Training Area Concept in a Two-Phase Biomass Inventory Using Airborne Laser Scanning and RapidEye Satellite Data
Abstract
:1. Introduction
2. Materials
2.1. Study Area and Field Data
2.2. Remote Sensing Data
3. Methods
3.1. Preprocessing and Estimation of ALS Predictors
3.2. RapidEye Image Preprocessing
3.3. Estimation of RapidEye predictors
3.4. Two-Phase Sampling Method
3.5. Statistical Modeling
3.6. Model Accuracy Assessment
4. Results
4.1. Model Building and Accuracy at Phase I (ALS data)
4.2. Model Building and Accuracy at Phase II (RapidEye Data)
4.3. Accuracy at the Independent Validation Plots
5. Discussion and Conclusion
5.1. Statistical Modelling
5.2. Method Pros and Cons
5.3. Model Calibration and Validation
5.4. Sensor Pros and Cons
5.5. Concluding Remarks
Acknowledgments
Conflicts of Interest
References
- Winjum, J.K.; Dixon, R.K.; Schroeder, P.E. Forest management and carbon storage: An analysis of 12 key forest nations. Water Air Soil Pollut 1993, 70, 239–257. [Google Scholar]
- United Nations Framework Convention on Climate Change (UNFCCC). Kyoto Protocol Reference Manual on Accounting of Emissions and Assigned Amounts. Available online: http://unfccc.int/files/national_reports/accounting_reporting_and_review_under_the_kyoto_protocol/application/pdf/rm_final.pdf (accessed on 10 June 2013).
- Rosenqvist, A.; Milne, A.; Lucas, R.; Imhoff, M.; Dobson, C. A review of remote sensing technology in support of the Kyoto protocol. Environ. Sci. Policy 2003, 6, 441–455. [Google Scholar]
- Intergovernmental Panel on Climate Change (IPCC). Good Practice Guidance for Land Use, Land-Use Change and Forestry; IPCC National Greenhouse Gas Inventories Programme: Hayama, Japan, 2003. [Google Scholar]
- Streck, C.; Scholz, S.M. The role of forests in global climate change: Whence we come and where we go. Int. Aff 2006, 82, 861–879. [Google Scholar]
- Southworth, J.; Gibbes, C. Digital remote sensing within the field of land change science: Past, present and future directions. Geogr. Compass 2010, 4, 1695–1712. [Google Scholar]
- Houghton, R.A. Aboveground forest biomass and the global carbon balance. Glob. Chang. Biol 2005, 11, 945–958. [Google Scholar]
- Saatchi, S.; Houghton, R.A.; Dos-Santos-Alvala, R.C.; Soares, J.V.; Yu, Y. Distribution of aboveground live biomass in the Amazon basin. Glob. Chang. Biol 2007, 13, 816–837. [Google Scholar]
- Global Observation of Forest and Land Cover Dynamics (GOFC-GOLD). Reducing Greenhouse Gas Emissions from Deforestation and Degradation in Developing Countries: A Sourcebook of Methods and Procedures for Monitoring, Measuring and Reporting; GOFC-GOLD Report Version COP14-2; GOFC-GOLD Project Office, Natural Resources Canada: Edmonton, AB, Canada, 2009. [Google Scholar]
- Wilkie, M.L. Global Forest Resource Assessment Report, Finland; FRA Report No. 69; FAO Forestry Department VialedelleTerme di Caracalla: Rome, Italy, 2010. [Google Scholar]
- Myneni, R.B.; Keeling, C.D.; Tucker, C.J.; Asrar, G.; Nemani, R.R. Increased plant growth in the northern high latitudes from 1981 to 1991. Nature 1997, 386, 698–702. [Google Scholar]
- Hame, T.; Salli, A.; Andersson, K.; Lohi, A. A new methodology for the estimation of biomass of conifer dominated boreal forest using NOAA AVHRR data. Int. J. Remote Sens 1997, 18, 3211–3243. [Google Scholar]
- Kauppi, P.E.; Mielikäinen, K.; Kuusela, K. Biomass and carbon budget of European forests, 1971 to 1990. Science 1992, 256, 70–74. [Google Scholar]
- Næsset, E.; Gobakken, T.; Holmgren, J.; Hyyppä, H.; Hyyppä, J.; Maltamo, M.; Nilsson, M.; Olsson, H.K.; Persson, Å S.; Derman, U.S. US Laser scanning of forest resources: The Nordic experience. Scand. J. For. Res 2004, 19, 482–499. [Google Scholar]
- Drake, J.B.; Knox, R.G.; Dubayah, R.O.; Clark, D.B.; Condit, R.; Blair, J.B.; Hofton, M. Above-ground biomass estimation in closed canopy neotropical forests using ALS remote sensing: Factors affecting the generality of relationships. Glob. Ecol. Biogeogr 2003, 12, 147–159. [Google Scholar]
- Asner, G.P.; Hughes, R.F.; Varga, T.A.; Knapp, D.E.; Kennedy-Bowdoin, T. Environmental and biotic controls over aboveground biomass throughout a rain forest. Ecosystems 2009, 12, 261–278. [Google Scholar]
- Hou, Z.; Xu, Q.; Tokola, T. Use of ALS, Airborne CIR and ALOS AVNIR-2 data for estimating tropical forest attributes in Lao PDR. ISPRS J. Photogramm. Remote Sens 2011, 66, 776–786. [Google Scholar]
- Maltamo, M.; Bollandäs, O.M.; Næsset, E.; Gobakken, T.; Packalén, P. Different plot selection strategies for field training data in ALS-assisted forest inventory. Forestry 2011, 84, 23–31. [Google Scholar]
- Watt, P.; Watt, M. Applying satellite imagery for forest planning. NZ J. For 2011, 56, 23–25. [Google Scholar]
- Ozdemir, I.; Ozkan, K.; Mert, A.; Ozkan, U.Y.; Senturk, O.; Alkan, O. Mapping Forest Stand Structural Diversity Using Rapideye Satellite Data. Available online: http://congrexprojects.com/docs/12c04_docs2/poster2_6_ozdemir.pdf (accessed on 11 December 2012).
