Estimating the Above-Ground Biomass in Miombo Savanna Woodlands (Mozambique, East Africa) Using L-Band Synthetic Aperture Radar Data
Abstract
:1. Introduction
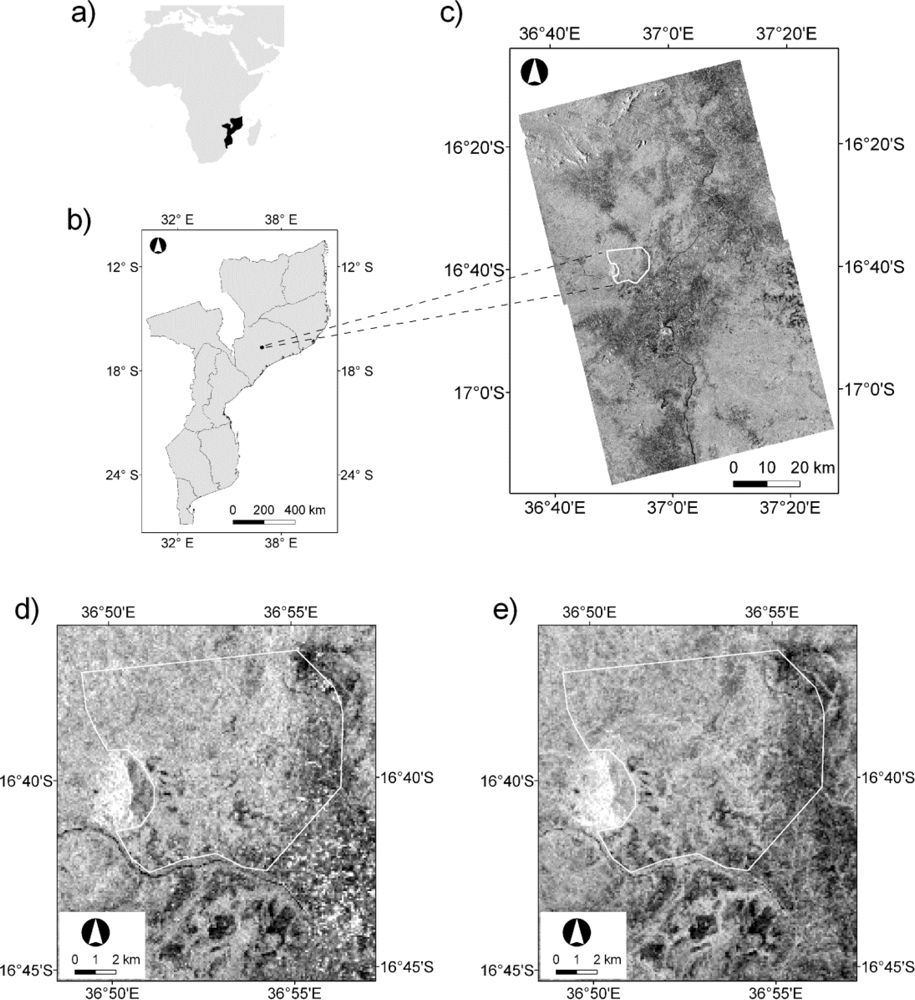
2. Background and Study Area
3. Data
3.1. Field Measurements
- (1)
- The allometric equation developed by Ryan et al.[25] for estimating the C content of the AGB of each tree is given by Equation (1):where Bstem is the tree AGB (kg·C), DBH the diameter at breast height (cm) and ln the natural logarithm. C content was estimated by using a conversion factor of 0.47 [25]. The coefficient of determination (R2) of this fitted equation was 0.93 and the root mean square error was 0.52 ln(kg C) (n = 29).
- (2)
- Chidumayo [26] developed relationships that relate AGB as a function of DBH (in Williams et al.[29]) and are presented in Equations (2) and (3):where AGB is in kg and DBH in cm.
- (3)
- Separate equations for the estimation of AGB as a function of climate and primarily the mean monthly evapotranspiration (ET) and rainfall (R) were developed by Chave et al.[27], with these being wet (ET > R in less than one month per year), moist (ET > R more than one month and less than five months per year) and dry (ET > R more than five months per year) forests. These criteria basically defined the extent of the growing period (when R is greater than ET) as a proportion of the year and on a monthly basis. Thus, the same criteria can be rearranged in terms of growing period (GP) as (i) wet (GP > 11/12), (ii) moist (11/12 > GP > 7/12) and (iii) dry (GP < 7/12). Using the GP dataset produced by Silva et al.[14], the forests of the study area could be considered as belonging to the dry category. Therefore, Equation 4 was used:where AGB is in kg, ρ is the wood specific density (oven-dried on a green volume basis, g·cm−3) and DBH is in cm.
- (4)
- Brown et al.[28] developed another equation (Equation (5)) for the dry forest life zone:where AGB is in kg and DBH is in cm. The model has a R2 = 0.67 and a mean square error of 0.02208 (n = 32).
