Comparison of Airborne LiDAR and Satellite Hyperspectral Remote Sensing to Estimate Vascular Plant Richness in Deciduous Mediterranean Forests of Central Chile
Abstract
:1. Introduction
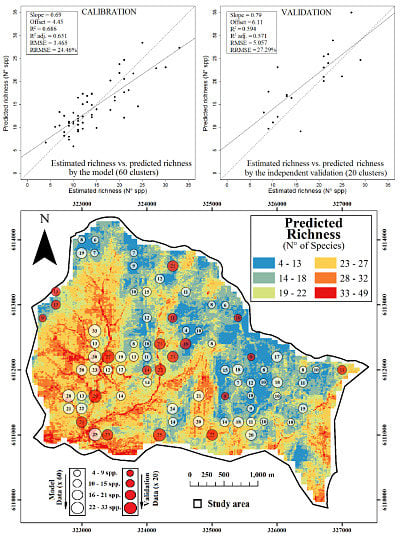
2. Results and Discussion
2.1. Final Predictive Model



| Linear Model Assumptions | Value | p-value | ||||
|---|---|---|---|---|---|---|
| Global Stat | 4.242 | 0.3742 * | ||||
| Skewness | 3.389 | 0.0657 * | ||||
| Kurtosis | 0.001 | 0.9728 * | ||||
| Nonlinear link function | 0.635 | 0.4257 * | ||||
| Heteroscedasticity | 0.218 | 0.6407 * | ||||
| Outlier Evaluation | Cluster | p-value | Bonfer. p | |||
| Bonferroni test | 25 § | 0.0068 ¥ | 0.4091 | |||
| Residuals Normality | W | p-value | ||||
| Shapiro–Wilk normality test | 0.9698 | 0.1435 * | ||||
| Co-linearity Evaluation | MCS60 | DCM90 | MRT10 | SDS15 | MAl30 | MSl30 |
| Variance Inflex Factors (VIF) | 2.935 € | 1.302 € | 1.394 € | 1.426 € | 1.306 € | 2.69 € |
| Validation regression analysis | Adj R2 | p-value | RMSE | |||
| 0.5714 | 6 × 10−5 ¥ | 5.057 | ||||
| Autocorrelation Test | Index | Expect. | z-score | p-value | pattern | |
| Global Moran’s I | −0.0851 | −0.0169 | −08067 | 0.4197 * | random |
2.2. Ecological Implications

2.3. Spatial Prediction

3. Experimental Section
3.1. Study Area

3.2. Field Data
3.3. Remote Sensing Data
3.3.1. Hyperspectral Information
| Predictors | Type | Name | Reference |
|---|---|---|---|
| Hyperion Hyperspectral Image 155 reflectance bands | Vegetation Index (non–photosynthetic vegetation) | Cellulose absorption index (CAI) | [75] |
| Plant Senescence Reflectance Index (PSRI) | [76] | ||
| Vegetation Index (live vegetation, vigor, greenness) | Enhanced vegetation index (EVI) | [77] | |
| Modified red edge Simple Ratio Index (mSR705 ) | [78] | ||
| Canopy Single Ratio (SRcanopy) | [79] | ||
| Single Ratio (SR705) | [79] | ||
| Simple Ratio Vogelmann index (SRv) | [80] | ||
| Modified Normalized Difference Vegetation Index (mND705) | [79] | ||
| Canopy Normalized Difference Vegetation Index (NDcanopy) | [79] | ||
| Normalized Difference Vegetation Index (ND705) | [79] | ||
| Modified Single Ratio (MSR) | [81] | ||
| Ratio TCARI/OSAVI | [82] | ||
| Vegetation Index (canopy structural water) | Moisture Stress Index (MSI ) | [83] | |
| Normalized Difference Infrared Index (NDII) | [84] | ||
| Normalized Difference Water Index (NDWI ) | [85] | ||
| Water Band Index (WBI) | [86] | ||
| Water Index (WI1180 ) | [87] | ||
| Vegetation Index (photosynthetic activity) | Photochemical Reflectance Index (PRI) | [88] | |
| Total Chlorophyll Concentration | [89] | ||
| Transform (Processing of dimensionality reduction) | Forward Principal component analysis (PCA) (first 6 components) | [73] | |
| Minimun Noise Fraction (MNF) (first 7 components) | [74] | ||
| Light Detection and Ranging (LiDAR) 3D Point Cloud and derived Digital Terrain Model (DTM) | DTM10 | Altitude | – |
| First topographic derived (from DTM) | Slope | [90] | |
| Aspect | [90] | ||
| Curvature | [90] | ||
| Plan Curvature | [90] | ||
| Profile Curvature | [90] | ||
| Landform variable (Hydrology from DTM) | Catchment Area | [91] | |
| Catchment Height | [91] | ||
| Catchment Slope | [91] | ||
| Flow Path Length | [92] | ||
| Valley Depth | [93] | ||
| Landform variable (Topo–hydrology from DTM) | SAGA Wetness Index | [94] | |
| Slope Length | [93] | ||
| Stream Power Index | [95] | ||
| Topographic Wetness Index (TWI) | [96] | ||
| TCI Low | [97] | ||
| LS–Factor | [96] | ||
| Landform variable (Topo–morphometry from DTM) | Topographic Position Index (TPI) | [98] | |