- Garcia, M.; Riano, R.; Chuvieco, E.; Danson, F.M. Estimating biomass carbon stocks for a Mediterranean forest in central Spain using LiDAR height and intensity data. Remote Sens. Environ 2010, 114, 816–830. [Google Scholar]
- Gautam, B.R. LiDAR-Assisted Multi-source Program (LAMP) for FRA Nepal; FRA Bulletin No. 1; MoFSC/Department of Forest Research and Survey: Kathmandu, Nepal, 2011. [Google Scholar]
- Tamura, H.; Mori, S.; Yamawaki, T. Textural features corresponding to visual perception. IEEE Trans. Syst. Man Cybern 1978, 8, 460–472. [Google Scholar]
- Haralick, M.R.; Shanmugam, K.; Dinstein, J. Textural features for image classification. IEEE Trans. Syst. Man Cybern 1973, 3, 610–621. [Google Scholar]
- Tuominen, S.; Pekkarinen, A. Performance of different spectral and textural aerial photograph features in multi-source forest inventory. Remote Sens. Environ 2005, 94, 256–268. [Google Scholar]
- Packalén, P.; Maltamo, M. The k-MSN method for the prediction of species specific stand attributes using airborne laser scanning and aerial photographs. Remote Sens. Environ 2007, 109, 328–341. [Google Scholar]
- Holopainen, M.; Talvitie, M. Forest inventory by means of tree-wise 3D-measurements of laser scanning data and digital aerial photographs. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci 2012, XXXVI-8/W2, 67–72. [Google Scholar]
- Tegel, K. A Comparison of Landsat-7 ETM+ and Terrasar-X Satellite Imagery in Estimating Forest Aboveground Biomass in a Two-Stage Sampling Procedure. M.Sc. Dissertation,. University of Helsinki: Helsinki, Finland, 2011. [Google Scholar]
- Gautam, B.; Peuhkurinen, J.; Kauranne, T.; Gunia, K.; Tegel, K.; Latva-Käyrä, P.; Rana, P.; Eivazi, A.; Kolesnikov, A.; Hämäläinen, J.; et al. Estimation of Forest Carbon Using LiDAR-Assisted Multi-Source Programme (LAMP) in Nepal. Proceedings of the International Conference on Advanced Geospatial Technologies for Sustainable Environment and Culture, Pokhara, Nepal, 12–13 September 2013.
- Laasasenaho, J. Taper curve and volume function for pine, spruce and birch. Commun. Instituti. For. Fenn 1982, 108, 1–74. [Google Scholar]
- Repola, J. Biomass equations for scots pine and norway spruce in Finland. Silva. Fenn 2009, 43, 625–647. [Google Scholar]
- Repola, J. Biomass equations for birch in Finland. Silva. Fenn 2008, 42, 605–624. [Google Scholar]
- RapidEye. RapidEye—Delivering the World. Available online: http://www.rapideye.de (accessed on 1 February 2013).
- Axelsson, P. DEM Generation from Laser Scanner Data Using Adaptive TIN Models. Proceedings of the XIXth ISPRS Conference, IAPRS, Amsterdam, The Netherlands, 16–22 July 2000.
