Individual Tree Crown Segmentation and Classification of 13 Tree Species Using Airborne Hyperspectral Data
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Remote Sensing Data
2.3. Forest Mask
2.4. Reference Data
2.5. Workflow Description
2.6. Noise Removal
2.7. Feature Extraction
2.8. Segmentation
2.8.1. Stratification
2.8.2. Segmentation of Individual Tree Crowns
2.9. Random Forest Classification
3. Results
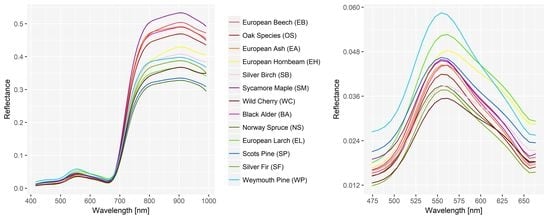
3.1. Spectral Signatures of Tree Species
3.2. Classification of Manually Delineated Reference Trees
3.3. Classification of Mean Shift-Segmented Reference Trees
3.4. Comparison of Classifications Results for Manually and Automatically Segmented Reference Trees
3.5. Importance of Wavelengths for Principal Components
3.6. Wall-to-Wall Mapping on Mean Shift-Segmented VNIR Image
4. Discussion
4.1. Classification Accuracies
4.2. Suitability of Hyperspectral Dataset and Segmentation Algorithms
4.3. Classification of Tree Species
4.4. Variable Importance
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Index | Equation | Reference |
|---|---|---|
| Ratio Vegetation Index | [81] | |
| Difference Index | [82] | |
| Normalized Difference Vegetation Index | [83] | |
| Green-Red Difference Index | [82,84] | |
| Difference Difference Vegetation Index | [85] | |
| Atmospherically Resistant Vegetation Index | [86] | |
| Green Atmospherically Resistant Vegetation Index | [87] | |
| Green Normalized Difference Vegetation Index | [88] | |
| Visible Atmospherically Resistant Index | [89] | |
| Enhanced Vegetation Index | [90] | |
| 2-band Enhanced Vegetation Index | [91] | |
| Photochemical Reflectance Index | [92,93] | |
| Red Edge Normalized Difference Vegetation Index | [94] | |
| Modified Simple Ratio | [95] | |
| Modified Normalized Difference Index | [95] | |
| Green Ratio | [15] | |
| Blue Ratio | [15] | |
| Red Ratio | [15] | |
| Infrared Percentage Vegetation Index | [96] | |
| Normalized Difference Red Edge Index | [97] | |
| Plant Senescence Reflectance Index | [98] | |
| Weighted Difference Vegetation Index | [99,100,101] |
References
- Fassnacht, F.E.; Latifi, H.; Stereńczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.T.; Straub, C.; Ghosh, A. Review of studies on tree species classification from remotely sensed data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- Wulder, M.A.; Franklin, S.E. Remote Sensing of Forest Environments: Concepts and Case Studies; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2012. [Google Scholar]
- Wulder, M.A.; Hall, R.J.; Coops, N.C.; Franklin, S.E. High spatial resolution remotely sensed data for ecosystem characterization. BioScience 2004, 54, 511–521. [Google Scholar] [CrossRef]
- Immitzer, M.; Atzberger, C.; Koukal, T. Tree species classification with Random Forest using very high spatial resolution 8-band WorldView-2 satellite data. Remote Sens. 2012, 4, 2661–2693. [Google Scholar] [CrossRef]
- Hantson, W.; Kooistra, L.; Slim, P.A. Mapping invasive woody species in coastal dunes in the Netherlands: a remote sensing approach using LIDAR and high-resolution aerial photographs. Appl. Veg. Sci. 2012, 15, 536–547. [Google Scholar] [CrossRef]
- Dalponte, M.; Bruzzone, L.; Gianelle, D. Fusion of hyperspectral and LIDAR remote sensing data for classification of complex forest areas. IEEE Trans. Geosci. Remote Sens. 2008, 46, 1416–1427. [Google Scholar] [CrossRef]
- Asner, G.P. Biophysical and Biochemical Sources of Variability in Canopy Reflectance. Remote Sens. Environ. 1998, 64, 234–253. [Google Scholar] [CrossRef]