| Morphometric Protection Index | [99] | ||
| Terrain Ruggedness Index (TRI) | [100] | ||
| Multi–resolution Index of Valley Bottom Flatness (MrVBF) | [48] | ||
| Multi–resolution Index of Ridge Top Flatness (MrRTF) | [48] | ||
| Convergence Index | [93] | ||
| Vector Ruggedness Measure (VRM) | [101] | ||
| Climate and Lighting (from DTM) | Diurnal Anisotropic Heating | [93] | |
| Total Insolation | [102] | ||
| Topographic Positive Openness | [99] | ||
| Analytical Hillshading | [103] | ||
| Structural variables | DSM10 | – | |
| DCM10 | – |
3.3.2. LiDAR
3.4. Statistical Analysis and Spatialization
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Duffy, J.E. Why biodiversity is important to the functioning of real-world ecosystems? Front. Ecol. Environ. 2006, 7, 437–444. [Google Scholar] [CrossRef]
- Balvanera, P.; Pfisterer, A.B.; Buchmann, N.; He, J.S.; Nakashizuka, T.; Raffaelli, D.; Schmid, B. Quantifying the evidence for biodiversity effects on ecosystem functioning and services. Ecol. Lett. 2006, 9, 1146–1156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carpenter, S.R.; Bennett, E.M.; Peterson, G.D. Scenarios for Ecosystem Services: An Overview. Available online: http://www.uvm.edu/giee/pubpdfs/Carpenter_2006_Ecology_and_Society.pdf (accessed on 1 September 2014).
- Díaz, S.; Fargione, J.; Chapin, F.S.; Tilman, D. Biodiversity loss threatens human well-being. PLoS Biol. 2006, 4, 1300–1305. [Google Scholar] [CrossRef]
- Myers, N.; Mittermeler, R.A.; Mittermeler, C.G.; da Fonseca, G.A.B.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Squeo, F.A.; Estévez, R.A.; Stolla, A.; Gaymerb, C.F.; Leteliera, L.; Sierralta, L. Towards the creation of an integrated system of protected areas in Chile: Achievements and challenges. Plant Ecol. Divers. 2012, 1, 1–11. [Google Scholar]
- Estrategia Y Plan De Acción Para La Biodiversidad En La VII Región Del Maule. Available online: www.sinia.cl/1292/articles-37025_pdf_maule.pdf (accessed on 2 March 2015).
- Luebert, F.; Pliscoff, P. Sinopsis Bioclimática y Vegetacional de Chile; Editorial Universitaria: Santiago, Chile, 1999. [Google Scholar]
- Altamirano, A.; Lara, A. Deforestation in temperate ecosystems of pre-Andean range of south-central Chile. Bosque 2010, 31, 53–64. [Google Scholar] [CrossRef]
- Echeverria, C.; Coomesa, D.; Salas, J.; Rey–Benayas, J.M.; Lara, A.; Newtone, A. Rapid deforestation and fragmentation of Chilean Temperate Forests. Biol. Conserv. 2006, 130, 481–494. [Google Scholar] [CrossRef]
- González, M.E.; Lara, A.; Urrutia, R.; Bosnich, J. Climatic change and its potential impact on forest fire occurrence in south-central Chile (33°–42° S). Bosque 2011, 32, 215–219. [Google Scholar] [CrossRef]
- Bellard, C.; Bertelsmeier, C.; Leadley, P.; Thuiller, W.; Courchamp, F. Impacts of climate change on the future of biodiversity. Ecol. Lett. 2012, 15, 365–377. [Google Scholar] [CrossRef]
- Thomas, C.D.; Cameron, A.; Green, R.E.; Bakkenes, M.; Beaumont, L.J.; Collingham, Y.C.; Erasmus, B.F.N.; de Siqueira, M.F.; Grainger, A.; Hannah, L.; et al. Extinction risk from climate change. Nature 2004, 427, 145–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armesto, J.J.; Rozzi, R.; Smith-Ramirez, C.; Arroyo, M.T.K. Conservation targets in South American temperate forests. Science 1998, 282, 1271–1272. [Google Scholar] [CrossRef]
- Ceballos, G.; Vale, M.M.; Bonacic, C.; Calvo-Alvarado, J.; List, R.; Bynum, N.; Medellín, R.A.; Simonetti, J.A.; Rodríguez, J.P. Conservation challenges for the Austral and Neotropical America section. Conserv. Biol. 2009, 23, 811–817. [Google Scholar] [CrossRef] [PubMed]