- Næsset, E. Practical large-scale forest stand inventory using small-footprint airborne scanning laser. Scand. J. For. Res 2004, 19, 164–179. [Google Scholar]
- Junttila, V.; Kauranne, T.; Leppanen, V. Estimation of forest stand parameters from airborne laser scanning using calibrated plot databases. For. Sci 2010, 56, 257–270. [Google Scholar]
- Næsset, E. Predicting forest stand characteristics with airborne scanning laser using a practical two-stage procedure and field data. Remote Sens. Environ 2002, 80, 88–99. [Google Scholar]
- Song, C.; Woodcock, C.E.; Seto, K.C.; Lenney, M.P.; Macomber, S.A. Classification and change detection using Landsat TM data: When and how to correct atmospheric effects? Remote Sens. Environ 2001, 75, 230–244. [Google Scholar]
- Du, Y.; Teillet, P.M.; Cihlar, J. Radiometric normalization of multitemporal high-resolution satellite images with quality control for land cover change detection. Remote Sens. Environ 2002, 82, 123–134. [Google Scholar]
- Xu, Q.; Hou, Z.; Tokola, T. Relative radiometric correction of multi-temporal ALOS AVNIR-2 data for the estimation of forest attributes. ISPRS J. Photogramm. Remote Sens 2012, 68, 69–78. [Google Scholar]
- Baldi, G.; Nosetto, M.D.; Aragón, R.; Aversa, F.; Paruelo, J.M.; Jobbágy, E.G. Long-term satellite NDVI data sets: Evaluating their ability to detect ecosystem functional changes in South America. Sensors 2008, 8, 5397–5425. [Google Scholar]
- Seager, S.; Turner, E.L.; Schafer, J.; Ford, E.B. Vegetation’s red-edge: A possible spectroscopic bio signature of extraterrestrial plants. Astrobiology 2005, 5, 173–194. [Google Scholar]
- Barnes, E.M.; Clarke, T.R.; Richards, S.E.; Colaizzi, P.D.; Haberland, J.; Kostrzewski, M.; Waller, P.; Choi, C.; Riley, E.; Thompson, T.; et al. Coincident Detection of Crop Water Stress, Nitrogen Status and Canopy Density Using Ground-Based Multispectral Data. Proceedings of the Fifth International Conference on Precision Agriculture, Bloomington, MN, USA, 16–19 July 2000. [CD Rom].
- R Development Core Team. R: A Language and Environment for Statistical Computing. Available online: http://www.R-project.org/ (accessed on 2 June 2013).
- Næsset, E.; Gobakken, T. Estimation of above- and below-ground biomass across regions of the boreal forest zone using airborne laser. Remote Sens. Environ 2008, 112, 3079–3090. [Google Scholar]
- Latifi, H.; Nothdurft, A.; Koch, B. Non-parametric prediction and mapping of standing timber volume and biomass in a temperate forest: Application of multiple optical/ALS-derived predictors. Forestry 2010, 83, 395–407. [Google Scholar]
- Heiskanen, J. Estimating aboveground tree biomass and leaf area index in a mountain birch forest using ASTER satellite data. Int. J. Remote Sens 2006, 27, 1135–1158. [Google Scholar]
- Haara, A.; Korhonen, K.T. Kuvioittaisen arvioinnin luotettavuus. Metsätieteenaikakauskirja 2004, 4, 489–508. [Google Scholar]
- Kangas, A.; Heikkinen, E.; Maltamo, M. Accuracy of partially visually assessed stand characteristics—A case study of Finnish forest inventory by compartments. Can. J. For. Res 2004, 34, 916–930. [Google Scholar]
- Montgomery, D.C.; Peck, E.A.; Vining, G.G. Introduction to Linear Regression Analysis; John Wiley & Sons, Inc: Hoboken, NJ, USA, 2006. [Google Scholar]
- Nelson, R.; Hyde, P.; Johnson, P.; Emessiene, B.; Imhoff, M.L.; Campbell, R.; Edwards, W. Investigating RaDAR-LiDAR synergy in a North Carolina pine forest. Remote Sens. Environ 2007, 110, 98–108. [Google Scholar]
- Patenaude, G.; Hill, R.A.; Milne, R.; Gaveau, D.L.A.; Briggs, B.B.J.; Dawson, T.P. Quantifying forest above ground carbon content using LiDAR remote sensing. Remote Sens. Environ 2004, 93, 368–380. [Google Scholar]
- Næsset, E.; Bollandäs, O.M.; Gobakken, T. Comparing regression methods in estimation of biophysical properties of forest stands from two different inventories using laser scanner data. Remote Sens. Environ 2004, 94, 541–553. [Google Scholar]
- Packalén, P.; Suvanto, A.; Maltamo, M. A two stage method to estimate species-specific growing stock by combining ALS data and aerial photographs of known orientation parameters. Photogramm. Eng. Remote Sens 2009, 75, 1451–1460. [Google Scholar]
- Packalén, P.; Temesgen, H.; Maltamo, M. Variable selection strategies for nearest neighbor imputation methods used in remote sensing based forest inventory. Can. J. Remote Sens 2012, 38, 1–13. [Google Scholar]
- Rana, M.P. Effect of Field Plot Location on Estimating Tropical Forest Attributes of Nepal. University of Eastern Finland, Joensuu, Finland, 2012. [Google Scholar]
- Boudreau, J.; Nelson, R.F.; Margolis, H.A.; Beaudoin, A.; Guindon, L.; Kimes, D.S. An analysis of regional aboveground forest biomass using airborne and spaceborne LIDAR in Québec. Remote Sens. Environ 2008, 112, 3876–3890. [Google Scholar]
- Gautam, B.R.; Tokola, T.; Hamalainen, J.; Gunia, M.; Peuhkurinen, J.; Parviainen, H.; Leppanen, V.; Kauranne, T.; Havia, J.; Norjamaki, I.; et al. Integration of Airborne LiDAR, Satellite Imagery and Field Measurements Using A Two-Phase Sampling Method for Forest Biomass Estimation in Tropical Forests. Proceedings of the International Symposium on Benefiting from Earth Observation, 4–6 October 2010; Kathmandu, Nepal.
