Appling the One-Class Classification Method of Maxent to Detect an Invasive Plant Spartina alterniflora with Time-Series Analysis
Abstract
:1. Introduction
2. Materials and Data
2.1. Study Area
2.2. Remote Sensing Data
2.3. Reference Data
3. Methods
3.1. Biased Support Vector Machine
3.2. Maxent Model and Tuning of Complexity
3.3. Four Experimental Setups
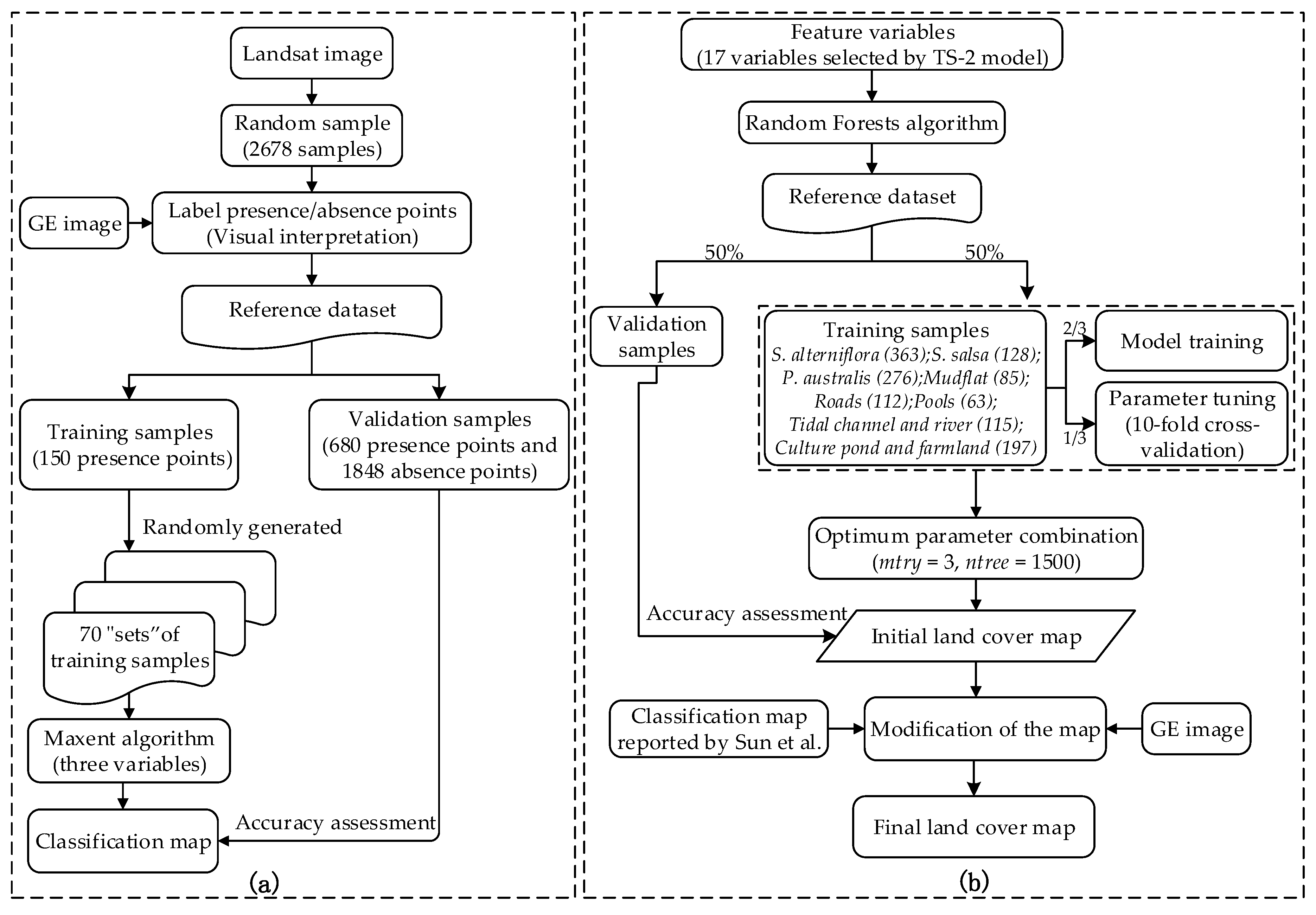