3.2. ALOS PALSAR
3.3. Extraction of ALOS PALSAR FBD Data at Field Plot Locations
4. Methods
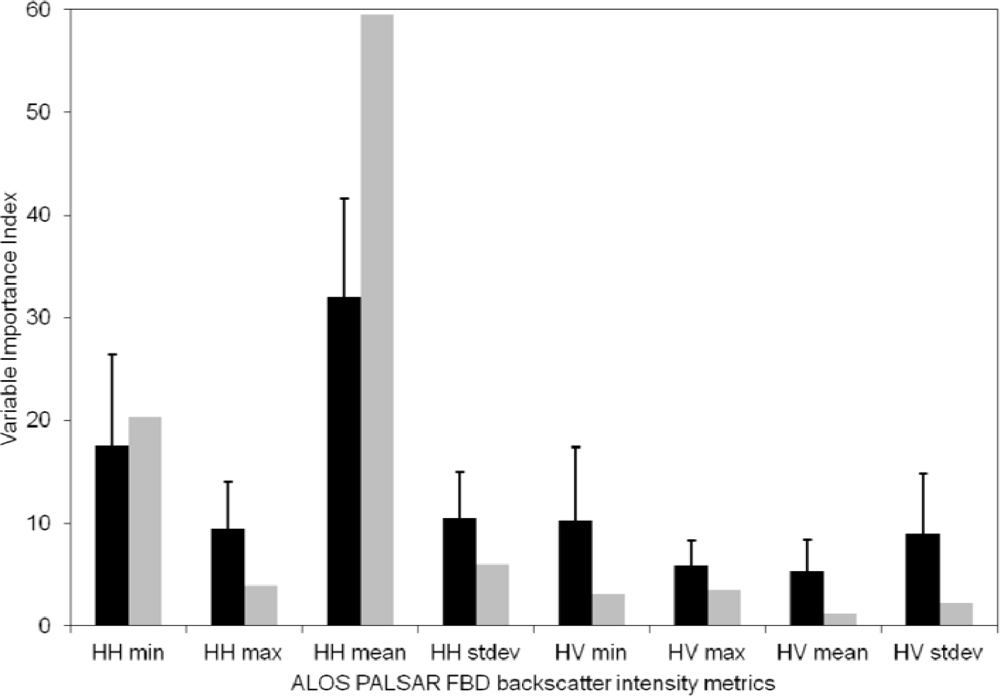
4.1. Contribution of Different ALOS PALSAR Polarizations and Metrics
4.2. Regression with Stochastic Gradient Boosting (SGB) and Bagging SGB (BagSGB)
4.3. C Stocks and Comparison with Published Biomass Maps
5. Results and Discussion
5.1. Forest AGB from Field Data
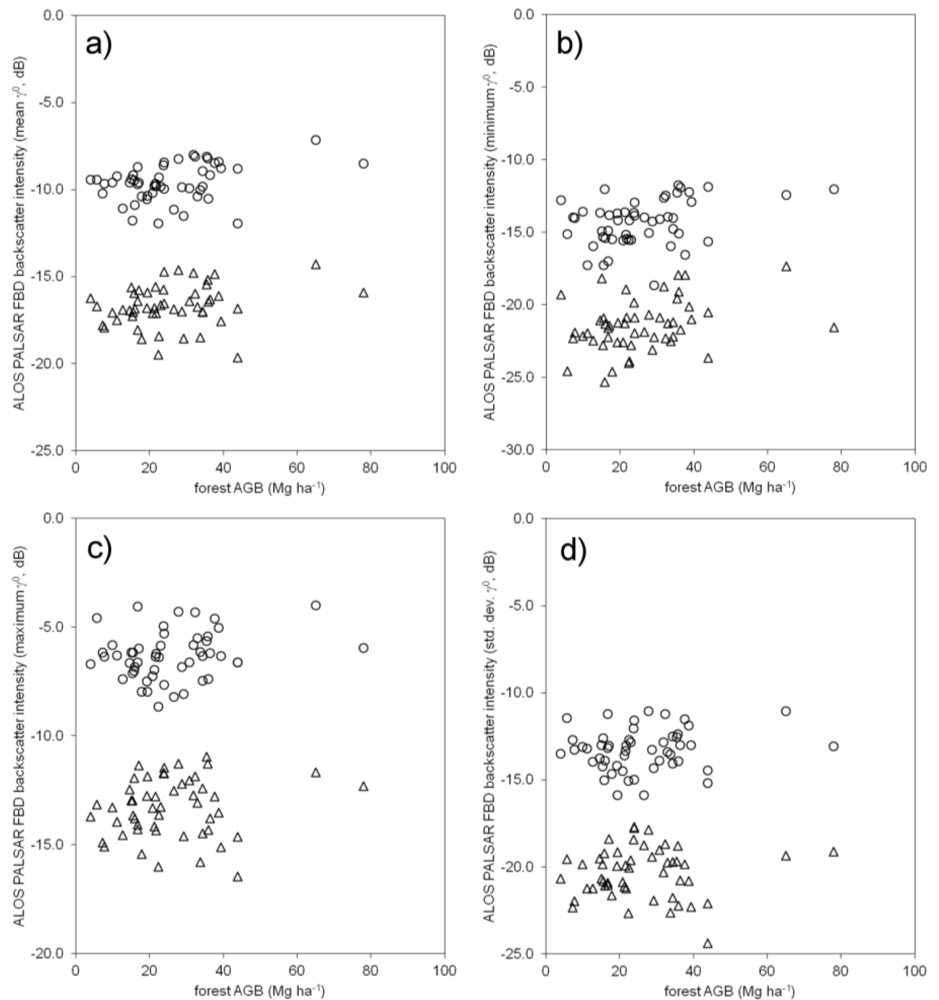
5.2. Contribution of ALOS PALSAR Polarizations and Metrics
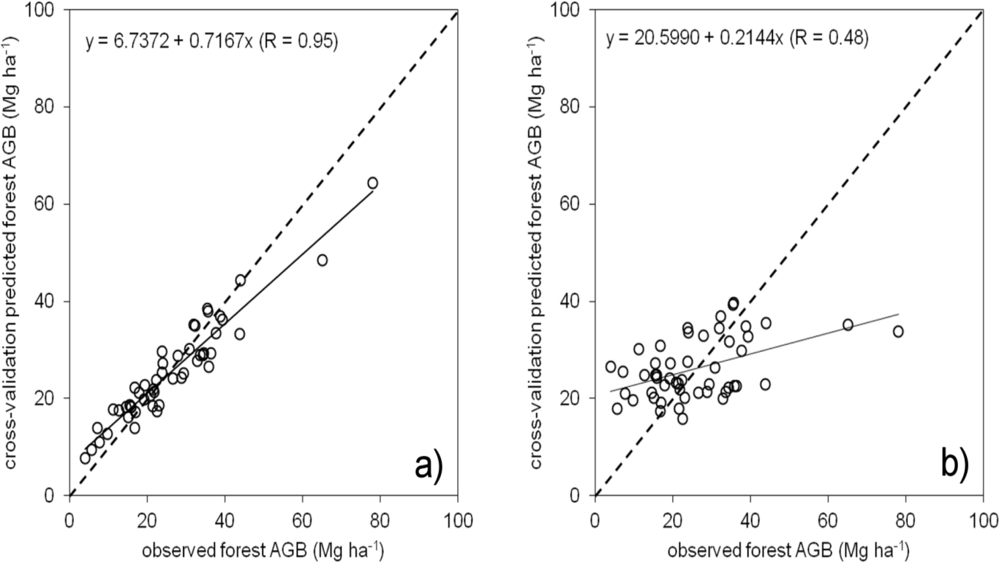
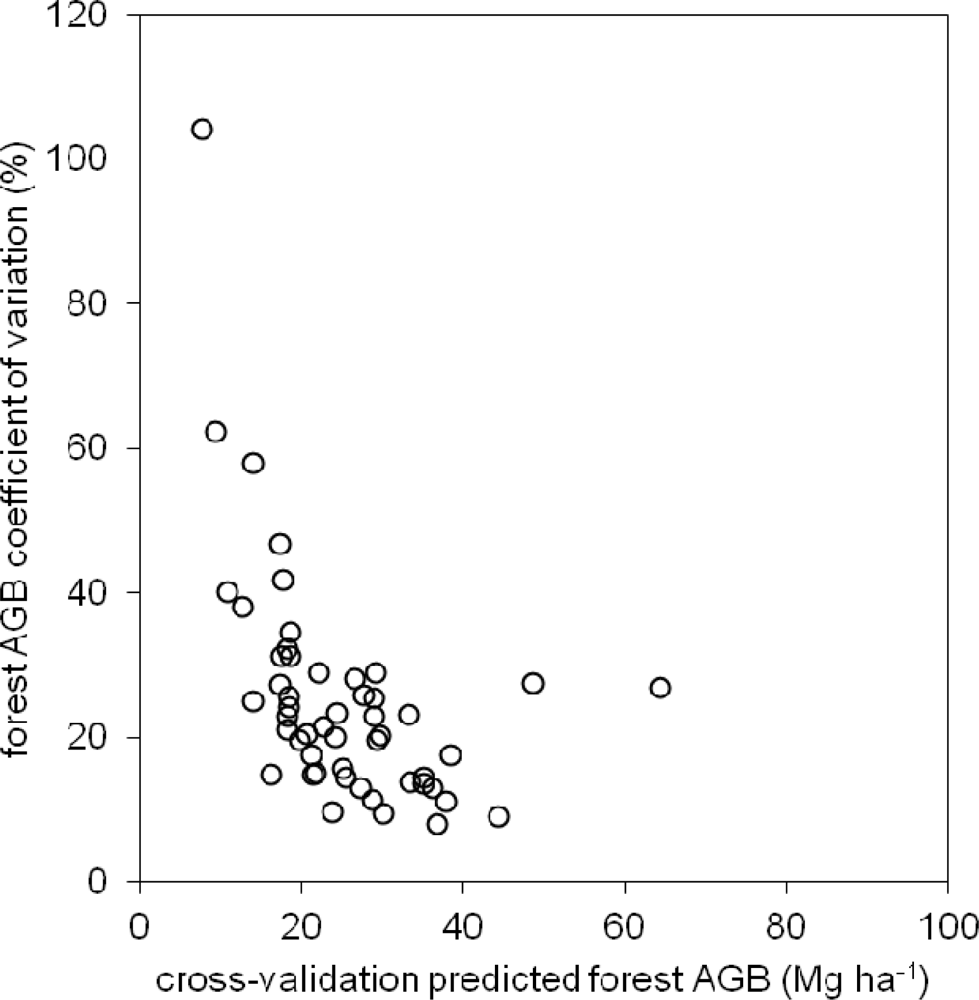
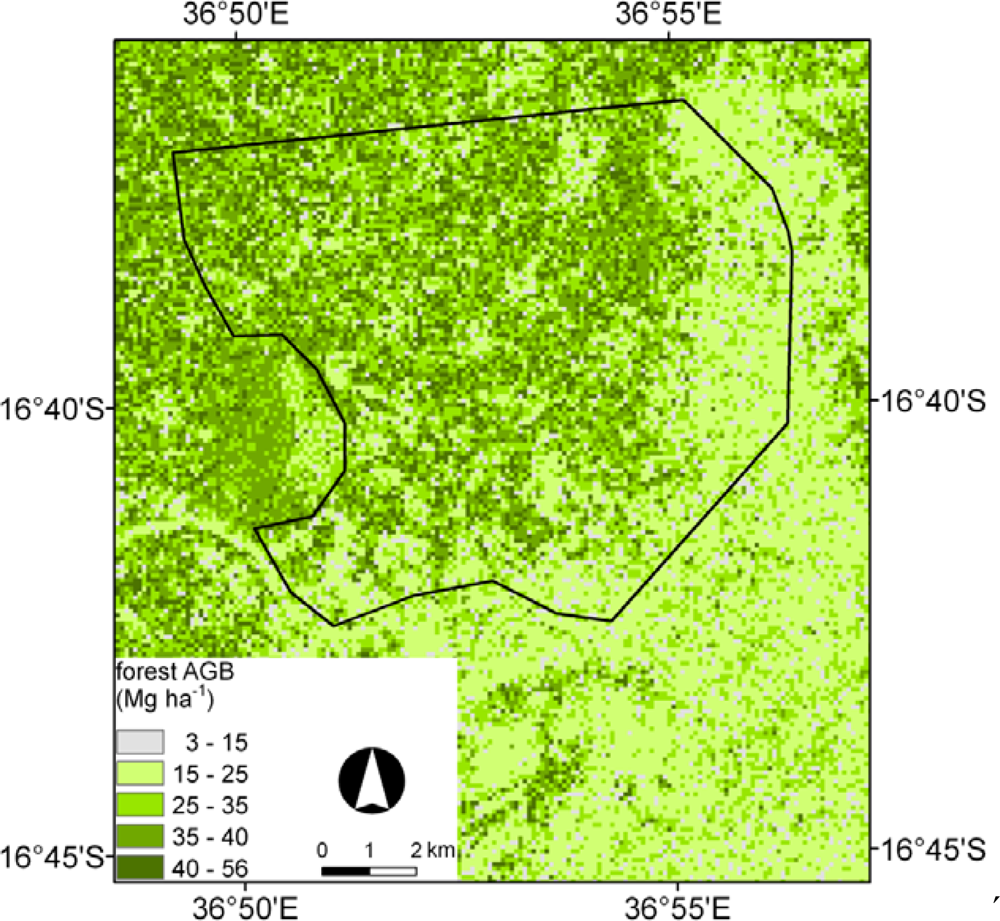
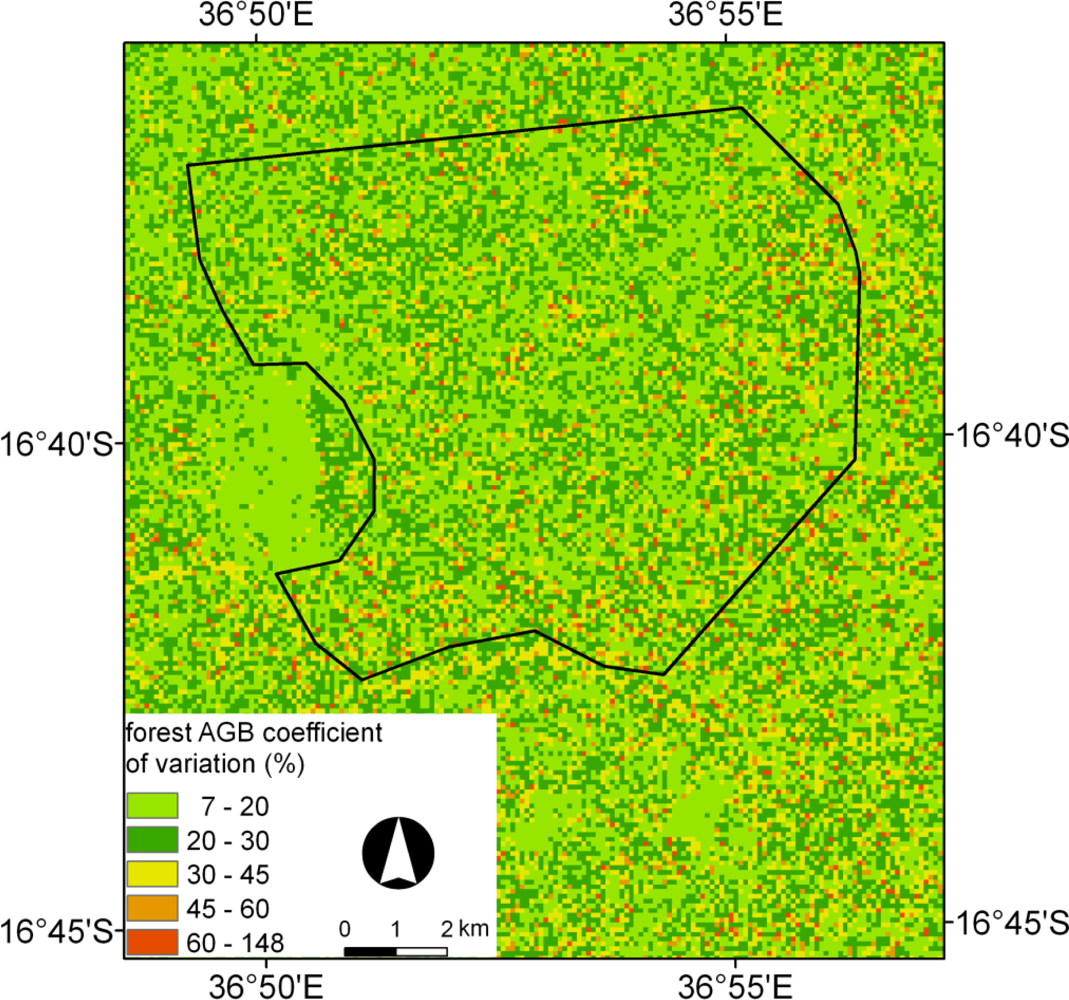
5.3. BagSGB Modeling
5.4. C Stocks and Comparison with Published Biomass Maps
6. Conclusions
Acknowledgments
References
- Pan, Y.; Birdsey, R.; Fang, J.; Houghton, R.; Kauppi, P.; Kurz, W.; Phillips, O.; Shvidenko, A.; Lewis, S.; Canadell, J.; et al. A Large and persistent carbon sink in the world’s forests. Science 2011, 333, 988–993. [Google Scholar]
- Le Quere, C.; Raupach, M.; Canadell, J.; Marland, G.; Bopp, L.; Ciais, P.; Conway, T.; Doney, S.; Feely, R.; Foster, P.; et al. Trends in the sources and sinks of carbon dioxide. Nat. Geosci. 2009. [Google Scholar] [CrossRef]
- Houghton, R.; Hall, F.; Goetz, S. Importance of biomass in the global carbon cycle. J. Geophys. Res.-Biogeosci. 2009, 114. [Google Scholar] [CrossRef]
- Gibbs, H.; Brown, S.; Niles, J.; Foley, J. Monitoring and estimating tropical forest carbon stocks: Making REDD a reality. Environ. Res. Lett. 2007, 2. [Google Scholar] [CrossRef]
- Campbell, B. Beyond Copenhagen: REDD plus, agriculture, adaptation strategies and poverty. Glob. Environ. Chang.-Human Policy Dimens 2009, 19, 397–399. [Google Scholar]
