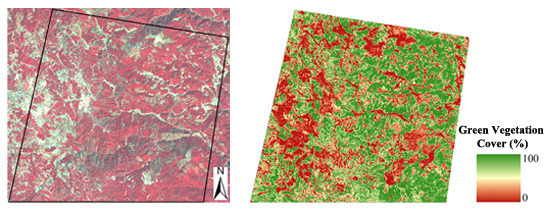
Remote Sensing of Fractional Green Vegetation Cover Using Spatially-Interpolated Endmembers
Abstract
:1. Introduction
2. Study Area and Data
3. Methods
3.1. Atmospheric and Geometric Correction
3.2. Calculation of Vegetation Indices
3.3. Calculation of VIs and VIv at Each Sample Location
3.4. Spatial Interpolation of VIs and VIv
3.5. Calculation of FVC Using Invariant and Interpolated VIs and VIv
3.6. Validation of ASTER FVC Estimates
4. Results and Discussion
5. Conclusions
Acknowledgments
References
- Adams, J.; Smith, M.; Johnson, P. Spectral mixture modeling: A new analysis of rock and soil types at the Viking Lander 1 Site. J. Geophys. Res 1986, 91, 8098–8112. [Google Scholar]
- de Asis, A.; Omasa, K.; Oki, K.; Shimizu, Y. Accuracy and applicability of linear spectral unmixing in delineating potential erosion areas in tropical watersheds. Int. J. Remote Sens 2008, 29, 4151–4171. [Google Scholar]
- Gutman, G.; Ignatov, A. The derivation of the green vegetation fraction from NOAA/AVHRR data for use in numerical weather prediction models. Int. J. Remote Sens 1998, 19, 1533–1543. [Google Scholar]
- Zeng, X.; Dickinson, R.E.; Walker, A.; Shaikh, M.; Defries, R.S.; Qi, J. Derivation and evaluation of global 1-km fractional vegetation cover data for land modeling. J. Appl. Meteorol 2000, 39, 826–839. [Google Scholar]
- Montandon, L.; Small, E. The impact of soil reflectance on the quantification of the green vegetation fraction from NDVI. Remote Sens. Environ 2008, 112, 1835–1845. [Google Scholar]
- James, K.; Stensrud, D.; Yussouf, N. Value of real-time vegetation fraction to forecasts of severe convection in high-resolution models. Weather Forecast 2009, 24, 187–210. [Google Scholar]
- Jiménez-Muñoz, J.; Sobrino, J.; Plaza, A.; Guanter, L.; Moreno, J.; Martinez, P. Comparison between fractional vegetation cover retrievals from vegetation indices and spectral mixture analysis: Case study of PROBA/CHRIS data over an agricultural area. Sensors 2009, 9, 768–793. [Google Scholar]
- Gilalabert, M.; Gonzalez-Piqueras, F.; Garcia-Haro, F.; Melia, J. A generalized soil-adjusted vegetation index. Remote Sens. Environ 2002, 82, 303–310. [Google Scholar]
- Gitelson, A.; Kaufman, Y.; Stark, R.; Rundquist, D. Novel algorithms for remote estimation of vegetation fraction. Remote Sens. Environ 2002, 80, 76–87. [Google Scholar]
- Xiao, J.; Moody, A. A comparison of methods for estimating fractional green vegetation cover within a desert-to-upland transition zone in central New Mexico, USA. Remote Sens. Environ 2005, 98, 237–250. [Google Scholar]
- Zhou, X.; Guan, H.; Xie, H.; Wilson, J. Analysis and optimization of NDVI definitions and areal fraction models in remote sensing of vegetation. Int. J. Remote Sens 2009, 30, 721–751. [Google Scholar]
- Qi, J.; Marsett, R.; Moran, M.; Goodrich, D.; Heilman, P.; Kerr, Y.; Dedieu, G. Spatial and temporal dynamics of vegetation in the San Pedro River basin area. Agr. For. Meteorol 2000, 105, 55–68. [Google Scholar]
- Guilfoyle, K.; Althouse, M.; Chang, C. A quantitative and comparative analysis of linear and nonlinear spectral mixture models using radial basis function neural networks. IEEE Trans. Geosci. Remote Sens 2001, 39, 2314–2318. [Google Scholar]