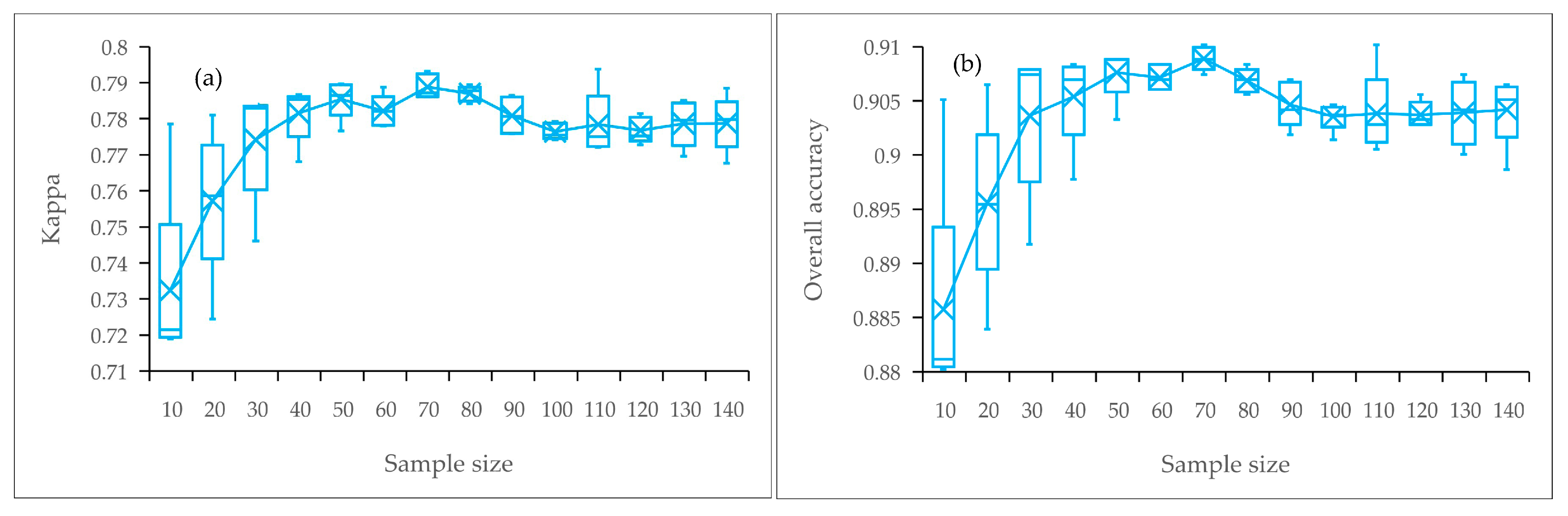
3.4. Evaluation of the Expansibility of the Method and Settings of the Training Sample Size
3.4.1. Reference Dataset and 70 “Sets” of Training Samples