- Danielsen, F.; Skutsch, M.; Burgess, N.D.; Jensen, P.M.; Andrianandrasana, H.; Karky, B.; Lewis, R.; Lovett, J.C.; Massao, J.; Ngaga, Y.; et al. At the heart of REDD: A role for local people in monitoring forests. Conserv. Lett 2011, 4, 158–167. [Google Scholar]
- Herold, M.; Verchot, L.; Angelsen, A.; Maniatis, D.; Bauch, S. A Step-Wise Framework for Setting REDD+ Forest Reference Emission Levels and Forest Reference Levels; Center for International Forestry Research (CIFOR): Bogor, Indonesia, 2012. [Google Scholar]
- Mora, B.; Herold, M.; De Sy, V.; Wijaya, A.; Verchot, L.; Penman, J. Capacity Development in National Forest Monitoring: Experiences and Progress for REDD+; Joint Report by CIFOR and GOFC-GOLD: Bogor, Indonesia, 2012. [Google Scholar]
- Chambers, J.; Asner, G.; Morton, D.; Anderson, L.; Saatch, S.; Espirito-Santo, F.; Palace, M.; Souza, C. Regional ecosystem structure and function: Ecological insights from remote sensing of tropical forests. Trends Ecol. Evol 2007, 22, 414–423. [Google Scholar]
- Goetz, S.J.; Baccini, A.; Laporte, N.T.; Johns, T.; Walker, W.; Kellndorfer, J.; Houghton, R.A.; Sun, M. Mapping and monitoring carbon stocks with satellite observations: A comparison of methods. Carbon Balan. Manag. 2009, 4. [Google Scholar] [CrossRef]
- Lucas, R.; Armston, J.; Fairfax, R.; Fensham, R.; Accad, A.; Carreiras, J.; Kelley, J.; Bunting, P.; Clewley, D.; Bray, S.; et al. An Evaluation of the ALOS PALSAR L-Band Backscatter-Above Ground Biomass Relationship Queensland, Australia: Impacts of Surface Moisture Condition and Vegetation Structure. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens 2010, 3, 576–593. [Google Scholar]