- Rocchini, D.; Balkenhol, N.; Carter, G.A.; Foody, G.M.; Gillespie, T.W.; He, K.S.; Kark, S.; Levin, N.; Lucas, K.; Luoto, K.; et al. Remotely sensed spectral heterogeneity as a proxy of species diversity: Recent advances and open challenges. Ecol. Inf. 2010, 5, 318–329. [Google Scholar] [CrossRef]
- Turner, W.; Spector, S.; Gardiner, N.; Fladeland, M.; Sterling, E.; Steininger, M. Remote sensing for biodiversity science and conservation. Trends Ecol. Evol. 2003, 18, 306–314. [Google Scholar] [CrossRef]
- Dufour, A.; Gadallah, F.; Wagner, H.H.; Guisan, A.; Buttler, A. Plant species richness and environmental heterogeneity in a mountain landscape: effects of variability and spatial configuration. Ecography 2006, 29, 573–584. [Google Scholar] [CrossRef]
- Parviainen, M.; Zimmermann, N.E.; Heikkinen, R.K.; Luoto, M. Using unclassified continuous remote sensing data to improve distribution models of red-listed plant species. Biodivers. Conserv. 2013, 22. [Google Scholar] [CrossRef]
- Bergen, K.M.; Goetz, S.J.; Dubayah, R.O.; Henebry, G.M.; Hunsaker, C.T.; Imhoff, M.L.; Nelson, R.F.; Parker, G.G.; Radeloff, V.C. Remote sensing of vegetation 3-D structure for biodiversity and habitat: Review and implications for Lidar and radar spaceborne missions. J. Geophys. Res. 2009, 114. [Google Scholar] [CrossRef]
- Gaston, K.J. Global patterns in biodiversity. Nature 2000, 405, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Pacini, A.; Mazzoleni, S.; Battisti, C.; Ricotta, C. More rich means more diverse: Extending the “environmental heterogeneity hypothesis” to taxonomic diversity. Ecol. Indic. 2009, 9, 1271–1274. [Google Scholar] [CrossRef]
- Carlson, K.; Asner, G.; Hughes, R.; Ostertag, R.; Martin, R. Hyperspectral remote sensing of canopy biodiversity in Hawaiian lowland rainforests. Ecosystems 2007, 10, 536–549. [Google Scholar] [CrossRef]
- Ewers, R.M.; Didham, R.K.; Wratten, S.D.D.; Tylianakis, J.M. Remotely sensed landscape heterogeneity as a rapid tool for assessing local biodiversity value in a highly modified New Zealand landscape. Biodivers. Conserv. 2005, 14, 1469–1485. [Google Scholar] [CrossRef]
- Kalacska, M.; Sanchez-Azofeifa, G.A.; Rivard, B.; Caelli, T.; White, H.P.; Calvo-Alvarado, J.C. Ecological fingerprinting of ecosystem succession: estimating secondary tropical dry forest structure and diversity using imaging spectroscopy. Remote Sens. Environ. 2007, 108, 82–96. [Google Scholar] [CrossRef]
- Leutner, B.; Reineking, B.; Müller, J.; Bachmann, M.; Beierkuhnlein, C.; Dech, S.; Wegmann, M. Modelling forest α-diversity and floristic composition—On the added value of LiDAR plus hyperspectral remote sensing. Remote Sens. 2012, 4, 2818–2845. [Google Scholar] [CrossRef]
- White, J.C.; Gómez, C.; Wulder, M.A.; Coops, N.C. Characterizing temperate forest structural and spectral diversity with Hyperion EO-1 data. Remote Sens. Environ. 2010, 114, 1576–1589. [Google Scholar] [CrossRef]
- Palmer, M.W.; Earls, P.G.; Hoagland, B.W.; White, P.S.; Wohlgemuth, T. Quantitative tools for perfecting species lists. Environmetrics 2002, 13, 121–137. [Google Scholar] [CrossRef]
- Gould, W. Remote sensing of vegetation, plant species richness, and regional biodiversity hotspots. Ecol. Appl. 2000, 10, 1861–1870. [Google Scholar] [CrossRef]
- Levin, N.; Shmida, A.; Levanoni, O.; Tamari, H.; Kark, S. Predicting mountain plant richness and rarity from space using satellite-derived vegetation indices. Divers. Distrib. 2007, 13, 692–703. [Google Scholar] [CrossRef]