- Asner, G.P.; Ustin, S.L.; Townsend, P.A.; Martin, R.E.; Chadwick, K.D. Forest biophysical and biochemical properties from hyperspectral and LiDAR remote sensing. In Land Resources Monitoring, Modeling and Mapping with Remote Sensing; Thenkabail, P.S., Ed.; CRC Press: Boca Raton, FL, USA, 2015; pp. 429–448. [Google Scholar]
- Clark, M.L.; Roberts, D.A.; Clark, D.B. Hyperspectral discrimination of tropical rain forest tree species at leaf to crown scales. Remote Sens. Environ. 2005, 96, 375–398. [Google Scholar] [CrossRef]
- Rautiainen, M.; Lukeš, P.; Homolová, L.; Hovi, A.; Pisek, J.; Mõttus, M. Spectral Properties of Coniferous Forests: A Review of In Situ and Laboratory Measurements. Remote Sens. 2018, 10, 207. [Google Scholar] [CrossRef]
- Einzmann, K.; Ng, W.; Immitzer, M.; Bachmann, M.; Pinnel, N.; Atzberger, C. Method analysis for collecting and processing in-situ hyperspectral needle reflectance data for monitoring Norway spruce. Photogramm. Fernerkund. Geoinf. 2014, 2014, 351–367. [Google Scholar] [CrossRef]
- Ghiyamat, A.; Shafri, H.Z.M.; Amouzad Mahdiraji, G.; Shariff, A.R.M.; Mansor, S. Hyperspectral discrimination of tree species with different classifications using single- and multiple-endmember. Int. J. Appl. Earth Obs. Geoinf. 2013, 23, 177–191. [Google Scholar] [CrossRef] [Green Version]
- Immitzer, M.; Atzberger, C. Early Detection of Bark Beetle Infestation in Norway Spruce (Picea abies, L.) using WorldView-2 Data. Photogramm. Fernerkund. Geoinf. 2014, 351–367. [Google Scholar] [CrossRef]
- Schlerf, M.; Atzberger, C.; Hill, J. Tree species and age class mapping in a Central European woodland using optical remote sensing imagery and orthophoto derived stem density–performance of multispectral and hyperspectral sensors. In Geoinformation for European-Wide Integration, Proceedings of the 22nd Symposium of the European Association of Remote Sensing Laboratories, Prague, Czech Republic, 4–6 June 2002; Millpress: Cape Town, Western Cape, South Africa, 2003; pp. 413–418. [Google Scholar]
- Waser, L.T.; Küchler, M.; Jütte, K.; Stampfer, T. Evaluating the Potential of WorldView-2 Data to Classify Tree Species and Different Levels of Ash Mortality. Remote Sens. 2014, 6, 4515–4545. [Google Scholar] [CrossRef] [Green Version]
- Jones, H.G.; Vaughan, R.A. Remote Sensing of Vegetation: Principles, Techniques, and Applications; Oxford University Press: New York, NY, USA, 2010. [Google Scholar]
- Spanner, M.A.; Pierce, L.L.; Peterson, D.L.; Running, S.W. Remote sensing of temperate coniferous forest leaf area index The influence of canopy closure, understory vegetation and background reflectance. Int. J. Remote Sens. 1990, 11, 95–111. [Google Scholar] [CrossRef]
- Schlerf, M.; Atzberger, C. Inversion of a forest reflectance model to estimate structural canopy variables from hyperspectral remote sensing data. Remote Sens. Environ. 2006, 100, 281–294. [Google Scholar] [CrossRef]
- Chang, C.-I. Hyperspectral Imaging: Techniques for Spectral Detection and Classification; Kluwer Academic Publishers: New York, NY, USA, 2003. [Google Scholar]
- Rock, G.; Gerhards, M.; Schlerf, M.; Hecker, C.; Udelhoven, T. Plant species discrimination using emissive thermal infrared imaging spectroscopy. Int. J. Appl. Earth Obs. Geoinf. 2016, 53, 16–26. [Google Scholar] [CrossRef]
- Lillesand, T.M.; Kiefer, R.W.; Chipman, J.W. Remote Sensing and Image Interpretation; John Wiley & Sons: New York, NY, USA, 2008. [Google Scholar]
- Ørka, H.O.; Hauglin, M. Use of Remote Sensing for Mapping of Non-Native Conifer Species. 2016. Available online: http://www.umb.no/statisk/ina/publikasjoner/fagrapport/if33.pdf (accessed on 25 June 2018).