- Saatchi, S.; Harris, N.; Brown, S.; Lefsky, M.; Mitchard, E.; Salas, W.; Zutta, B.; Buermann, W.; Lewis, S.; Hagen, S.; et al. Benchmark map of forest carbon stocks in tropical regions across three continents. Proc. Natl. Acad. Sci. USA 2011, 108, 9899–9904. [Google Scholar]
- Carreiras, J.M.B.; Vasconcelos, M.J.; Lucas, R.M. Understanding the relationship between aboveground biomass and ALOS PALSAR data in the forests of Guinea-Bissau (West Africa). Remote Sens. Environ 2012, 121, 426–442. [Google Scholar]
- Silva, J.M.N.; Carreiras, J.M.B.; Rosa, I.; Pereira, J.M.C. Greenhouse gas emissions from shifting cultivation in the tropics, including uncertainty and sensitivity analysis. J. Geophys. Res.-Atmos. 2011, 116. [Google Scholar] [CrossRef]
- Hall, F.; Bergen, K.; Blair, J.; Dubayah, R.; Houghton, R.; Hurtt, G.; Kellndorfer, J.; Lefsky, M.; Ranson, J.; Saatchi, S.; et al. Characterizing 3D vegetation structure from space: Mission requirements. Remote Sens. Environ 2011, 115, 2753–2775. [Google Scholar]
- Saatchi, S.; Halligan, K.; Despain, D.; Crabtree, R. Estimation of forest fuel load from radar remote sensing. IEEE Trans. Geosci. Remote Sens 2007, 45, 1726–1740. [Google Scholar]
- Popescu, S.; Zhao, K.; Neuenschwander, A.; Lin, C. Satellite lidar vs. small footprint airborne lidar: Comparing the accuracy of aboveground biomass estimates and forest structure metrics at footprint level. Remote Sens. Environ 2011, 115, 2786–2797. [Google Scholar]
- Lefsky, M.; Harding, D.; Keller, M.; Cohen, W.; Carabajal, C.; Espirito-Santo, F.; Hunter, M.; de Oliveira, R. Estimates of forest canopy height and aboveground biomass using ICESat. Geophys. Res. Lett. 2005, 32. [Google Scholar] [CrossRef]