- Waring, R.H.; Coops, N.C.; Fan, W.; Nightingale, J.M. MODIS enhanced vegetation index predicts tree species richness across forested ecoregions in the contiguous USA. Remote Sens. Environ. 2006, 103, 218–226. [Google Scholar] [CrossRef]
- Deutschewitz, K.; Lausch, A.; Kühn, I.; Klotz, S. Native and alien plant species richness in relation to spatial heterogeneity on a regional scale in Germany. Glob. Ecol. Biogeogr. 2003, 12, 299–311. [Google Scholar] [CrossRef]
- Gamon, J.A. Tropical remote sensing—Opportunities and challenges. In Hyperspectral Remote Sensing of Tropical and Sub–Tropical Forests; Kalacska, M., Sanchez-Azofeifa, A., Eds.; Taylor & Francis Group: New York, NY, USA, 2008. [Google Scholar]
- Thenkabail, P.S.; Lyon, J.G.; Huete, A. Advances in hyperspectral remote sensing of vegetation and agricultural croplands. In Hyperspectral Remote Sensing of Vegetation; Thenkabail, P.S., Lyon, J.G., Huete, A., Eds.; Taylor & Francis Group: New York, NY, USA, 2012; pp. 12–14. [Google Scholar]
- Dauber, J.; Hirsch, M.; Simmering, D.; Waldhardt, R.; Otte, A.; Wolters, V. Landscape structure as an indicator of biodiversity: matrix effects on species richness. Agric. Ecosyst. Environ. 2003, 98, 321–329. [Google Scholar] [CrossRef]
- Wohlgemuth, T.; Nobis, M.P.; Kienast, F.; Plattner, M. Modelling vascular plant diversity at the landscape scale using systematic samples. J. Biogeogr. 2008, 35, 1226–1240. [Google Scholar] [CrossRef]
- Lefsky, M.A.; Cohen, W.B.; Harding, D.J.; Parker, G.G.; Acker, S.A.; Gower, S.T. Lidar remote sensing of above-ground biomass in three biomes. Glob. Ecol. Biogeogr. 2002, 11, 393–399. [Google Scholar] [CrossRef]
- Dalponte, M.; Bruzzone, L.; Gianelle, D. Fusion of hyperspectral and Lidar remote sensing data for classification of complex forest areas. IEEE Trans. Geosci. Remote Sens. 2008, 46, 1416–1427. [Google Scholar] [CrossRef]
- Magurran, A.E. Ecological Diversity and Its Measurement; Princeton University Press: New Jersey, NJ, USA, 1988. [Google Scholar]
- Corvalán, P.; Galleguillos, M.; Hernández, J. Presencia, abundancia y asociatividad de Citronella mucronata en bosques secundarios dominados por Nothofagus obliqua de la precordillera de Curicó, región del Maule, Chile. Bosque 2014, 35, 269–278. [Google Scholar]
- Camathias, L.; Bergamini, A.; Küchler, M.; Stofer, S.; Baltensweiler, A. High-resolution remote sensing data improves models of species richness. Appl. Veg. Sci. 2013, 16. [Google Scholar] [CrossRef]
- Hernández-Stefanoni, J.L.; Gallardo-Cruz, J.A.; Meaveb, J.A.; Rocchini, D.; Bello-Pineda, J.; López-Martíneza, J.O. Modeling α- and β-diversity in a tropical forest from remotely sensed and spatial data. Int. J. Appl. Earth Obs. Geoinf. 2012, 19, 359–368. [Google Scholar] [CrossRef]
- Féret, J.B.; Asner, G.P. Mapping tropical forest canopy diversity using high-fidelity imaging spectroscopy. Ecol. Appl. 2014, 24, 1289–1296. [Google Scholar] [CrossRef]
- Vaglio, G.L.; Cheung-Wai, J.C.; Chen, Q.; Lindsell, J.A.; Coomes, D.A.; Guerriero, L.; Del Frate, F.; Miglietta, F.; Valentini, R. Biodiversity mapping in a tropical west African forest with airborne hyperspectral data. PloS One 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Simonson, W.D.; Allen, H.D.; Coomes, D.A. Use of an airborne Lidar system to model plant species composition and diversity of mediterranean oak forests. Conserv. Biol. 2012, 26, 840–850. [Google Scholar] [CrossRef] [PubMed]
- Bacaro, G.; Rocchini, D.; Bonini, I.; Marignani, M.; Maccherini, S.; Chiarucci, A. The role of regional and local scale predictors for plant species richness in Mediterranean forests. Plant Biosyst. 2008, 142, 630–642. [Google Scholar] [CrossRef]