- Dalponte, M.; Ørka, H.O.; Gobakken, T.; Gianelle, D.; Næsset, E. Tree Species Classification in Boreal Forests With Hyperspectral Data. IEEE Trans. Geosci. Remote Sens. 2013, 51, 2632–2645. [Google Scholar] [CrossRef]
- Wang, Z.; Jensen, J.R.; Im, J. An automatic region-based image segmentation algorithm for remote sensing applications. Environ. Model. Softw. 2010, 25, 1149–1165. [Google Scholar] [CrossRef]
- Ke, Y.; Quackenbush, L.J. A review of methods for automatic individual tree-crown detection and delineation from passive remote sensing. Int. J. Remote Sens. 2011, 32, 4725–4747. [Google Scholar] [CrossRef]
- Fu, G.; Zhao, H.; Li, C.; Shi, L. Segmentation for High-Resolution Optical Remote Sensing Imagery Using Improved Quadtree and Region Adjacency Graph Technique. Remote Sens. 2013, 5, 3259–3279. [Google Scholar] [CrossRef] [Green Version]
- Richter, R.; Reu, B.; Wirth, C.; Doktor, D.; Vohland, M. The use of airborne hyperspectral data for tree species classification in a species-rich Central European forest area. Int. J. Appl. Earth Obs. Geoinf. 2016, 52, 464–474. [Google Scholar] [CrossRef]
- James, G.; Witten, D.; Hastie, T.; Tibshirani, R. An Introduction to Statistical Learning: With Applications in R; Springer: New York, NY, USA, 2013. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Naidoo, L.; Cho, M.A.; Mathieu, R.; Asner, G. Classification of savanna tree species, in the Greater Kruger National Park region, by integrating hyperspectral and LiDAR data in a Random Forest data mining environment. ISPRS J. Photogramm. Remote Sens. 2012, 69, 167–179. [Google Scholar] [CrossRef]
- Ballanti, L.; Blesius, L.; Hines, E.; Kruse, B. Tree Species Classification Using Hyperspectral Imagery: A Comparison of Two Classifiers. Remote Sens. 2016, 8, 445. [Google Scholar] [CrossRef]
- Kilian, W.; Müller, F.; Starlinger, F. Die forstlichen Wuchsgebiete Österreichs: Eine Naturraumgliederung Nach Waldökologischen Gesichtspunkten. 1994. Available online: https://bfw.ac.at/300/pdf/1027.pdf (accessed on 16 June 2018).
- Biosphärenpark Wienerwald Management GmbH Zonation. Available online: https://www.bpww.at/en/node/183 (accessed on 8 June 2018).
- Österreichische Bundesforste Biosphärenpark Wienerwald. Available online: http://www.bundesforste.at/natur-erlebnis/biosphaerenpark-wienerwald.html (accessed on 8 June 2018).
- Richter, R.A. spatially adaptive fast atmospheric correction algorithm. Int. J. Remote Sens. 1996, 17, 1201–1214. [Google Scholar] [CrossRef]
- Richter, R.; Richter, R.; Schläpfer, D. Geo-atmospheric processing of airborne imaging spectrometry data. Part 2: Atmospheric/topographic correction. Int. J. Remote Sens. 2002, 23, 2631–2649. [Google Scholar] [CrossRef]
- Eilers, P.H.C. A Perfect Smoother. Anal. Chem. 2003, 75, 3631–3636. [Google Scholar] [CrossRef] [PubMed]
- Atzberger, C.; Eilers, P.H.C. Evaluating the effectiveness of smoothing algorithms in the absence of ground reference measurements. Int. J. Remote Sens. 2011, 32, 3689–3709. [Google Scholar] [CrossRef]
- Atzberger, C.; Eilers, P.H.C. A time series for monitoring vegetation activity and phenology at 10-daily time steps covering large parts of South America. Int. J. Digit. Earth 2011, 4, 365–386. [Google Scholar] [CrossRef]
- Lobo, A.; Mattiuzzi, M. Modified Whittaker Smoother. 2012. Available online: https://github.com/MatMatt/MODIS/blob/master/R/miwhitatzb1.R (accessed on 22 June 2018).
- Lang, M.; Alleaume, S.; Luque, S.; Baghdadi, N.; Féret, J.-B. Monitoring and Characterizing Heterogeneous Mediterranean Landscapes with Continuous Textural Indices Based on VHSR Imagery. Remote Sens. 2018, 10, 868. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, 3, 610–621. [Google Scholar] [CrossRef] [Green Version]
- OTB Development Team the ORFEO Tool Box Software Guide Updated for OTB-6.4.0. Available online: https://www.orfeo-toolbox.org/SoftwareGuide/ (accessed on 12 Jun 2018).