- Neumann, M.; Saatchi, S.; Ulander, L.; Fransson, J. Assessing Performance of L- and P-Band polarimetric interferometric SAR data in estimating boreal forest above-ground biomass. IEEE Trans. Geosci. Remote Sens 2012, 50, 714–726. [Google Scholar]
- Edson, C.; Wing, M. Airborne Light Detection and Ranging (LiDAR) for Individual Tree Stem Location, Height, and Biomass Measurements. Remote Sens 2011, 3, 2494–2528. [Google Scholar] [Green Version]
- Saatchi, S.; Houghton, R.; Alvala, R.; Soares, J.; Yu, Y. Distribution of aboveground live biomass in the Amazon basin. Glob. Chang. Biol 2007, 13, 816–837. [Google Scholar]
- Campbell, B.M. Center for International Forestry Research. In The Miombo in Transition: Woodlands and Welfare in Africa; Center for International Forestry Research: Bogor, Indonesia, 1996. [Google Scholar]
- Mitchell, R.; Mitchell, M. Conservation for Future Generations: The Miombo Ecoregion; WWF-Southern Africa Regional Programme Office: Harare, Zimbabwe, 2001. [Google Scholar]
- European Parliament; Council of the European Union. Directive 2009/28/EC of 23 April 2009 on the promotion of the use of energy from renewable sources and amending and subsequently repealing Directives 2001/77/EC and 2003/30/EC. Official Journal of the European Union 2009, 140, 16–62. [Google Scholar]
- Ryan, C.; Williams, M.; Grace, J. Above- and Belowground Carbon Stocks in a Miombo Woodland Landscape of Mozambique. Biotropica 2011, 43, 423–432. [Google Scholar]
- Chidumayo, E.N.; Stockholm Environment Institute. Miombo Ecology and Management: An Introduction; IT Publications in association with the Stockholm Environment Institute: London, UK, 1997. [Google Scholar]
