propagation neural network with adaptive differential evolution algorithm for time series forecasting. Expert Syst. Appl. 2015, 42, 855–863. [Google Scholar] [CrossRef]
- Pandya, D.H.; Upadhyay, S.H.; Harsha, S.P. Fault diagnosis of rolling element bearing with intrinsic mode function of acoustic emission data using APF-KNN. Expert Syst. Appl. 2013, 40, 4137–4145. [Google Scholar] [CrossRef]
- Preacher, K.J.; Curran, P.J.; Bauer, D.J. Computational tools for probing interactions in multiple linear regression, multilevel modeling, and latent curve analysis. J. Educ. Behav. Stat. 2006, 31, 437–448. [Google Scholar] [CrossRef]
- Martinez, B.; Garcia-Haro, F.J.; Camacho-de Coca, F. Derivation of high-resolution leaf area index maps in support of validation activities: Application to the cropland Barrax site. Agric. For. Meteorol. 2009, 149, 130–145. [Google Scholar] [CrossRef]
- Fang, J.Y.; Wang, Z.M. Forest biomass estimation at regional and global levels, with special reference to China’s forest biomass. Ecol. Res. 2001, 16, 587–592. [Google Scholar] [CrossRef]
- Xing, Y.-Q.; Wang, L.-H. Compatible biomass estimation models of natural forests in Changbai Mountains based on forest inventory. Yingyong Shengtai Xuebao 2007, 18, 1–8. [Google Scholar] [PubMed]
- Ahmed, R.; Siqueira, P.; Hensley, S.; Bergen, K. Uncertainty of forest biomass estimates in north temperate forests due to allometry: Implications for remote sensing. Remote Sens. 2013, 5, 3007–3036. [Google Scholar] [CrossRef]
- Gobakken, T.; Naesset, E.; Nelson, R.; Bollandsas, O.M.; Gregoire, T.G.; Stahl, G.; Holm, S.; Orka, H.O.; Astrup, R. Estimating biomass in Hedmark County, Norway using national forest inventory field plots and airborne laser scanning. Remote Sens. Environ. 2012, 123, 443–456. [Google Scholar] [CrossRef]
- Kulawardhana, R.W.; Popescu, S.C.; Feagin, R.A. Fusion of LiDAR and multispectral data to quantify salt marsh carbon stocks. Remote Sens. Environ. 2014, 154, 345–357. [Google Scholar] [CrossRef]
- Kenzo, T.; Furutani, R.; Hattori, D.; Tanaka, S.; Sakurai, K.; Ninomiya, I.; Kendawang, J.J. Aboveground and belowground biomass in logged-over tropical rain forests under different soil conditions in borneo. J. For. Res. 2015, 20, 197–205. [Google Scholar] [CrossRef]
- Simard, M.; Zhang, K.; Rivera-Monroy, V.H.; Ross, M.S.; Ruiz, P.L.; Castañeda-Moya, E.; Twilley, R.R.; Rodriguez, E. Mapping height and biomass of mangrove forests in Everglades National Park with SRTM elevation data. Photogramm. Eng. Remote Sens. 2006, 72, 299–311. [Google Scholar] [CrossRef]
- Lefsky, M.A.; Cohen, W.B.; Harding, D.J.; Parker, G.G.; Acker, S.A.; Gower, S.T. Lidar remote sensing of above-ground biomass in three biomes. Glob. Ecol. Biogeogr. Lett. 2002, 11, 393–399. [Google Scholar] [CrossRef]
- Lefsky, M.A.; Harding, D.; Cohen, W.B.; Parker, G.; Shugart, H.H. Surface LiDAR remote sensing of basal area and biomass in deciduous forests of Eastern Maryland, USA. Remote Sens. Environ. 1999, 67, 83–98. [Google Scholar]
- Chen, G.; Ozelkan, E.; Singh, K.K.; Zhou, J.; Brown, M.R.; Meentemeyer, R.K. Uncertainties in mapping forest carbon in urban ecosystems. J. Environ. Manag. 2017, 187, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Townsend, P.A. Mapping forest structure and uncertainty in an urban area using leaf-off LiDAR data. Urban Ecosyst. 2017, 20, 497–509. [Google Scholar] [CrossRef]
- Hultquist, C.; Chen, G.; Zhao, K. A comparison of Gaussian process regression, random forests and support vector regression for burn severity assessment in diseased forests. Remote Sens. Lett. 2014, 5, 723–732. [Google Scholar] [CrossRef]