- Maaten, V.D.L.; Postma, E.; Van Den Herik, J. Dimensionality Reduction: A Comparative Review; Tilburg Centre for Creative Computing, Tilburg University: Tilburg, The Netherlands, 2009. [Google Scholar]
- Abdi, H. Normalizing data. In Encyclopedia of Research Design; Salkind, N.S., Ed.; Sage Publications: Thousand Oaks, CA, USA, 2010; pp. 935–938. [Google Scholar]
- Comaniciu, D.; Meer, P. Mean shift: A robust approach toward feature space analysis. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 603–619. [Google Scholar] [CrossRef]
- Fukunaga, K.; Hostetler, L. The estimation of the gradient of a density function, with applications in pattern recognition. IEEE Trans. Inf. Theory 1975, 21, 32–40. [Google Scholar] [CrossRef]
- Michel, J.; Youssefi, D.; Grizonnet, M. Stable Mean-Shift Algorithm and Its Application to the Segmentation of Arbitrarily Large Remote Sensing Images. IEEE Trans. Geosci. Remote Sens. 2015, 53, 952–964. [Google Scholar] [CrossRef]
- Chehata, N.; Orny, C.; Boukir, S.; Guyon, D.; Wigneron, J.P. Object-based change detection in wind storm-damaged forest using high-resolution multispectral images. Int. J. Remote Sens. 2014, 35, 4758–4777. [Google Scholar] [CrossRef]
- Einzmann, K.; Immitzer, M.; Böck, S.; Bauer, O.; Schmitt, A.; Atzberger, C. Windthrow Detection in European Forests with Very High-Resolution Optical Data. Forests 2017, 8, 21. [Google Scholar] [CrossRef]
- Immitzer, M.; Vuolo, F.; Atzberger, C. First Experience with Sentinel-2 Data for Crop and Tree Species Classifications in Central Europe. Remote Sens. 2016, 8, 166. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction; Springer: New York, NY, USA, 2009. [Google Scholar]
- Breiman, L. Manual on Setting up, Using, and Understanding Random Forests V3.1. 2002. Available online: http://oz.berkeley.edu/users/breiman/Using_random_forests_V3.1.pdf (accessed on 3 May 2012).
- Tso, B.; Mather, P.M. Classification Methods for Remotely Sensed Data; CRC Press: Boca Raton, FL, USA, 2009. [Google Scholar]
- Toscani, P.; Immitzer, M.; Atzberger, C. Texturanalyse mittels diskreter Wavelet Transformation für die objektbasierte Klassifikation von Orthophotos. Photogramm. Fernerkund. Geoinf. 2013, 2, 105–121. [Google Scholar] [CrossRef]
- Vuolo, F.; Neuwirth, M.; Immitzer, M.; Atzberger, C.; Ng, W.-T. How much does multi-temporal Sentinel-2 data improve crop type classification? Int. J. Appl. Earth Obs. Geoinf. 2018, 72, 122–130. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2/3, 18–22. [Google Scholar]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Kuhn, M.; Wing, J.; Weston, S.; Williams, A.; Keefer, C.; Engelhardt, A.; Cooper, T.; Mayer, Z.; Kenkel, B.; Benesty, M.; et al. Caret: Classification and Regression Training, R Package. 2017. Available online: http://CRAN.R-project.org/package=caret (accessed on 18 June 2018).
- Hijmans, R.J. Raster: Geographic Data Analysis and Modeling, R Package Version 2.6-7. 2017. Available online: http://CRAN.R-project.org/package=raster (accessed on 10 January 2018).