- Chave, J.; Andalo, C.; Brown, S.; Cairns, M.; Chambers, J.; Eamus, D.; Folster, H.; Fromard, F.; Higuchi, N.; Kira, T.; et al. Tree allometry and improved estimation of carbon stocks and balance in tropical forests. Oecologia 2005, 145, 87–99. [Google Scholar]
- Brown, S.; Gillespie, A.; Lugo, A. Biomass estimation methods for tropical forests with applications to forest inventory data. For. Sci 1989, 35, 881–902. [Google Scholar]
- Williams, M.; Ryan, C.; Rees, R.; Sarnbane, E.; Femando, J.; Grace, J. Carbon sequestration and biodiversity of re-growing miombo woodlands in Mozambique. For. Ecol. Manag 2008, 254, 145–155. [Google Scholar]
- Rosenqvist, A.; Shimada, M.; Ito, N.; Watanabe, M. ALOS PALSAR: A Pathfinder mission for global-scale monitoring of the environment. IEEE Trans. Geosci. Remote Sens 2007, 45, 3307–3316. [Google Scholar]
- Ulaby, F.T.; Dobson, M.C. Handbook of Radar Scattering Statistics for Terrain; Artech House: Norwood, MA, USA, 1989. [Google Scholar]
- Meier, E.; Frei, U.; Nüesch, D. Precise Terrain Corrected Geocoded Images. In SAR Geocoding: Data and Systems; Schreier, G., Ed.; Herbert Wichmann Verlag GmbH: Karlsruhe, Germany, 1993; pp. 173–186. [Google Scholar]
- Oliver, C.; Quegan, S. Understanding Synthetic Aperture Radar Images; SciTech Publishing: Raleigh, NC, USA, 2004. [Google Scholar]
- Woodhouse, I.H. Introduction to Microwave Remote Sensing; Taylor Francis: Boca Raton, FL, USA, 2006. [Google Scholar]
- Holecz, F.; Meier, E.; Piesbergen, J.; Nüesch, D. Topographic Effects on Radar Cross Section. Proceedings of the CEOS SAR Calibration Workshop, Noordwijk, The Netherlands, 20–24 September 1993; pp. 23–28.