- Gobakken, T.; Naesset, E. Assessing effects of laser point density, ground sampling intensity, and field sample plot size on biophysical stand properties derived from airborne laser scanner data. Can. J. For. Res.-Rev. Can. Rech. For. 2008, 38, 1095–1109. [Google Scholar] [CrossRef]
- Strunk, J.; Temesgen, H.; Andersen, H.-E.; Flewelling, J.P.; Madsen, L. Effects of LiDAR pulse density and sample size on a model-assisted approach to estimate forest inventory variables. Can. J. Remote Sens. 2012, 38, 644–654. [Google Scholar] [CrossRef]
- Paine, C.E.T.; Baraloto, C.; Diaz, S. Optimal strategies for sampling functional traits in species-rich forests. Funct. Ecol. 2015, 29, 1325–1331. [Google Scholar] [CrossRef]
- Gutjahr, A. Geostatistics for sampling designs and analysis. ACS Symp. Ser. 1990, 465, 48–90. [Google Scholar]
- Hilbert, C.; Schmullius, C. Influence of surface topography on ICESat/GLAS forest height estimation and waveform shape. Remote Sens. 2012, 4, 2210–2235. [Google Scholar] [CrossRef]

| Variables | Equations | Description |
|---|---|---|
| Ratio Vegetation Index (RVI) | bandNir/bandR | Reduces the effects of atmosphere and topography |
| Normalized Difference Index (NDVI) | (bandNir − bandR)/(bandNir + bandR) | Enhances the responsiveness to the vegetation |
| Soil-Adjusted Vegetation Index (SAVI) | The value of L can be automatically adjusted following the density of the vegetation | |
| Modified Soil-Adjusted Vegetation Index (MSAVI) | Reduces the disturbances of soil background |
| Metric ID | Metric | Description |
|---|---|---|
| LiDAR-derived variables | Hmax | Maximum of LiDAR height |
| Hmean | Mean of LiDAR height | |
| HSD | Standard deviation of LiDAR height | |
| HCV | Coefficient of variation of LiDAR height | |
| Canopy cover (Cc) | Canopy cover derived from LiDAR returns number | |
| Terrain variables | Topographic Wetness Index (TWI) | Reflects the condition of soil |
| Hillshade | Generalized directional topographic exposure | |
| Profile Curvature (PC) | Describes variation of slope | |
| Slope | Reflects tilt of local topography |
| Methods | Package | Parameters |
|---|---|---|
| Random Forest (RF) | ‘randomforest’ | Ntree with 500 and mtry with 25 |
| Support Vector Machines (SVM) | ‘kernlab’ | Penalty factor with 2^(−6:2) and kernel parameters with 2^(−4:4) |
| Back Propagational Neural Networks (BPNN) | ‘amore’ | Number of iterations with 16 |
| K-Nearest Neighbor (KNN) | ‘knn’ | Number of adjacent sample sizes, which was set from 2 to 5 |
| Generalized Linear Mixed Model (GLMM) | ‘lme4’ | Logistic regression function was used and the growth parameter with 0.2 |
| Response Variable | df | R2 | RMSE | ||||
|---|---|---|---|---|---|---|---|
| SumSq | F | Sig. | SumSq | F | Sig. | ||
| Data types | 4 | 0.082 | 17.603 | 0.000 | 1230.246 | 25.513 | 0.000 |
| Methods | 4 | 0.065 | 13.798 | 0.000 | 148.086 | 3.071 | 0.047 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, L.; Pan, J.; Li, R.; Li, J.; Li, Z. Integrating Airborne LiDAR and Optical Data to Estimate Forest Aboveground Biomass in Arid and Semi-Arid Regions of China. Remote Sens. 2018, 10, 532. https://doi.org/10.3390/rs10040532
Cao L, Pan J, Li R, Li J, Li Z. Integrating Airborne LiDAR and Optical Data to Estimate Forest Aboveground Biomass in Arid and Semi-Arid Regions of China. Remote Sensing. 2018; 10(4):532. https://doi.org/10.3390/rs10040532
Chicago/Turabian StyleCao, Luodan, Jianjun Pan, Ruijuan Li, Jialin Li, and Zhaofu Li. 2018. "Integrating Airborne LiDAR and Optical Data to Estimate Forest Aboveground Biomass in Arid and Semi-Arid Regions of China" Remote Sensing 10, no. 4: 532. https://doi.org/10.3390/rs10040532






