1. Introduction
Hyperspectral imaging (HSI) allows one to provide information on the spatial and spectral distributions of a target scene simultaneously by acquiring up to hundreds of narrow and adjacent spectral band images ranging from ultraviolet to far-infrared electromagnetic spectrum [
1,
2]. To do this, an imaging sensor such as a charged coupled device collects the different wavelengths dispersed from the incoming light. Then, the signals captured by the imaging sensor are digitized and arranged into pixels of a two-dimensional image
, where
I denotes the size of
X-directional spatial information, and
is the number of quantized spectra of the signals. The procedure of capturing the pixels as an
X-directional single line continues until the spatial range reaches
J, which is the
Y-directional size of the entire target scene. Finally, HSI constructs a three-dimensional data
. Once the HSI data are obtained, they can be used in many applications, such as detecting and identifying objects at a distance in environmental monitoring [
3] or medical image processing [
4], finding anomaly in automatic visual inspection [
5], or detecting and identifying targets of interest [
6,
7]. However, as the area of the target scene
or the number of quantized spectra
increase, the manipulation of
demands prohibitively large computational resources and storage space. To overcome this, efficient compression techniques must be employed as preprocessing for applications to filter out some redundancy along their adjacent spectral bands or spatial information, thereby reducing the size of
.
Owing to the shape of HSI data, the typical mathematical basis for compression techniques is based on tensor decompositions, because it facilitates the simultaneous preservation and analysis of the spatial and spectral structures of data [
8]. Fang et al. used canonical polyadic decomposition (CPD) for dimension reduction of HSI data, where CPD decomposes a tensor into several rank-one tensors [
9,
10]. De Lathauwer et al. suggested a Tucker decomposition-based low rank approximation algorithm of a tensor, where Tucker decomposition decomposes a tensor into a smaller sized core tensor multiplied by a factor matrix along each mode [
11]. In this study, we considered Tucker decomposition for compressing HSI data. Specifically, we focused on developing an efficient algorithm to compute a higher order singular value decomposition (HOSVD), which is a special case of Tucker decomposition with orthogonal constraints on the factor matrices. Subsequently, we applied it to compression problems from real world HSI data.
The remainder of this paper is organized as follows.
Section 2 defines the notations and preliminaries frequently used in this paper.
Section 3 briefly explains the well-known algorithms for computing HOSVD for a compression.
Section 4 introduces the algorithm we propose.
Section 5 provides the experimental results, and
Section 6 concludes the paper.
2. Notations and Preliminaries
Here, we define the symbols and terminology for the simplicity of notation and presentation. We use calligraphic letters to denote tensors, e.g.,
; boldface capital letters for matrices, e.g.,
; boldface lowercase letters for vectors, e.g.,
; and lowercase letters for scalars, e.g.,
a. We define an operation of tensor-matrix multiplication between an arbitrary
N-th order tensor
and a matrix
along mode-
n, such that
where
and the
-th element of
is computed by
for arbitrary
and
. Note that the values
and
are the elements of
and
, respectively [
12].
A tensor
can be matricized with mode-
n, after rearranging each element of
appropriately. For example, a third order tensor
is matricized along each mode, as shown in
Figure 1.
The Frobenius norm of a tensor is the square root of the sum of the squares of all elements in , such that
3. Related Works
We briefly revisit here, well-known algorithms to compute HOSVD of a tensor. HOSVD, which is a special case of Tucker decomposition that has an orthogonal constraint, decomposes an
N-th order tensor
into factor matrices
and a core tensor
such that
while satisfying a constraint
, where a matrix
represents an identity matrix [
11]. Here, a factor matrix
is considered as the principal components in each mode, and elements of a core tensor
reveal the level of interactions between the different components. Note that
, and we denote
as multilinear ranks of
. The representation of
with the form of (
1) is not unique. Thus, there are many algorithms to compute (
1). One of the simplest methods to obtain
and
is computing leading singular vectors of each unfolding matrix of
such that
where
indicates the mode-
n unfolding matrix of
. Then a core tensor
is obtained from
, and
is regarded as a compressed tensor of
. Despite its simple procedure of computation, a single step of applying singular value decomposition (SVD) is insufficient to improve approximations of
in many contexts. De Lathauwer et al. proposed a more accurate algorithm of computing HOSVD via iterative computations of SVD, called higher order orthogonal iterations (HOOI) [
13]. HOOI is designed to solve the optimization problem of finding
and
, such that
Since
, the minimization problem (
3) is identical to finding
; thus, by definition,
Therefore, HOOI obtains each factor matrix
independently from the
, leading singular vectors of the unfolding matrix matricized from
along mode-
k while fixing the other factor matrices. The iterations continue until the output converges. The procedure of HOOI is summarized in Algorithm 1 for
. In practice, HOOI produces more accurate outputs than those from the algorithm based on (
2); it is considered one of the most accurate algorithms for obtaining HOSVD from a tensor. Therefore, many hyperspectral compression techniques have been developed based on HOOI. For example, Zhang et. al applied HOOI to the compression of HSI [
14]. An et al. suggested the method based on [
11] with an adaptive, multilinear rank estimation, and applied it to HSI compression [
15]. However, iterative computations of SVD require huge computational resources. Additionally, the number of iterations for convergence is difficult to predict.