- Junttila, V.; Maltamo, M.; Kauranne, T. Sparse Bayesian estimation of forest stand characteristics from airborne laser scanning. For. Sci 2008, 54, 543–552. [Google Scholar]
- Latifi, H.; Koch, B. Evaluation of most similar neighbor and random forest methods for imputing forest inventory variables using data from target and auxiliary stands. Int. J. Remote Sens 2012, 33, 6668–6694. [Google Scholar]
- Dalponte, M.; Martinez, C.; Rodeghiero, M.; Gianelle, D. The role of ground reference data collection in the prediction of stem volume with ALS data in mountain areas. ISPRS J. Photogramm. Remote Sens 2011, 66, 787–797. [Google Scholar]
- Hall, S.A.; Burke, I.C.; Box, D.O.; Kaufmann, M.R.; Stoker, J.M. Estimating stand structure using discrete-return lidar: An example from low density, fire prone ponderosa pine forests. For. Ecol. Manag 2005, 208, 189–209. [Google Scholar]
- Bright, B.C.; Hicke, J.A.; Hudak, A.T. Estimating aboveground carbon stocks of a forest affected by mountain pine beetle in Idaho using lidar and multispectral imagery. Remote Sens. Environ 2012, 124, 270–281. [Google Scholar]
- Fu, A.; Sun, G.; Guo, Z. Estimating forest biomass with GLAS samples and MODIS imagery in northeastern China. Proc. SPIE 2009. [Google Scholar] [CrossRef]
- Gobakken, T.; Næsset, E.; Nelson, R.; Bollandäs, O.M.; Gregoire, T.G.; Ståhl, G.; Holm, S.; Ørka, H.O.; Astrup, R. Estimating biomass in Hedmark County, Norway using national forest inventory field plots and airborne laser scanning. Remote Sens. Environ 2012, 123, 443–456. [Google Scholar]
- Hawbaker, T.J.; Keuler, N.S.; Lesak, A.A.; Gobakken, T.; Contrucci, K.; Radeloff, V.C. Improved estimates of forest vegetation structure and biomass with a LiDAR optimized sampling design. J. Geophy. Res.: Biogeosci 2009. [Google Scholar] [CrossRef]
- Ferster, C.J.; Coops, N.C.; Trofymow, J.A. Aboveground large tree mass estimation in a coastal forest in British Columbia using plot-level metrics and individual tree detection from lidar. Can. J. Remote Sens 2009, 35, 270–275. [Google Scholar]
- Næsset, E. Effects of different sensors, flying altitudes, and pulse repetition frequencies on forest canopy metrics and biophysical stand properties derived from small-footprint airborne laser data. Remote Sens. Environ 2009, 113, 148–159. [Google Scholar]
- Gómez, C.; Wulder, M.A.; Montes, F.; Delgado, J.A. Modeling forest structural parameters in the Mediterranean pines of central Spain using QuickBird-2 imagery and Classification and Regression Tree Analysis (CART). Remote Sens 2012, 4, 135–159. [Google Scholar]
- Eckert, S. Improved forest biomass and carbon estimations using texture measures from WorldView-2 satellite data. Remote Sens 2012, 4, 810–829. [Google Scholar]