- Bässler, C.; Stadler, J.; Müller, J.; Förster, B.; Göttlein, A.; Brandl, R. LiDAR as a rapid tool to predict forest habitat types in Natura 2000 networks. Biodivers. Conserv. 2011, 20, 465–481. [Google Scholar] [CrossRef]
- Gallant, J.C.; Dowling, T.I. A multiresolution index of valley bottom flatness for mapping depositional areas. Water Resour. Res. 2003, 39. [Google Scholar] [CrossRef]
- Yuan, Z.; Gazol, A.; Wang, X.; Lin, F.; Ye, J.; Bai, X.; Li, B.; Hao, Z. Scale specific determinants of tree diversity in an old growth temperate forest in China. Basic Appl. Ecol. 2011, 12, 488–495. [Google Scholar] [CrossRef]
- Silvertown, J.; Dodd, M.E.; Gowing, D.J.G.; Mountford, J.O. Hydrologically defined niches reveal a basis for species richness in plant communities. Nature 1999, 400, 61–63. [Google Scholar] [CrossRef]
- Moeslund, J.E.; Arge, L.; Bocher, P.K.; Dalgaard, T.; Odgaard, M.V.; Nygaard, B.; Svenning, J.C. Topographically Controlled Soil Moisture is the Primary Driver of Local Vegetation Patterns across a Lowland Region. Available online: http://www.esajournals.org/doi/abs/10.1890/ES13-00134.1 (accessed on 1 September 2014).
- Everson, D.A.; Boucher, H. Tree species-richness and topographic complexity along the riparian edge of the Potomac River. For. Ecol. Manag. 1998, 109, 305–314. [Google Scholar] [CrossRef]
- Hoersch, B.; Braun, G.; Schmid, U. Relation between landform and vegetation in alpine regions of Wallis, Switzerland. A multiscale remote sensing and GIS approach. Comput. Environ. Urban Syst. 2002, 26, 113–139. [Google Scholar] [CrossRef]
- Lindeman, R.H.; Merenda, P.F.; Gold, R.Z. Introduction to Bivariate and Multivariate Analysis; Scott, Foresman: Glenview, IL, USA, 1980. [Google Scholar]
- Simonson, W.D.; Allen, H.D.; Coomes, D.A. Applications of airborne Lidar for the assessment of animal species diversity. Methods Ecol. Evol. 2014, 5, 719–729. [Google Scholar] [CrossRef]
- Oldeland, J.; Wesuls, D.; Rocchini, D.; Schmidt, M.; Jürgens, N. Does using species abundance data improve estimates of species diversity from remotely sensed spectral heterogeneity? Ecol. Indic. 2010, 10, 390–396. [Google Scholar] [CrossRef]
- John, R.; Chen, J.; Lu, N.; Guo, K.; Liang, C.; Wei, Y.; Noormets, A.; Ma, K.; Han, X. Predicting plant diversity based on remote sensing products in the semi-arid region of Inner Mongolia. Remote Sens. Environ. 2008, 112, 2018–2032. [Google Scholar] [CrossRef]
- Dalponte, M.; Bruzzone, L.; Gianelle, D. Tree species classification in the Southern Alps based on the fusion of very high geometrical resolution multispectral/hyperspectral images and Lidar data. Remote Sens. Environ. 2012, 123, 258–270. [Google Scholar]
- Naidoo, L.; Cho., M.A.; Mathieu, R.; Asner, G. Classification of savanna tree species, in the Greater Kruger National Park region, by integrating hyperspectral and LiDAR data in a Random Forest data mining environment. ISPRS J. Photogramm. 2012, 69, 167–179. [Google Scholar] [CrossRef]
- Ginzburg, L.R.; Jensen, C. Rules of thumb for judging ecological theories. Trends Ecol. Evol. 2004, 19, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Rey-Benayas, J.M. Patterns of diversity in the strata of boreal montane forest in British Columbia. J. Veg. Sci. 1995, 6, 95–98. [Google Scholar] [CrossRef]
- Stevens, G.C. The elevational gradient in altitudinal range: An extension of Rapoport’s latitudinal rule to altitude. Am. Nat. 1992, 140, 893–911. [Google Scholar] [CrossRef] [PubMed]