- Dahinden, C. An improved Random Forests approach with application to the performance prediction challenge datasets. In Hands-on Pattern Recognition, Challenges in Machine Learning; Guyon, I., Cawley, G., Dror, G., Saffari, A., Eds.; Microtome Publishing: Brookline, MA, USA, 2011; Volume 1, pp. 223–230. [Google Scholar]
- Guyon, I.; Weston, J.; Barnhill, S.; Vapnik, V. Gene Selection for Cancer Classification using Support Vector Machines. Mach. Learn. 2002, 46, 389–422. [Google Scholar] [CrossRef] [Green Version]
- Granitto, P.M.; Furlanello, C.; Biasioli, F.; Gasperi, F. Recursive feature elimination with random forest for PTR-MS analysis of agroindustrial products. Chemom. Intell. Lab. Syst. 2006, 83, 83–90. [Google Scholar] [CrossRef]
- Immitzer, M.; Böck, S.; Einzmann, K.; Vuolo, F.; Pinnel, N.; Wallner, A.; Atzberger, C. Fractional cover mapping of spruce and pine at 1ha resolution combining very high and medium spatial resolution satellite imagery. Remote Sens. Environ. 2018, 204, 690–703. [Google Scholar] [CrossRef]
- Schultz, B.; Immitzer, M.; Formaggio, A.R.; Sanches, I.D.A.; Luiz, A.J.B.; Atzberger, C. Self-Guided Segmentation and Classification of Multi-Temporal Landsat 8 Images for Crop Type Mapping in Southeastern Brazil. Remote Sens. 2015, 7, 14482–14508. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, A.; Fassnacht, F.E.; Joshi, P.K.; Koch, B. A framework for mapping tree species combining hyperspectral and LiDAR data: Role of selected classifiers and sensor across three spatial scales. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 49–63. [Google Scholar] [CrossRef]
- Fassnacht, F.; Neumann, C.; Forster, M.; Buddenbaum, H.; Ghosh, A.; Clasen, A.; Joshi, P.; Koch, B. Comparison of Feature Reduction Algorithms for Classifying Tree Species with Hyperspectral Data on Three Central European Test Sites. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7. [Google Scholar] [CrossRef]
- Dalponte, M.; Ørka, H.O.; Ene, L.T.; Gobakken, T.; Næsset, E. Tree crown delineation and tree species classification in boreal forests using hyperspectral and ALS data. Remote Sens. Environ. 2014, 140, 306–317. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Mangold, D.; Schäfer, J.; Immitzer, M.; Kattenborn, T.; Koch, B.; Latifi, H. Estimating stand density, biomass and tree species from very high resolution stereo-imagery-towards an all-in-one sensor for forestry applications? Forestry 2017, 90, 613–631. [Google Scholar] [CrossRef]
- Zhen, Z.; Quackenbush, L.J.; Zhang, L. Trends in Automatic Individual Tree Crown Detection and Delineation—Evolution of LiDAR Data. Remote Sens. 2016, 8, 333. [Google Scholar] [CrossRef]
- Lamprecht, S.; Stoffels, J.; Dotzler, S.; Haß, E.; Udelhoven, T. aTrunk—An ALS-Based Trunk Detection Algorithm. Remote Sens. 2015, 7, 9975–9997. [Google Scholar] [CrossRef] [Green Version]
- Vaughn, N.R.; Asner, G.P.; Brodrick, P.G.; Martin, R.E.; Heckler, J.W.; Knapp, D.E.; Hughes, R.F. An Approach for High-Resolution Mapping of Hawaiian Metrosideros Forest Mortality Using Laser-Guided Imaging Spectroscopy. Remote Sens. 2018, 10, 502. [Google Scholar] [CrossRef]
- Böck, S.; Immitzer, M.; Atzberger, C. On the Objectivity of the Objective Function—Problems with Unsupervised Segmentation Evaluation Based on Global Score and a Possible Remedy. Remote Sens. 2017, 9, 769. [Google Scholar] [CrossRef]
- Weinmann, M.; Weinmann, M.; Mallet, C.; Brédif, M. A Classification-Segmentation Framework for the Detection of Individual Trees in Dense MMS Point Cloud Data Acquired in Urban Areas. Remote Sens. 2017, 9, 277. [Google Scholar] [CrossRef]
- Clark, M.L.; Roberts, D.A. Species-Level differences in hyperspectral metrics among tropical rainforest trees as determined by a tree-based classifier. Remote Sens. 2012, 4, 1820–1855. [Google Scholar] [CrossRef]
- Ellenberg, H.; Leuschner, C. Vegetation Mitteleuropas Mit Den Alpen: in Ökologischer, Dynamischer und Historischer Sicht, 6th ed.; UTB: Stuttgart, Germany, 2010. [Google Scholar]
- Chen, C.; Liaw, A.; Breiman, L. Using Random Forest to Learn Imbalanced Data; University of California, Berkeley: Berkeley, CA, USA, 2004; pp. 1–12. [Google Scholar]
- Cho, M.A.; Debba, P.; Mathieu, R.; Naidoo, L.; Aardt, J.V.; Asner, G.P. Improving discrimination of savanna tree species through a multiple-endmember spectral angle mapper approach: Canopy-level analysis. IEEE Trans. Geosci. Remote Sens. 2010, 48, 4133–4142. [Google Scholar] [CrossRef]
- Fern, C.J.; Warner, T.A. Scale and texture in digital image classification. Photogramm. Eng. Remote Sens. 2002, 68, 51–63. [Google Scholar]
- Atzberger, C.; Guérif, M.; Baret, F.; Werner, W. Comparative analysis of three chemometric techniques for the spectroradiometric assessment of canopy chlorophyll content in winter wheat. Comput. Electron. Agric. 2010, 73, 165–173. [Google Scholar] [CrossRef]
- Jordan, C.F. Derivation of leaf area index from quality of light on the forest floor. Ecology 1969, 50, 663–666. [Google Scholar] [CrossRef]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef] [Green Version]
- Rouse, J.; Haas, R.; Schell, J.; Deering, D. Monitoring Vegetation Systems in the Great Plains with ERTS. 1974. Available online: https://ntrs.nasa.gov/archive/nasa/casi.ntrs.nasa.gov/19740022614.pdf (accessed on 25 June 2018).