- Muukkonen, P.; Heiskanen, J. Estimating biomass for boreal forests using ASTER satellite data combined with standwise forest inventory data. Remote Sens. Environ 2005, 99, 434–447. [Google Scholar]
- Tokola, T.; Heikkilä, J. Improving satellite image based forest inventory by using a priori site quality information. Silva. Fenn 1997, 31, 67–78. [Google Scholar]
- Tomppo, E.; Nilsson, M.; Rosengren, M.; Aalto, P.; Kennedy, P. Simultaneous use of Landsat-TM and IRS-1c WiFS data in estimating large area tree stem volume and aboveground biomass. Remote Sens. Environ 2002, 82, 156–171. [Google Scholar]
- Kilpeläinen, P.; Tokola, T. Gain to be achieved from stand delineation in Landsat TM image-based estimates of stand volume. For. Ecol. Manag 1999, 124, 105–111. [Google Scholar]
- Hyyppä, J.; Hyyppä, H.; Inkinen, M.; Engdahl, M.; Linko, S.; Zhu, Y.H. Accuracy comparison of various remote sensing data sources in the retrieval of forest stand attributes. For. Ecol. Manag 2000, 128, 109–120. [Google Scholar]
- Makela, H.; Pekkarinen, A. Estimation of forest stand volumes by Landsat TM imagery and stand-level field-inventory data. For. Ecol. Manag 2004, 196, 245–255. [Google Scholar]
- Hyvönen, P. Kuvioittaisten puustotunnusten ja toimenpide-ehdotusten estimointi k-lähimmän naapurin menetelmälläLandsatTM-satelliittikuvan, vanhan inventointitiedon ja kuviotason tukiaineiston avulla. Metsätieteen Aikakauskirja 2002, 3, 363–379. [Google Scholar]
- Koch, B. Status and future of laser scanning, synthetic aperture radar and hyper-spectral remote sensing data for forest biomass assessment. ISPRS J. Photogramm. Remote Sens 2010, 65, 581–590. [Google Scholar]
- Treitz, P.; Lim, K.; Woods, M.; Pitt, D.; Nesbitt, D.; Etheridge, D. LiDAR sampling density for forest resource inventories in Ontario, Canada. Remote Sens 2012, 4, 830–848. [Google Scholar]
- Kankare, V.; Vastaranta, M.; Holopainen, M.; Räty, M.; Yu, X.; Hyyppä, J.; Hyyppä, H.; Alho, P.; Viitala, R. Retrieval of forest aboveground biomass and stem volume with airborne scanning LiDAR. Remote Sens 2013, 5, 2257–2274. [Google Scholar]
Appendix
| Models | Predictors | Estimate | S.E. | t Value | Pr(>|t|) |
|---|---|---|---|---|---|
| ALS AGB (50 training plots) | Intercept | 455.1 | 27.0 | 16.8 | *** |
| Dhlp4 | −397.0 | 31.2 | −12.7 | *** | |
| ALS volume (50 training plots) | Intercept | 859.4 | 49.7 | 17.2 | *** |
| Dhlp4 | −754.4 | 57.5 | −13.1 | *** | |
| ALS AGB (25 training plots) | Intercept | 462.5 | 51.8 | 8.9 | *** |
| Dhlp4 | −406.8 | 59.8 | −6.8 | *** | |
| ALS volume (25 training plots) | Intercept | 822.2 | 97.9 | 8.4 | *** |