- Naksuntorn, N. Nonlinear spectral mixture analysis for hyperspectral imagery in an unknown environment. IEEE Geosci. Remote Sens. Lett 2010, 7, 836–840. [Google Scholar]
- Heylen, R.; Burazerovic, D.; Scheunders, P. Non-linear spectral unmixing by geodesic simplex volume maximization. IEEE J. Sel. Top. Signal Process 2011, 5, 534–542. [Google Scholar]
- Obata, K.; Yoshioka, H. Relationships between errors propogated in fraction of vegetation cover by algorithms based on a two-endmember linear mixture model. Remote Sens 2010, 2, 2680–2699. [Google Scholar]
- Theseira, M.; Thomas, G.; Sannier, A. An evaluation of spectral mixture modeling applied to a semi-arid environment. Int. J. Remote Sens 2002, 23, 687–700. [Google Scholar]
- Obata, K.; Yoshioka, H. Inter-algorithm relationships for the estimation of the fraction of vegetation cover based on a two endmember linear mixture model with the VI constraint. Remote Sens 2010, 2, 1680–1701. [Google Scholar]
- Merlin, O.; Duchemin, B.; Hagolle, O.; Jacob, F.; Coudert, B.; Chehbouni, G.; Dedieu, G.; Garatuza, J.; Kerr, Y. Disaggregation of MODIS surface temperature over an agricultural area using a time series of Formosat-2 images. Remote Sens. Environ 2010, 114, 2500–2512. [Google Scholar] [Green Version]
- Baumgardner, M.; Silva, L.; Biehl, L.; Stoner, E. Reflectance Properties of Soils. In Advances in Agronomy; Brady, N., Ed.; America Press Inc.: Orlando, FL, USA, 1985. [Google Scholar]
- Jensen, J. Introductory Digital Image Processing: A Remote Sensing Perspective, 3rd ed.; Pearson Prentice Hall: Upper Saddle River, NJ, USA, 2004. [Google Scholar]
- Richter, R.; Kellenberger, T.; Kaufmann, H. Comparison of topographic correction methods. Remote Sens 2009, 1, 184–196. [Google Scholar]
- Huete, A. A soil-adjusted vegetation index (SAVI). Remote Sens. Environ 1988, 25, 295–309. [Google Scholar]
- Matsui, T.; Lakshmi, V.; Small, E. The effects of satellite-derived vegetation cover variability on simulated land-atmosphere interactions in the NAMS. J. Climate 2005, 18, 21–40. [Google Scholar]
- Johnson, B.; Tateishi, R.; Xie, Z. Using geographically weighted variables for image classification. Remote Sens. Lett 2012, 3, 491–499. [Google Scholar]
- Rouse, J.; W.; Haas, R.; Schell, J.; Deering, D.; Harlan, J. Monitoring the Vernal Advancement of Retrogradation of Natural Vegetation; Type III Final Report; NASA/GSFC: Greenbelt, MD, USA, 1974; p. 371. [Google Scholar]
- Qi, J.; Chehbouni, A.; Huete, A.; Kerr, Y.; Sorooshian, S. A modified soil adjusted vegetation index (MSAVI). Remote Sens. Environ 1994, 48, 119–126. [Google Scholar]
- Kodani, J. Ecology and management of conifer plantations in Japan: control of tree growth and maintenance of biodiversity. J. Forest Res 2006, 11, 267–274. [Google Scholar]
- Seiwa, K.; Eto, Y.; Masaka, K. Effects of thinning intensity on species diversity and timber production in a conifer (Cryptomeria japonica) plantation in Japan. J. Forest. Res 2011. [Google Scholar] [CrossRef]
- Japan Ministry of Agriculture, Forestry and Fisheries. Annual Report on Forest and Forestry in Japan: Fiscal Year 2010 (Summary). 2010. Available online: http://www.rinya.maff.go.jp/j/kikaku/hakusyo/22hakusho/pdf/22_e.pdf (accessed on 20 June 2012).