- Chamard, P.; Courel, M.F.; Guenegou, M.; Lerhun, J.; Levasseur, J.; Togola, M. Utilisation des bandes spectrales du vert et du rouge pour une meilleure évaluation des formations végétales actives. In Télédétection et Cartographie; AUPELF-UREF: Sherbrooke, QC, Canada, 1991; pp. 203–209. [Google Scholar]
- Jackson, R.D.; Slater, P.N.; Pinter, P.J. Discrimination of Growth and Water Stress in Wheat by Various Vegetation Indices through Clear and Turbid Atmospheres. Remote Sens. Environ. 1983, 13, 187–208. [Google Scholar] [CrossRef]
- Kaufman, Y.J.; Tanre, D. Atmospherically resistant vegetation index (ARVI) for EOS-MODIS. IEEE Trans. Geosci. Remote Sens. 1992, 30, 261–270. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Merzlyak, M.N. Use of a green channel in remote sensing of global vegetation from EOS-MODIS. Remote Sens. Environ. 1996, 58, 289–298. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Merzlyak, M.N. Signature Analysis of Leaf Reflectance Spectra: Algorithm Development for Remote Sensing of Chlorophyll. J. Plant Physiol. 1996, 148, 494–500. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Stark, R.; Rundquist, D. Novel algorithms for remote estimation of vegetation fraction. Remote Sens. Environ. 2002, 80, 76–87. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.Q.; Huete, A.R. A feedback based modification of the NDVI to minimize canopy background and atmospheric noise. IEEE Trans. Geosci. Remote Sens. 1995, 33, 457–465. [Google Scholar] [CrossRef]
- Jiang, Z.; Huete, A.R.; Didan, K.; Miura, T. Development of a two-band enhanced vegetation index without a blue band. Remote Sens. Environ. 2008, 112, 3833–3845. [Google Scholar] [CrossRef]
- Gamon, J.A.; Serrano, L.; Surfus, J.S. The photochemical reflectance index: an optical indicator of photosynthetic radiation use efficiency across species, functional types, and nutrient levels. Oecologia 1997, 112, 492–501. [Google Scholar] [CrossRef] [PubMed]
- Gamon, J.A.; Peñuelas, J.; Field, C.B. A narrow-waveband spectral index that tracks diurnal changes in photosynthetic efficiency. Remote Sens. Environ. 1992, 41, 35–44. [Google Scholar] [CrossRef]
- Gitelson, A.; Merzlyak, M.N. Spectral reflectance changes associated with autumn senescence of Aesculus hippocastanum L. and Acer platanoides L. leaves. Spectral features and relation to chlorophyll estimation. J. Plant Physiol. 1994, 143, 286–292. [Google Scholar] [CrossRef]
- Sims, D.A.; Gamon, J.A. Relationships between leaf pigment content and spectral reflectance across a wide range of species, leaf structures and developmental stages. Remote Sens. Environ. 2002, 81, 337–354. [Google Scholar] [CrossRef]
- Crippen, R. Calculating the vegetation index faster. Remote Sens. Environ. 1990, 34, 71–73. [Google Scholar] [CrossRef]
- Barnes, E.M.; Clarke, T.R.; Richards, S.E.; Colaizzi, P.D.; Haberland, J.; Kostrzewski, M.; Waller, P.; Choi, C.; Riley, E.; Thompson, T.; et al. Coincident detection of crop water stress, nitrogen status and canopy density using ground-based multispectral data. In Proceedings of the Fifth International Conference on Precision Agriculture, Bloomington, MN, USA, 16–19 July 2000. [Google Scholar]