| Dhlp4 | −716.3 | 113.0 | −6.3 | *** | |
| ALS AGB (10 training plots) | Intercept | 430.9 | 48.1 | 8.9 | *** |
| Dhlp4 | −371.9 | 54.9 | −6.7 | *** | |
| ALS volume (10 training plots) | Intercept | 864.7 | 92.5 | 9.3 | *** |
| Dhlp4 | −763.6 | 105.6 | −7.2 | *** | |
| RapidEye AGB (50 training plots and 200 surrogate plots) | Intercept | 3,356.7 | 345.3 | 9.7 | *** |
| B2 | −0.52 | 0.07 | −7.0 | *** | |
| B3 | −0.06 | 0.03 | −2.1 | * | |
| B4 | 0.74 | 0.09 | 8.5 | *** | |
| B5 | −0.07 | 0.01 | −6.4 | *** | |
| NDVI2 | −33.6 | 3.8 | −8.7 | *** | |
| HR5 | −181.6 | 34.4 | −5.2 | *** | |
| RapidEye volume (50 training plot and 200 surrogate plot) | Intercept | 5,952.4 | 633.3 | 9.4 | *** |
| B2 | −1.0 | 0.14 | −7.3 | *** | |
| B4 | 1.2 | 0.15 | 8.4 | *** | |
| B5 | −0.10 | 0.02 | −6.6 | *** | |
| NDVI2 | −56.8 | 6.5 | −8.6 | *** | |
| HR5 | −345.1 | 66.0 | −5.2 | *** | |
| RapidEye AGB (25 training plots and 200 surrogate plots) | Intercept | 3,435.1 | 353.7 | 9.7 | *** |
| B2 | −0.53 | 0.07 | −7.0 | *** | |
| B3 | −0.07 | 0.03 | −2.1 | * | |
| B4 | 0.75 | 0.08 | 8.5 | *** | |
| B5 | −0.07 | 0.01 | −6.4 | *** | |
| NDVI2 | −34.4 | 3.9 | −8.7 | *** | |
| HR5 | −186.0 | 35.2 | −5.2 | *** | |
| RapidEye volume (25 training plots and 200 surrogate plots) | Intercept | 5,658.5 | 601.3 | 9.4 | *** |
| B2 | −0.97 | 0.13 | −7.3 | *** | |
| B4 | 1.2 | 0.14 | 8.4 | *** | |
| B5 | −0.10 | 0.02 | −6.6 | *** | |
| NDVI2 | −53.9 | 6.2 | −8.6 | *** | |
| HR5 | −327.7 | 62.6 | −5.2 | *** | |
| RapidEye AGB (10 training plots and 200 surrogate plots) | Intercept | 3,148.3 | 323.3 | 9.7 | *** |
| B2 | −0.48 | 0.07 | −7.0 | *** | |
| B3 | −0.06 | 0.03 | −2.1 | * | |
| B4 | 0.69 | 0.08 | 8.5 | *** | |
| B5 | −0.07 | 0.01 | −6.4 | *** | |
| NDVI2 | −31.5 | 3.5 | −8.7 | *** | |
| HR5 | −170.1 | 32.2 | −5.2 | *** | |
| RapidEye volume (10 training plots and 200 surrogate plots) | Intercept | 6,020.3 | 641.0 | 9.3 | *** |
| B2 | −1.0 | 0.14 | −7.3 | *** | |
| B4 | 1.2 | 0.15 | 8.4 | *** | |
| B5 | −0.11 | 0.02 | −6.6 | *** | |
| NDVI2 | −57.5 | 6.6 | −8.6 | *** | |
| HR5 | −349.3 | 66.8 | −5.2 | *** |







| Parameters | Scots Pine | Norway Spruce | Deciduous |
|---|---|---|---|
| Fixed effects | |||
| b0 | −3.1 | −1.8 | −3.6 |
| b1 | 9.5 | 9.4 | 10.5 |
| b2 | 3.2 | 0.4 | 3.0 |
| Random effects | |||
| uk | 0.009 | 0.006 | 0.00068 |
| eki | 0.010 | 0.013 | 0.000727 |
| Minimum | Maximum | Mean | SD | |
|---|---|---|---|---|
| Training plots, n = 50 | ||||
| Total volume (m3/ha) | 96.1 | 433.8 | 209.3 | 74.9 |
| AGB (ton/ha) | 51.5 | 226.6 | 113.0 | 39.7 |
| Validation plots, n = 28 | ||||
| Total volume (m3/ha) | 103.6 | 382.5 | 219.1 | 69.0 |
| AGB (ton/ha) | 55.9 | 182.2 | 115.7 | 31.5 |