- Kolasa, J.; Rollo, C.D. The heterogeneity of heterogeneity: A glossary. In Ecological heterogeneity; Kolasa, J., Pickett, S.T.A., Eds.; Springer: New York, NY, USA, 1991. [Google Scholar]
- Rahbek, C.; Graves, G.R. Multiscale assessment of patterns of avian species richness. Proc. Natl. Acad. Sci. USA 2001, 98, 4534–4539. [Google Scholar] [CrossRef] [PubMed]
- Rahbek, C. The role of spatial scale and the perception of large-scale species-richness patterns. Ecol. Lett. 2005, 8, 224–239. [Google Scholar] [CrossRef]
- Turner, M.G.; O’Neill, R.V.; Gardner, R.H.; Milne, B.T. Effects of changing spatial scale on the analysis of landscape pattern. Landsc. Ecol. 1989, 3, 153–162. [Google Scholar] [CrossRef]
- Stohlgren, T.J. Measuring Plant Diversity: Lessons from the Field; Oxford University Press Inc.: New York, NY, USA, 2007. [Google Scholar]
- Pearlman, J.; Barry, P.; Segal, C.; Shepanski, D.B.; Carman, S. Hyperion, a space-based imaging spectrometer. IEEE Trans. Geosci. Remote Sens. 2003, 41, 1160–1173. [Google Scholar] [CrossRef]
- Datt, B.; McVicar, T.; van Niel, T.; Jupp, D.; Pearlman, J. Preprocessing EO-1 hyperion hyperspectral data to support the application of agricultural indexes. IEEE Trans. Geosci. Remote Sens. 2003, 41, 1246–1259. [Google Scholar] [CrossRef]
- Matthew, M.W.; Adler-Golden, S.M.; Berk, A.; Richtsmeier, S.C.; Levine, R.Y.; Bernstein, L.S.; Acharya, P.K.; Anderson, G.P.; Felde, G.W.; Hoke, M.P.; et al. Status of atmospheric correction using a MODTRAN 4-based algorithm. Proc. SPIE 2000, 4049. [Google Scholar] [CrossRef]
- Lucas, R.; Mitchell, A.; Bunting, P. Hyperspectral data for assessing carbon dynamics and biodiversity of forests. In Hyperspectral Remote Sensing of Tropical and Sub-Tropical Forests; Kalacska, M., Sanchez-Azofeifa, A., Eds.; Taylor & Francis Group: New York, NY, USA, 2008; pp. 47–86. [Google Scholar]
- Roberts, D.A.; Roth, K.L.; Perroy, R.L. Hyperspectral vegetation indices. In Hyperspectral Remote Sensing of Vegetation; Thenkabail, P.S., Lyon, J.G., Huete, A., Eds.; Taylor & Francis Group: New York, NY, USA, 2012; pp. 309–328. [Google Scholar]
- Richards, J.A. Remote Sensing Digital Image Analysis: An Introduction; Springer-Verlag: Berlin, Germany, 1999. [Google Scholar]
- Green, A.A.; Berman, M.; Switzer, P.; Craig, M.D. A transformation for ordering multispectral data in terms of image quality with implications for noise removal. IEEE Trans. Geosci. Remote Sens. 1988, 26, 65–74. [Google Scholar] [CrossRef]
- Daughtry, C.T.S. Discriminating crop residues from soil by shortwave infrared reflectance. Agron. J. 2001, 93, 125–131. [Google Scholar] [CrossRef]
- Merzlyak, M.N.; Gitelson, A.; Chivkunova, O.B.; Rakitin, V.Y. Non-destructive optical detection of pigment changes during leaf senescence and fruit ripening. Physiol. Plant 1999, 106, 135–141. [Google Scholar] [CrossRef]
- Huete, A.; Didan, K.; Miura, T.; Rodriguez, E.P.; Gao, X.; Ferreira, L.G. Overview of the radiometric and biophysical performance of the MODIS vegetation indices. Remote Sens. Environ. 2002, 83, 195–213. [Google Scholar] [CrossRef]
- Datt, B. Visible/near infrared reflectance and chlorophyll content in Eucalyptus leaves. Int. J. Remote Sens. 1999, 20, 2741–2759. [Google Scholar] [CrossRef]
- Sims, D.A.; Gamon, J.A. Relationships between leaf pigment content and spectral reflectance across a wide range of species, leaf structures and developmental stages. Remote Sens. Environ. 2002, 81, 337–354. [Google Scholar] [CrossRef]
- Vogalmann, J.E.; Rock, B.N.; Moss, D.M. Red edge spectral measurements from sugar maple leaves. Int. J. Remote Sens. 1993, 14, 1563–1575. [Google Scholar] [CrossRef]