- Holecz, F.; Meier, E.; Piesbergen, J.; Nüesch, D.; Moreira, J. In Rigorous Derivation of Backscattering Coefficient. Proceedings of the CEOS SAR Calibration Workshop, Ann Arbor, MI, USA, 28–30 September 1994; pp. 143–155.
- Steel, R.G.D.; Torrie, J.H. Principles and Procedures of Statistics: A Biometrical Approach, 2nd ed.; McGraw-Hill: New York, NY, USA, 1980. [Google Scholar]
- Hastie, T.; Tibshirani, R.; Friedman, J.H. The Elements of Statistical Learning: Data Mining, Inference, and Prediction, 2nd ed.; Springer: New York, NY, USA, 2009. [Google Scholar]
- Moisen, G.; Freeman, E.; Blackard, J.; Frescino, T.; Zimmermann, N.; Edwards, T. Predicting tree species presence and basal area in Utah: A comparison of stochastic gradient boosting, generalized additive models, and tree-based methods. Ecol. Model 2006, 199, 176–187. [Google Scholar]
- Elith, J.; Leathwick, J.; Hastie, T. A working guide to boosted regression trees. J. Anim. Ecol 2008, 77, 802–813. [Google Scholar]
- Carreiras, J.; Pereira, J.; Campagnolo, M.; Shimabukuro, Y. Assessing the extent of agriculture/pasture and secondary succession forest in the Brazilian Legal Amazon using SPOT VEGETATION data. Remote Sens. Environ 2006, 101, 283–298. [Google Scholar]
- Friedl, M.; Sulla-Menashe, D.; Tan, B.; Schneider, A.; Ramankutty, N.; Sibley, A.; Huang, X. MODIS Collection 5 global land cover: Algorithm refinements and characterization of new datasets. Remote Sens. Environ 2010, 114, 168–182. [Google Scholar]
- Friedman, J. Greedy function approximation: A gradient boosting machine. Ann. Stat 2001, 29, 1189–1232. [Google Scholar]
- Friedman, J. Stochastic gradient boosting. Comput. Stat. Data Anal 2002, 38, 367–378. [Google Scholar]
- Breiman, L.; Friedman, J.H.; Olshen, R.A.; Stone, C.J. Classification and Regression Trees; CRC Press: Boca Raton, FL, USA, 1984. [Google Scholar]
- Safavian, S.; Landgrebe, D. A survey of decision tree classifier methodology. IEEE Trans. Syst. Man Cybern 1991, 21, 660–674. [Google Scholar]
- Hansen, M.; Dubayah, R.; DeFries, R. Classification trees: An alternative to traditional land cover classifiers. Int. J. Remote Sens 1996, 17, 1075–1081. [Google Scholar]
- Friedl, M.; Brodley, C. Decision tree classification of land cover from remotely sensed data. Remote Sens. Environ 1997, 61, 399–409. [Google Scholar]
- Breiman, L. Bagging predictors. Mach. Learn 1996, 24, 123–140. [Google Scholar]
- Breiman, L. Arcing classifiers. Ann. Stat 1998, 26, 801–824. [Google Scholar]
- Bauer, E.; Kohavi, R. An empirical comparison of voting classification algorithms: Bagging, boosting, and variants. Mach. Learn 1999, 36, 105–139. [Google Scholar]
- De'ath, G. Boosted trees for ecological modeling and prediction. Ecology 2007, 88, 243–251. [Google Scholar]
- Sankaran, M.; Ratnam, J.; Hanan, N. Woody cover in African savannas: The role of resources, fire and herbivory. Glob. Ecol. Biogeogr 2008, 17, 236–245. [Google Scholar]
- Jalabert, S.; Martin, M.; Renaud, J.; Boulonne, L.; Jolivet, C.; Montanarella, L.; Arrouays, D. Estimating forest soil bulk density using boosted regression modelling. Soil Use Manag 2010, 26, 516–528. [Google Scholar]
- Lawrence, D.; Radel, C.; Tully, K.; Schmook, B.; Schneider, L. Untangling a decline in tropical forest resilience: Constraints on the sustainability of shifting cultivation across the globe. Biotropica 2010, 42, 21–30. [Google Scholar]
- Suen, Y.; Melville, P.; Mooney, R.; Gama, J.; Camacho, R.; Brazdil, P.; Jorge, A.; Torgo, L. Combining Bias and Variance Reduction Techniques for Regression Trees. Proceedings of the 2005 European Conference on Machine Learning, Porto, Portugal, 3–7 October 2005; 3720, pp. 741–749.
- R Development Core Team. R: A Language and Environment for Statistical Computing; 2.14.1; R Foundation for Statistical Computing: Vienna, Austria, 2011. [Google Scholar]
- Ridgeway, G. Generalized Boosted Models: A Guide to the GBM package. Available online: http://cran.open-source-solution.org/web/packages/gbm/vignettes/gbm.pdf (accessed on 1 March 2012).
- Intergovernmental Panel on Climate Change. 2006 IPCC Guidelines for National Greenhouse Gas Inventories; Eggleston, H.S., Buendia, L., Miwa, K., Ngara, T., Tanabe, K., Eds.; Institute for Global Environmental Strategies: Kanagawa, Japan, 2006. Available online: http://www.ipcc-nggip.iges.or.jp/public/2006gl/index.html (accessed on 1 March 2012).