| Predictors Number | ALS Predictors | Description |
|---|---|---|
| 1...10 | Hfp10–100 | Height for which the cumulative sum of ordered first and single echo heights is closest to 10%, 20%, 30%...100% of the total height sum. |
| 11...20 | Hlp10–100 | Height for which the cumulative sum of ordered last and single echo heights is closest to 10%, 20%, 30%...100% of the total height sum. |
| 21...23 | Ifp30–90 | Intensity for which the cumulative sum of ordered first and single echo intensities is closest to 30%, 60% and 90% of the total intensity sum. |
| 24...26 | Ilp30–90 | Intensity for which the cumulative sum of ordered last and single echo intensities is closest to 30%, 60% and 90% of the total intensity sum. |
| 27 | Hmean | Mean height of first and single echo vegetation points (points over high vegetation threshold 5 m). |
| 28 | Hstd | Standard deviation of first and single echo heights. |
| 29 | Dfp | Ratio of the number of first and single echoes below 5 m (low vegetation) and the total number of first and single echoes. |
| 30 | Dlp | Ratio of the number of last and single echoes below 5 m (low vegetation) and the total number of last and single echoes. |
| 31...38 | Dhlp0–7 | Ratio of last and single echoes with height lower than 1.5 m + i × 3 m for i = 0.7 and the total number of last and single echoes. |
| 39...41 | Dfp10,30,50 | Ratio of first and single echoes with intensity I ≤ 0.5+i for i = 10, 30, 50 and the total number of first and single echoes. |
| 42...44 | Dlp10,30,50 | Ratio of last and single echoes with intensity I ≤ 0.5+i for i = 10, 30, 50 and the total number of last and single echoes. |
| 45 | Dflog | Logarithm of the ratio of the number of first and single echoes below 5 m (low vegetation) and the total number of first and single echoes. |
| 46 | Hf3mean | Mean of the largest three heights within first and single echoes. |
| RapidEye Predictors | Description |
|---|---|
| B1 | Blue (mean) |
| B2 | Green (mean) |
| B3 | Red (mean) |
| B4 | Red-edge (mean) |
| B5 | NIR (mean) |
| NDVI1 | First NDVI (mean) |
| NDVI2 | Second NDVI (mean) |
| NDVI3 | Third NDVI (mean) |
| HR1 | Angular second moment |
| HR2 | Contrast |
| HR3 | Correlation |
| HR4 | Sum of squares |
| HR5 | Inverse difference moment |
| HR6 | Sum average |
| HR7 | Sum variance |
| HR8 | Sum entropy |
| HR9 | Entropy |
| HR10 | Difference variance |
| HR11 | Difference entropy |
| HR12 | Information measures of correlation |
| HR13 | Information measures of correlation |
| HR14 | Maximum correlation coefficient |
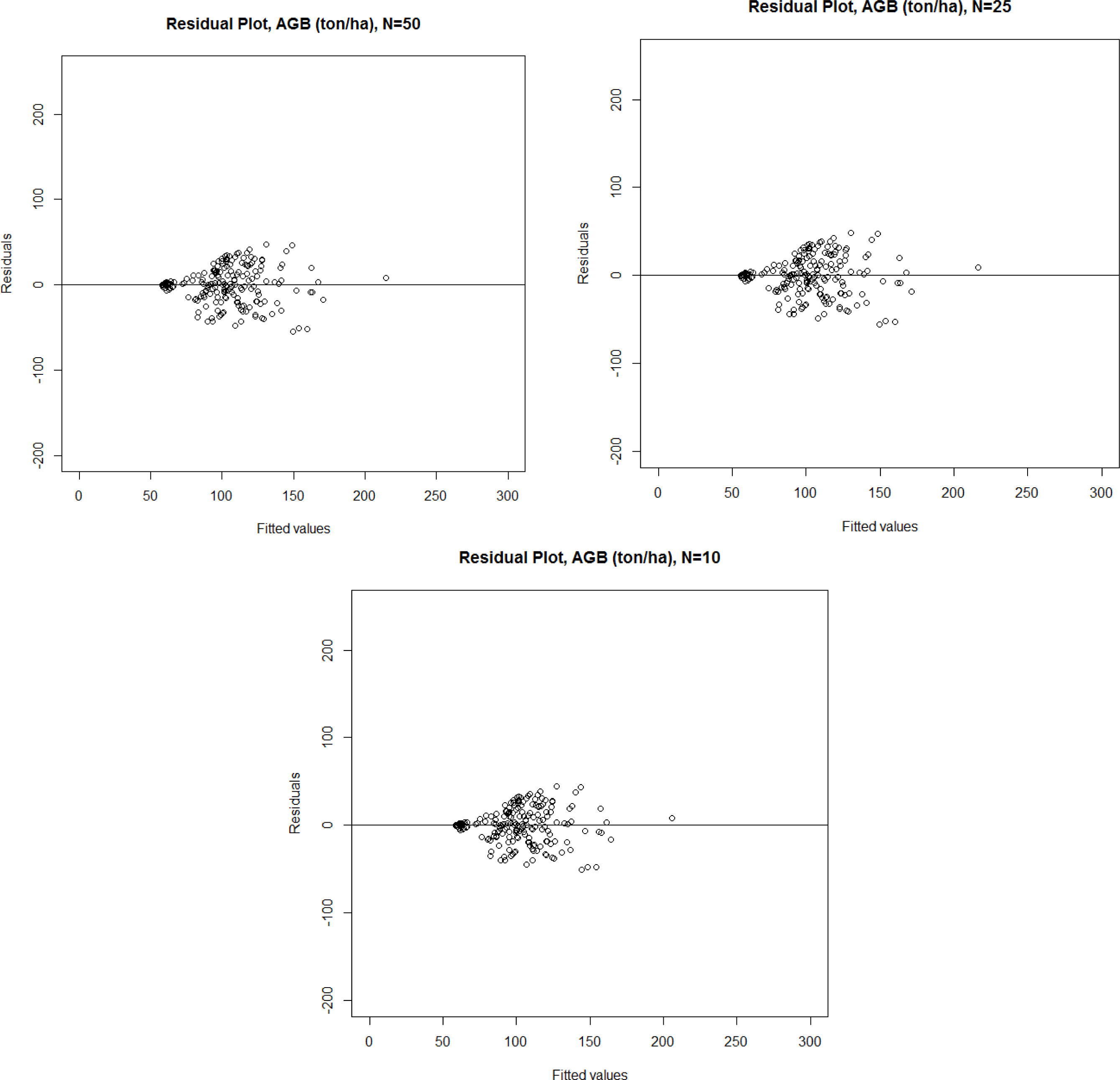
| Parameter | Number of Training Plots (n) | Mean of Estimates | Standard Deviation of Estimates | RMSE | RMSE % | Bias | Bias % | R2 Adj. |