- Merzlyak, M.N.; Gitelson, A.A.; Chivkunova, O.B.; Rakitin, V. Non-destructive optical detection of pigment changes during leaf senescence and fruit ripening. Physiol. Plant. 1999, 106, 135–141. [Google Scholar] [CrossRef]
- Clevers, J.G.P.W. Application of a weighted infrared-red vegetation index for estimating leaf Area Index by Correcting for Soil Moisture. Remote Sens. Environ. 1989, 29, 25–37. [Google Scholar] [CrossRef]
- Clevers, J.G.P.W. The derivation of a simplified reflectance model for the estimation of leaf area index. Remote Sens. Environ. 1988, 25, 53–69. [Google Scholar] [CrossRef]
- Richardson, A.; Wiegand, C. Distinguishing Vegetation from Soil Background Information. Photogramm. Eng. Remote Sens. 1977, 43, 1541–1552. [Google Scholar]











| Common Name | Acronym | Scientific Name | Type | Number |
|---|---|---|---|---|
| European beech | EB | Fagus sylvatica | Broadleaf | 99 |
| Oak species * | OS | Quercus sp. | Broadleaf | 64 |
| European ash | EA | Fraxinus excelsior | Broadleaf | 44 |
| European hornbeam | EH | Carpinus betulus | Broadleaf | 43 |
| Silver birch | SB | Betula pendula | Broadleaf | 41 |
| Sycamore maple | SM | Acer pseudoplatanus | Broadleaf | 40 |
| Wild cherry | WC | Prunus avium | Broadleaf | 37 |
| Black alder | BA | Alnus glutinosa | Broadleaf | 33 |
| Norway spruce | NS | Picea abies | Conifer | 83 |
| European larch | EL | Larix decidua | Conifer | 83 |
| Scots pine | SP | Pinus sylvestris | Conifer | 78 |
| Silver fir | SF | Abies alba | Conifer | 30 |
| Weymouth pine | WP | Pinus strobus | Conifer | 24 |
| Reflectance Values | 1st Derivatives | Vegetation Indices | Textural Metrics | Principal Components | |
|---|---|---|---|---|---|
| Number of Variables | 80 | 4 | 22 | 16 | 80 |
| Purpose | Parameter | Height Separation | |
|---|---|---|---|
| Spatial Radius | 12 | ||
| Strata definition | Range Radius | 3 | |
| Minimum Object Size | 20,000 | ||
| Conifer or small broadleaf | High broadleaf | ||
| Spatial Radius | 2 | 2 | |
| Tree crown delineation | Range Radius | 1 | 2 |
| Minimum Object Size | 5 | 10 | |
| Reference | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EB | OS | EA | EH | SB | SM | WC | BA | NS | EL | SP | SF | WP | ∑ | UA [%] | ||
| Classification | EB | 88 | 5 | 3 | 4 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 104 | 84.6 |
| OS | 4 | 55 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 62 | 88.7 | |
| EA | 2 | 1 | 32 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 36 | 88.9 | |
| EH | 1 | 1 | 0 | 39 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 42 | 92.9 | |
| SB | 1 | 0 | 0 | 0 | 41 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 44 | 93.2 | |
| SM | 2 | 2 | 4 | 0 | 0 | 37 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 47 | 78.7 | |
| WC | 0 | 0 | 0 | 0 | 0 | 0 | 33 | 0 | 0 | 0 | 0 | 0 | 0 | 33 | 100.0 | |
| BA | 1 | 0 | 4 | 0 | 0 | 1 | 0 | 27 | 0 | 0 | 0 | 0 | 0 | 33 | 81.8 | |
| NS | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 78 | 2 | 1 | 0 | 0 | 81 | 96.3 | |
| EL | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 3 | 81 | 0 | 0 | 0 | 85 | 95.3 | |
| SP | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 77 | 0 | 0 | 79 | 97.5 | |
| SF | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 29 | 0 | 29 | 100.0 | |