- Baccini, A.; Laporte, N.; Goetz, S.; Sun, M.; Dong, H. A first map of tropical Africa’s above-ground biomass derived from satellite imagery. Environ. Res. Lett. 2008, 3. [Google Scholar] [CrossRef]
- Bevington, P.R.; Robinson, D.K. Data Reduction and Error Analysis for the Physical Sciences, 3rd ed.; McGraw-Hill: Boston, MA, USA, 2003. [Google Scholar]
- Richards, J.A. Remote Sensing with Imaging Radar; Springer-Verlag: Berlin, Germany, 2009. [Google Scholar]
- Mitchard, E.; Saatchi, S.; Lewis, S.; Feldpausch, T.; Gerard, F.; Woodhouse, I.; Meir, P. Comment on “A first map of tropical Africa’s above-ground biomass derived from satellite imagery”. Environ. Res. Lett. 2011, 6. [Google Scholar] [CrossRef]
- Glenday, J. Carbon storage and emissions offset potential in an African dry forest, the Arabuko-Sokoke Forest, Kenya. Environ. Monit. Assess 2008, 142, 85–95. [Google Scholar]
- Malimbwi, R.E.; Solberg, B.; Luoga, E. Estimation of biomass and volume in miombo woodland at Kitulangalo Forest Reserve, Tanzania. J. Trop. For. Sci 1994, 7, 230–242. [Google Scholar]
- Shirima, D.; Munishi, P.; Lewis, S.; Burgess, N.; Marshall, A.; Balmford, A.; Swetnam, R.; Zahabu, E. Carbon storage, structure and composition of miombo woodlands in Tanzania’s Eastern Arc Mountains. Afr. J. Ecol 2011, 49, 332–342. [Google Scholar]
- Romijn, H. Land clearing and greenhouse gas emissions from Jatropha biofuels on African Miombo Woodlands. Energy Policy 2011, 39, 5751–5762. [Google Scholar]
- Ryan, C.M.; Hill, T.; Woollen, E.; Ghee, C.; Mitchard, E.; Cassells, G.; Grace, J.; Woodhouse, I.H.; Williams, M. Quantifying small-scale deforestation and forest degradation in African woodlands using radar imagery. Glob. Chang. Biol 2012, 18, 243–257. [Google Scholar]
| Tree Canopy Cover (%) | # Plots | AGB (tC·ha−1) |
|---|---|---|
| 10–30 | 23 | 10.8 (±1.4) |
| 30–40 | 16 | 11.6 (±1.2) |
| 40–50 | 12 | 15.4 (±2.1) |
| total | 51 | 12.1 (±0.9) |
| Polarization | ALOS PALSAR FBD Backscatter Intensity Metrics | |||||||
|---|---|---|---|---|---|---|---|---|
| Mean | Minimum | Maximum | Standard Deviation | |||||
| R | Rrank | R | Rrank | R | Rrank | R | Rrank | |
| HH | 0.36 * | 0.30 * | 0.36 * | 0.31 * | 0.22 | 0.21 | 0.14 | 0.14 |
| HV | 0.24 | 0.25 | 0.33 * | 0.33 * | 0.13 | 0.13 | 0.03 | 0.06 |
| Model | RMSE | s2 | b | R |
|---|---|---|---|---|
| BagSGB | 5.03 | 24.96 | 0.58 | 0.95 |
| SGB | 12.04 | 144.81 | −0.32 | 0.48 |
| Reference | Tree Canopy Cover (%) | Mean AGB (Mg ha−1) | Total AGB C Stock (Mg C) | Uncertainty1 | |
|---|---|---|---|---|---|
| This study | 10–30 | 27.2 | 7,910 | 14,155 | 0.75 |
| 30–40 | 30.1 | 3,390 | 1.25 | ||
| 40–50 | 31.3 | 2,856 | 1.35 | ||
| Baccini et al.[60] | 10–30 | 120.3 | 38,064 | 66,377 | n.a. |
| 30–40 | 126.4 | 15,945 | n.a. | ||
| 40–50 | 133.2 | 12,367 | n.a. | ||
| Saatchi et al.[12] | 10–30 | 91.4 | 28,629 | 49,704 | 3.55 |
| 30–40 | 95.1 | 11,891 | 5.54 | ||
| 40–50 | 95.5 | 9,184 | 5.29 | ||
Share and Cite
Carreiras, J.M.B.; Melo, J.B.; Vasconcelos, M.J. Estimating the Above-Ground Biomass in Miombo Savanna Woodlands (Mozambique, East Africa) Using L-Band Synthetic Aperture Radar Data. Remote Sens. 2013, 5, 1524-1548. https://doi.org/10.3390/rs5041524
Carreiras JMB, Melo JB, Vasconcelos MJ. Estimating the Above-Ground Biomass in Miombo Savanna Woodlands (Mozambique, East Africa) Using L-Band Synthetic Aperture Radar Data. Remote Sensing. 2013; 5(4):1524-1548. https://doi.org/10.3390/rs5041524
Chicago/Turabian StyleCarreiras, João M. B., Joana B. Melo, and Maria J. Vasconcelos. 2013. "Estimating the Above-Ground Biomass in Miombo Savanna Woodlands (Mozambique, East Africa) Using L-Band Synthetic Aperture Radar Data" Remote Sensing 5, no. 4: 1524-1548. https://doi.org/10.3390/rs5041524