- Smith, M.; Milton, E. The use of the empirical line method to calibrate remotely sensed data to reflectance. Int. J. Remote Sens 1999, 20, 2653–2662. [Google Scholar]
- Chavez, P. Image-based atmospheric corrections—Revisited and improved. Photogramm. Eng. Remote Sensing 1996, 62, 1025–1036. [Google Scholar]
- Baldridge, A.; Hook, S.; Grove, C.; Rivera, G. The ASTER Spectral Library Version 2.0. Remote Sens. Environ 2009, 113, 711–715. [Google Scholar]
- Moran, P. Notes on continuous stochastic phenomena. Biometrika 1950, 37, 17–23. [Google Scholar]
- Lam, N. Spatial interpolation methods: A review. Cartogr. Geogr. Inform 1983, 10, 129–150. [Google Scholar]
- O’Sullivan, D.; Unwin, D. Geographic Information Analysis; John Wiley & Sons, Inc: Hoboken, NJ, USA, 2003. [Google Scholar]
- Li, J.; Heap, A. A review of comparative studies of spatial interpolation methods in environmental sciences: Performance and impact factors. Ecol. Inform 2011, 6, 228–241. [Google Scholar]
- Kleijnen, J. Kriging metamodeling in simulation: A review. Eur. J. Oper. Res 2009, 192, 707–716. [Google Scholar]
- Kuemmerle, T.; Roder, A.; Hill, J. Separating grassland and shrub vegetation by multidate pixel-adaptive spectral mixture analysis. Int. J. Remote Sens 2006, 27, 3251–3271. [Google Scholar]
- Boardman, J. Geometric Mixture Analysis of Imaging Spectrometry Data. Proceedings of International Geoscience and Remote Sensing Symposium, Pasadena, CA, USA, 8–12 August 1994; 4, pp. 2369–2371.







| NDVI | MSAVI | |||
|---|---|---|---|---|
| MI | p | MI | p | |
| Vis | 0.23 | 0.03 | 0.30 | 0.004 |
| VIv | 0.15 | 0.07 | 0.10 | 0.20 |
| FVC Estimation Method | MAE | Relative Change in MAE (%) | RMSE | Relative Change in RMSE (%) |
|---|---|---|---|---|
| NDVI (invariant) | 0.136 | 0.182 | ||
| OK-NDVI | 0.129 | −5.1% | 0.177 | −2.7% |
| IDW-NDVI | 0.131 | −3.7% | 0.179 | −1.6% |
| MSAVI (invariant) | 0.164 | 0.200 | ||
| OK-MSAVI | 0.16 | −2.4% | 0.196 | −2.0% |
| IDW-MSAVI | 0.164 | 0% | 0.202 | +1.0% |
| Pixel Type | FVC Estimation Method | MAE | Relative Change in MAE (%) | RMSE | Relative Change in RMSE (%) |
|---|---|---|---|---|---|
| Edge | NDVI | 0.178 | 0.221 | ||
| OK-NDVI | 0.175 | −1.7% | 0.219 | −0.9% | |
| IDW-NDVI | 0.174 | −2.2% | 0.22 | −0.5% | |
| MSAVI | 0.183 | 0.218 | |||
| OK-MSAVI | 0.182 | −0.5% | 0.219 | +0.5% | |
| IDW-MSAVI | 0.186 | +1.6% | 0.226 | +3.6% | |
| Non-edge | NDVI | 0.104 | 0.145 | ||
| OK-NDVI | 0.095 | −8.7% | 0.136 | −6.2% | |
| IDW-NDVI | 0.098 | −5.8% | 0.139 | −4.1% | |
| MSAVI | 0.149 | 0.184 | |||
| OK-MSAVI | 0.144 | −3.4% | 0.178 | −3.3% | |
| IDW-MSAVI | 0.147 | −1.3% | 0.182 | −1.1% | |
Share and Cite
Johnson, B.; Tateishi, R.; Kobayashi, T. Remote Sensing of Fractional Green Vegetation Cover Using Spatially-Interpolated Endmembers. Remote Sens. 2012, 4, 2619-2634. https://doi.org/10.3390/rs4092619
Johnson B, Tateishi R, Kobayashi T. Remote Sensing of Fractional Green Vegetation Cover Using Spatially-Interpolated Endmembers. Remote Sensing. 2012; 4(9):2619-2634. https://doi.org/10.3390/rs4092619
Chicago/Turabian StyleJohnson, Brian, Ryutaro Tateishi, and Toshiyuki Kobayashi. 2012. "Remote Sensing of Fractional Green Vegetation Cover Using Spatially-Interpolated Endmembers" Remote Sensing 4, no. 9: 2619-2634. https://doi.org/10.3390/rs4092619