| WP | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 24 | 24 | 100.0 | |
| ∑ | 99 | 64 | 44 | 43 | 41 | 40 | 37 | 33 | 83 | 83 | 78 | 30 | 24 | 699 | ||
| PA [%] | 88.9 | 85.9 | 72.7 | 90.7 | 100.0 | 92.5 | 89.2 | 81.8 | 94.0 | 97.6 | 98.7 | 96.7 | 100.0 | |||
| OA [%] | 91.7 | |||||||||||||||
| κ | 0.909 | |||||||||||||||
| Reference | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EB | OS | EA | EH | SB | SM | WC | BA | NS | EL | SP | SF | WP | ∑ | UA [%] | ||
| Classification | EB | 84 | 5 | 3 | 3 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 2 | 0 | 100 | 84.0 |
| OS | 6 | 54 | 3 | 1 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 68 | 79.4 | |
| EA | 2 | 1 | 33 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 38 | 86.8 | |
| EH | 1 | 1 | 0 | 38 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 41 | 92.7 | |
| SB | 1 | 0 | 0 | 0 | 40 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 42 | 95.2 | |
| SM | 3 | 2 | 2 | 0 | 0 | 31 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 39 | 79.5 | |
| WC | 1 | 0 | 0 | 0 | 0 | 1 | 33 | 0 | 0 | 0 | 0 | 0 | 0 | 35 | 94.3 | |
| BA | 1 | 1 | 3 | 0 | 0 | 2 | 0 | 28 | 0 | 0 | 0 | 0 | 0 | 35 | 80.0 | |
| NS | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 77 | 2 | 2 | 1 | 0 | 82 | 93.9 | |
| EL | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 4 | 81 | 0 | 1 | 0 | 89 | 91.0 | |
| SP | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 76 | 0 | 0 | 78 | 97.4 | |
| SF | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 26 | 0 | 28 | 92.9 | |
| WP | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 24 | 24 | 100.0 | |
| ∑ | 99 | 64 | 44 | 43 | 41 | 40 | 37 | 33 | 83 | 83 | 78 | 30 | 24 | 699 | ||
| PA [%] | 84.8 | 84.4 | 75.0 | 88.4 | 97.6 | 77.5 | 89.2 | 84.8 | 92.8 | 97.6 | 97.4 | 86.7 | 100.0 | |||
| OA [%] | 89.4 | |||||||||||||||
| κ | 0.883 | |||||||||||||||
| Classification Designation | Variables | OA [%] (CI [%]) | Κ |
|---|---|---|---|
| VNIR (bands) | Bands | 58.1 (54.3, 61.8) | 0.536 |
| VNIR (bands, 1st derivative) | Bands, 1st Deriv. | 59.1 (55.3, 62.8) | 0.547 |
| VNIR (bands, texture) | Bands, Text. | 59.4 (55.6, 63.0) | 0.550 |
| VNIR (bands, indices) | Bands, Ind. | 75.5 (72.2, 78.7) | 0.730 |
| VNIR (bands, PCs) | Bands, PCs | 89.3 (86.7, 91.5) | 0.882 |
| VNIR (bands, indices, PCs) | Bands, Ind., PCs | 91.0 (88.6, 93.0) | 0.901 |
| VNIR (all) | Bands, 1st Deriv., Text., Ind., PCs, | 91.7 (89.4, 93.6) | 0.909 |
| VNIR (all—mean shift) | Bands, 1st Deriv., Text., Ind., PCs, | 89.4 (86.9, 91.6) | 0.883 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maschler, J.; Atzberger, C.; Immitzer, M. Individual Tree Crown Segmentation and Classification of 13 Tree Species Using Airborne Hyperspectral Data. Remote Sens. 2018, 10, 1218. https://doi.org/10.3390/rs10081218
Maschler J, Atzberger C, Immitzer M. Individual Tree Crown Segmentation and Classification of 13 Tree Species Using Airborne Hyperspectral Data. Remote Sensing. 2018; 10(8):1218. https://doi.org/10.3390/rs10081218
Chicago/Turabian StyleMaschler, Julia, Clement Atzberger, and Markus Immitzer. 2018. "Individual Tree Crown Segmentation and Classification of 13 Tree Species Using Airborne Hyperspectral Data" Remote Sensing 10, no. 8: 1218. https://doi.org/10.3390/rs10081218