|---|---|---|---|---|---|---|---|---|
| AGB *, ton/ha | 50 | 112.9 | 34.8 | 18.7 | 16.6 | 0.0 | 0.0 | 76.6 |
| Volume, m3/ha | 209.2 | 66.2 | 34.5 | 16.5 | 0.0 | 0.0 | 77.7 | |
| AGB *, ton/ha | 25 | 111.5 | 31.9 | 22.0 | 19.7 | 0.0 | 0.0 | 65.3 |
| Volume, m3/ha | 204.1 | 56.2 | 41.7 | 20.4 | 0.0 | 0.0 | 61.9 | |
| AGB *, ton/ha | 10 | 106.6 | 32.9 | 13.0 | 12.2 | 0.0 | 0.0 | 83.2 |
| Volume, m3/ha | 198.8 | 67.7 | 25.1 | 12.6 | 0.0 | 0.0 | 85.0 |
| Parameter | Number of Training Plots (n) ǂ | Number of Surrogate Plots | Mean of Estimates | Standard Deviation of Estimates | RMSE | RMSE % | Bias | Bias % | R2 Adj. |
|---|---|---|---|---|---|---|---|---|---|
| AGB *, ton/ha | 50 | 200 | 102.7 | 26.2 | 21.0 | 20.4 | 0.0 | 0.0 | 59.5 |
| Volume, m3/ha | 189.7 | 49.3 | 40.6 | 21.3 | 0.0 | 0.0 | 58.4 | ||
| AGB *, ton/ha | 25 | 101.4 | 26.8 | 21.5 | 21.2 | 0.0 | 0.0 | 59.5 | |
| Volume, m3/ha | 186.3 | 46.8 | 38.5 | 20.6 | 0.0 | 0.0 | 58.4 | ||
| AGB *, ton/ha | 10 | 100.8 | 24.5 | 19.6 | 19.5 | 0.0 | 0.0 | 59.5 | |
| Volume, m3/ha | 186.8 | 49.9 | 41.0 | 21.9 | 0.0 | 0.0 | 58.4 |
| Parameter | Number of Training Plots (n) ǂ | Number of Validation Plots (n) | Mean of Estimates | Standard Deviation of Estimates | RMSE | RMSE % | Bias | Bias % | R2 Adj. |
|---|---|---|---|---|---|---|---|---|---|
| AGB *, ton/ha | 50 | 28 | 112.5 | 32.5 | 23.6 | 20.4 | −3.1 | −2.7 | 50.7 |
| Volume, m3/ha | 207.5 | 59.4 | 43.2 | 19.7 | −11.4 | −5.2 | 54.0 | ||
| AGB *, ton/ha | 25 | 111.5 | 33.3 | 24.1 | 20.8 | −4.1 | −3.5 | 50.7 | |
| Volume, m3/ha | 203.2 | 56.4 | 44.3 | 20.2 | −15.7 | −7.1 | 54.0 | ||
| AGB *, ton/ha | 10 | 110.0 | 30.4 | 23.3 | 20.2 | −5.6 | −4.8 | 50.7 | |
| Volume, m3/ha | 204.9 | 60.1 | 44.3 | 20.1 | −14.1 | −6.4 | 54.0 |
© 2014 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Rana, P.; Tokola, T.; Korhonen, L.; Xu, Q.; Kumpula, T.; Vihervaara, P.; Mononen, L. Training Area Concept in a Two-Phase Biomass Inventory Using Airborne Laser Scanning and RapidEye Satellite Data. Remote Sens. 2014, 6, 285-309. https://doi.org/10.3390/rs6010285
Rana P, Tokola T, Korhonen L, Xu Q, Kumpula T, Vihervaara P, Mononen L. Training Area Concept in a Two-Phase Biomass Inventory Using Airborne Laser Scanning and RapidEye Satellite Data. Remote Sensing. 2014; 6(1):285-309. https://doi.org/10.3390/rs6010285
Chicago/Turabian StyleRana, Parvez, Timo Tokola, Lauri Korhonen, Qing Xu, Timo Kumpula, Petteri Vihervaara, and Laura Mononen. 2014. "Training Area Concept in a Two-Phase Biomass Inventory Using Airborne Laser Scanning and RapidEye Satellite Data" Remote Sensing 6, no. 1: 285-309. https://doi.org/10.3390/rs6010285