- Chen, J.M. Evaluation of vegetation indices and a modified simple ratio for Boreal applications. Can. J. Remote Sens. 1996, 22, 229–241. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Tremblay, N.; Zarco-Tejada, P.J.; Dextraze, L. Integrated narrow-band vegetation indices for prediction of crop chlorophyll content for application to precision agriculture. Remote Sens. Environ. 2002, 81, 416–426. [Google Scholar] [CrossRef]
- Hunt, E.R.; Rock, B.N. Detection of changes in leaf water content using near- and middle-infrared reflectances. Remote Sens. Environ. 1989, 30, 43–54. [Google Scholar] [CrossRef]
- Hardisky, M.A.; Klemas, V.; Daiber, F.D. Remote sensing salt marsh biomass and stress detection. Adv. Space Res. 1983, 2, 219–229. [Google Scholar] [CrossRef]
- Gao, B.C. NDWI: A normalized difference water index for remote sensing of vegetation liquid water from space. Remote Sens. Environ. 1996, 58, 257–266. [Google Scholar] [CrossRef]
- Peñuelas, J.; Pinol, J.; Ogaya, R.; Filella, I. Estimation of plant water concentration by the reflectance water index (R900/R970). Int. J. Remote Sens. 1997, 18, 2869–2875. [Google Scholar] [CrossRef]
- Sims, D.A.; Gamon, J.A. Estimation of vegetation water content and photosynthetic tissue area from spectral reflectance: A comparison of indices based on liquid water and chlorophyll absorption features. Remote Sens. Environ. 2003, 84, 526–537. [Google Scholar] [CrossRef]
- Gamon, J.A.; Peñuelas, J.; Field, C.B. A narrow-waveband spectral index that tracks diurnal changes in photosynthetic efficiency. Remote Sens. Environ. 1992, 41, 35–44. [Google Scholar] [CrossRef]
- Gitelson, A.; Merzlyak, M.N. Remote estimation of chlorophyll content in higher plant leaves. Int. J. Remote Sens. 1997, 18, 2691–2697. [Google Scholar] [CrossRef]
- Zevenbergen, L.W.; Thorne, C.R. Quantitative analysis of land surface topography. Earth Surf. Proc. Land. 1987, 12, 47–56. [Google Scholar] [CrossRef]
- Seibert, J.; McGlynn, B. A new triangular multiple flow direction algorithm for computing upslope areas from gridded digital elevation models. Water Resour. Res. 2007, 43, W04501. [Google Scholar] [CrossRef]
- Quinn, P.F.; Beven, K.J.; Chevallier, P.; Planchon, O. The prediction of hillslope flow paths for distributed hydrological modelling using digital terrain models. Hydrol. Process. 1991, 5, 59–79. [Google Scholar] [CrossRef]
- Conrad, O. System for Automated Geoscientific Analyses (SAGA). Version: 2.1.2. Available online: http://www.saga–gis.org (accessed on 29 July 2014).
- Boehner, J.; Koethe, R.; Conrad, O.; Gross, J.; Ringeler, A.; Selige, T. Soil Regionalisation by Means of Terrain Analysis and Process Parameterisation. Available online: http://www.scilands.de/referenzen/veroeffentlichung/601Bohner.pdf (accessed on 1 September 2014).
- Moore, I.D.; Grayson, R.B.; Ladson, A.R. Digital terrain modelling: A review of hydrogical, geomorphological, and biological applications. Hydrol. Process. 1991, 5, 3–30. [Google Scholar] [CrossRef]
- Boehner, J.; Selige, T. Spatial prediction of soil attributes using terrain analysis and climate regionalisation. In SAGA—Analysis and Modelling Applications; Boehner, J., McCloy, K.R., Strobl, J., Eds.; Goettinger Geographische Abhandlungen: Goettinger, Germany, 2006; pp. 13–27. [Google Scholar]
- Bock, M.; Boehner, J.; Conrad, O.; Koethe, R.; Ringeler, A. Methods for Creating Functional Soil Databases and applying Digital Soil Mapping with SAGA GIS. Available online: http://www.scilands.de/referenzen/veroeffentlichung/EUR22646EN_Bock_B%F6hner_Conrad_K%F6the_Ringeler.pdf (accessed on 1 September 2014).