To overcome these limitations, Elden et al. proposed a Newton–Grassman based algorithm that guaranteed quadratic convergence and fewer iteration steps than HOOI [
16]. However a single iteration of the algorithm is much more expensive owing to the computation of the Hessian. Sorber et al. introduced a quasi-Newton optimization algorithm that iteratively improves the initial guess by using a quasi-Newton method from the nonlinear cost function [
17]. Hassanzadeh and karami proposed a block coordinate descent search based algorithm [
18], which updates the factor matrices initialized by using compressed sensing. Instead of employing SVD for unfolding matrices, Phan et al. proposed a fast algorithm based on a Crank–Nicholson-like technique, which has a lower computational cost in a single step compared with HOOI [
19]. Lee proposed a HOSVD algorithm based on an alternating least squares method that recycles the intermediate results of computations for one factor matrix to the other computations [
20]. In contrast to the algorithm proposed by [
20], which computes the Tucker-1 model optimization individually along each mode, the proposed algorithm considers sub-problems of computing a factor matrix simultaneously in a single iteration. This approach enables more accurate computation of the intermediate results in each iteration.
| Algorithm 1 |
| Input: |
| Output: |
| 1: for l = 1, 2, 3, … do |
| 2: for n = 1, 2, 3 do |
| 3: |
| 4: |
| 5: = leading singular vectors of |
| 6: end for |
| 7: |
| 8: if then |
| 9: break |
| 10: end if |
| 11: end for |
4. Sequential Computations of Alternating Least Squares for Efficient HOSVD
In the proposed algorithm, we sequentially compute low multilinear rank factor matrices from the Tucker-1 model optimization problems via an alternating least squares approach. For simplicity, and to consider the shape of a HSI data, we only used a third order tensor in this study. However, the extension of the algorithm for application to any dimensional tensors without loss of generality is straightforward.
Assume that we have a third order tensor
. Our goal is to decompose
to
where
represents an orthogonal factor matrix along mode-
n,
is the appropriate truncation level or
n-multilinear rank, and
denotes the core tensor. Then, we rewrite the formulation of the optimization problem of finding
and
in (
3) as
Before starting the explanation, we note that the order of computation for each factor matrix does not need to be fixed. However, for convenience we will present the procedure for solving our optimization problems from the order of (1,2,3) mode. Let
. Then, the problem of finding
in (
6) is equivalent to the problem of finding the solution from the Tucker-1 model optimization of
, such that
where
denotes an identity matrix. Here, we can see that the tensor
is a Tucker-2 model, but it can be used to formulate another Tucker-1 optimization problem of finding
, such that
where
. Similarly, to find
, we use the optimization problem of Tucker-1 model, which is given by
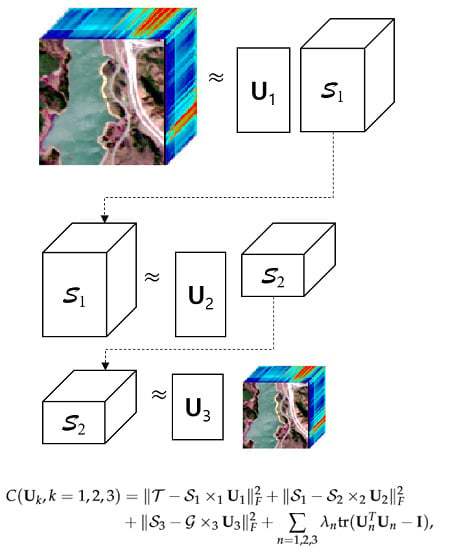
We illustrate the sequential procedure for the Tucker-1 model optimization problems in
Figure 2. Let us consider the optimization problems (
7)–(
9) simultaneously. Then, the optimal factor matrices of the tensor
are computed by solving the minimization problem of the cost function
, which is defined as
where the parameters
represent the Lagrangian multipliers, and the function
computes the trace of an arbitrary matrix
. Because the cost function
in (
10) has too many unknown variables, an alternating least squares method that optimizes one variable while leaving the others fixed, is a proper approach. First, by taking a derivative of
with respect to
while regarding the other variables as constant values, and matricizing those along mode-1, we obtain
where
is the unfolding matrix of
along mode-1. Thus, from (
11), we can compute
as follows:
After
is obtained, the next step is to compute
while fixing
and the others to recycle the intermediate results for computing the others. After updating
in (
12) and by taking a derivative to (
10) with respect to
, we can compute
such that
Because the orthogonal constraint must be satisfied, we reorthogonalize by simply applying QR-decomposition.
Next, we find
. After updating
and
, we take a derivative of
with respect to
and rearrange the terms similar to the procedure of computing
. Then, we obtain
where
and
are the unfolding matrices of
and
along mode-2, respectively. Then, we can compute
with fixed
such that
Finally,
is obtained by solving
where
and
are the unfolding matrices from the tensor
and
along mode-3, respectively.
Algorithm 2 summarizes the procedure explained in this section. Note that the function in steps 6, 10, and 14, returns the unfolding matrix from an arbitrary tensor along mode-i, and the function in steps 9, 13, and 16 returns the reorthogonalized matrix from by applying QR decomposition. If we assume that the size of an input tensor is , and its initial multilinear rank is , then the most expensive step in Algorithm 2 occurs at the computation of in step 7, and its computational complexity is approximately operations, which is similar to those of the other HOSVD algorithms. Additionally, unlike HOOI, we eliminate independence from the computation of each factor matrix by reusing intermediate tensors and factor matrices to find a specific factor matrix. Thus, we expected the proposed algorithm to achieve better convergence to the solution.
| Algorithm 2 |
| Input: |
| Output: |
| 1: for l = 1, 2, 3, … do |
| 2: for n = 1 to 3 do |
| 3: Rearrange the order such that |
| 4: |
| 5: |
| 6: , and |
| 7: |
| 8: |
| 9: |
| 10: , and |
| 11: |
| 12: |
| 13: |
| 14: , and |
| 15: |
| 16: |
| 17: |
| 18: if then |
| 19: break |
| 20: end if |
| 21: end for |
| 22: end for |