- Guisan, A.; Weiss, S.B.; Weiss, A.D. GLM versus CCA spatial modeling of plant species distribution. Plant Ecol. 1999, 143, 107–122. [Google Scholar] [CrossRef]
- Yokoyama, R.; Shirasawa, M.; Pike, R.J. Visualizing topography by openness: A new application of image processing to digital elevation models. Photogramm. Eng. Remote Sens. 2002, 68, 251–266. [Google Scholar]
- Riley, S.J.; De Gloria, S.D.; Elliot, R. A terrain ruggedness that quantifies topographic heterogeneity. Intermount. J. Sci. 1999, 5, 23–27. [Google Scholar]
- Sappington, J.M.; Longshore, K.M.; Thompson, D.B. Quantifying landscape ruggedness for animal habitat analysis: A Case Study Using Bighorn Sheep in the Mojave Desert. J. Wildl. Manag. 2007, 71, 1419–1426. [Google Scholar] [CrossRef]
- Boehner, J.; Antonic, O. Land surface parameters specific to topo-climatology. In Geomorphometry: Concepts, Software, Applications; Hengl, T., Reuter, H.I., Eds.; Elsevier: Amsterdam, The Netherlands, 2009; pp. 195–226. [Google Scholar]
- Tarini, M.; Cignoni, P.; Montani, C. Ambient occlusion and edge cueing to enhance real time molecular visualization. IEEE Trans. Vis. Comput. Gr. 2006, 12, 1237–1244. [Google Scholar] [CrossRef]
- Isenburg, M. LAStools-Efficient Tools for LiDAR Processing, Software for Rapid Converting, Filtering, Viewing, Gridding, and Compressing of Lidar. Version: 140221. Available online: http://lastools.org (accessed on 30 July 2014).
- Clark, M.L.; Clark, D.B.; Roberts, D.A. Small-footprint Lidar estimation of subcanopy elevation and tree height in a tropical rain forest landscape. Remote Sens. Environ. 2004, 91, 68–89. [Google Scholar] [CrossRef]
- R Core Team. R: A language and environment for statistical computing. In R Foundation for Statistical Computing, Version 3.1.1; R Foundation for Statistical Computing: Vienna, Austria, 2012. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Grossmann, E.; Ohmann, J.; Kagan, J.; May, H.; Gregory, M. Mapping ecological systems with a random forest model: Tradeoffs between errors and bias. Gap Anal. Bull. 2010, 17, 16–22. [Google Scholar]
- Moeslund, J.E.; Arge, L.; Bocher, P.K.; Nygaard, B.; Svenning, J.C. Geographically comprehensive assessment of salt-meadow vegetation-elevation relations using Lidar. Wetlands 2011, 31, 471–482. [Google Scholar] [CrossRef]
- Lumley, T. Regression subset selection: Package “leaps”. Available online: http://cran.r-project.org/web/packages/leaps/leaps.pdf (accessed on 16 October 2014).
- Pena, E.A.; Slate, E.H. Global validation of linear model assumptions. J. Am. Statist. Assoc. 2006, 101, 341–354. [Google Scholar] [CrossRef]
- Royston, P. Algorithm AS 181: The W test for normality. Appl. Stat. 1995, 44, 547–551. [Google Scholar] [CrossRef]
- Fox, J.; Weisberg, S. An R Companion to Applied Regression, 2nd ed.; SAGE Publications Inc.: Minneapolis, MN, USA, 2011. [Google Scholar]
- Arthur, G.; Ord, J.K. The analysis of spatial association by use of distance statistics. Geogr. Anal. 1992, 24, 3, 189–206. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ceballos, A.; Hernández, J.; Corvalán, P.; Galleguillos, M. Comparison of Airborne LiDAR and Satellite Hyperspectral Remote Sensing to Estimate Vascular Plant Richness in Deciduous Mediterranean Forests of Central Chile. Remote Sens. 2015, 7, 2692-2714. https://doi.org/10.3390/rs70302692
Ceballos A, Hernández J, Corvalán P, Galleguillos M. Comparison of Airborne LiDAR and Satellite Hyperspectral Remote Sensing to Estimate Vascular Plant Richness in Deciduous Mediterranean Forests of Central Chile. Remote Sensing. 2015; 7(3):2692-2714. https://doi.org/10.3390/rs70302692
Chicago/Turabian StyleCeballos, Andrés, Jaime Hernández, Patricio Corvalán, and Mauricio Galleguillos. 2015. "Comparison of Airborne LiDAR and Satellite Hyperspectral Remote Sensing to Estimate Vascular Plant Richness in Deciduous Mediterranean Forests of Central Chile" Remote Sensing 7, no. 3: 2692-2714. https://doi.org/10.3390/rs70302692