3.4.2. Final Land Cover Map
3.5. Model Verification
4. Results
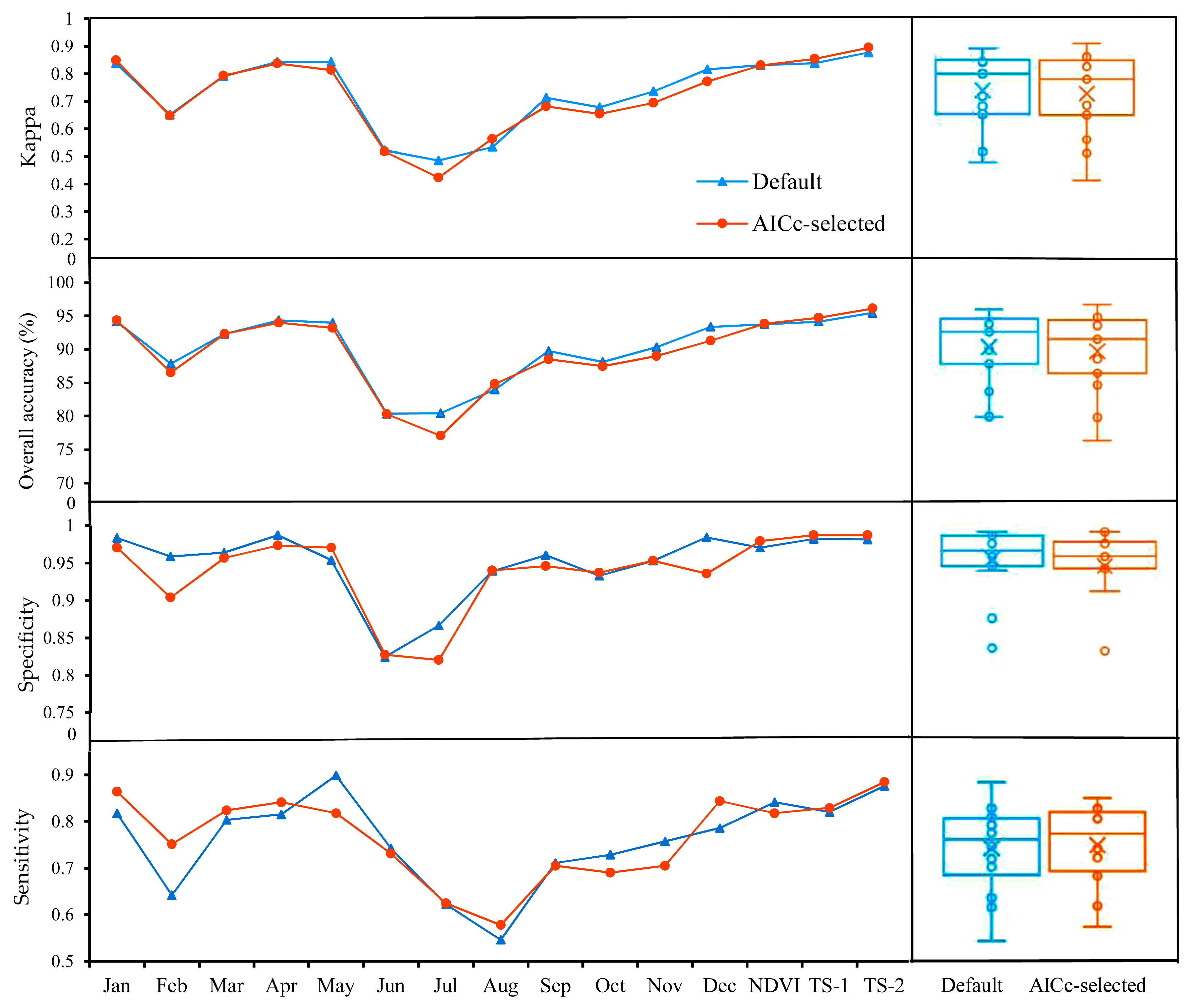
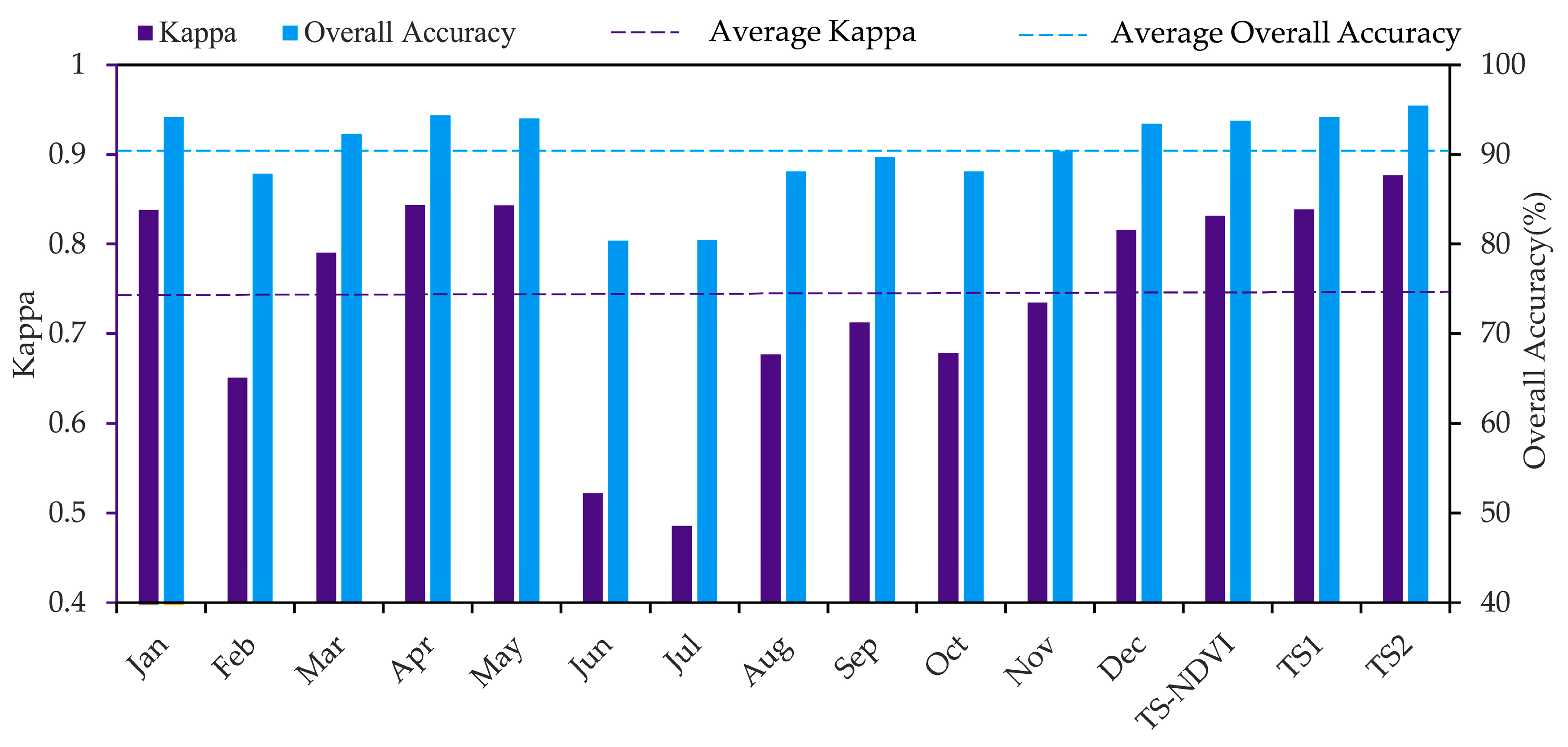
4.1. Model Comparison
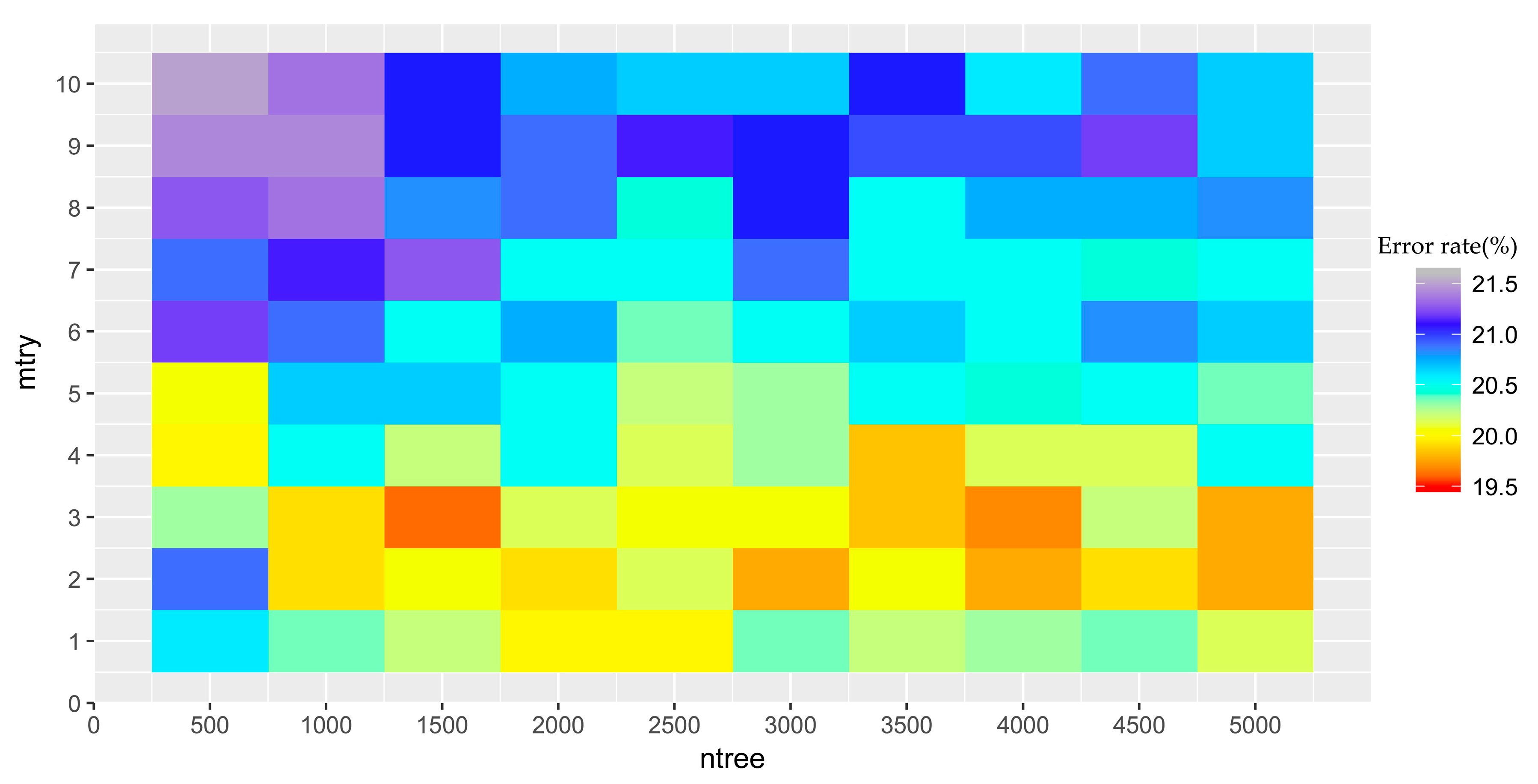
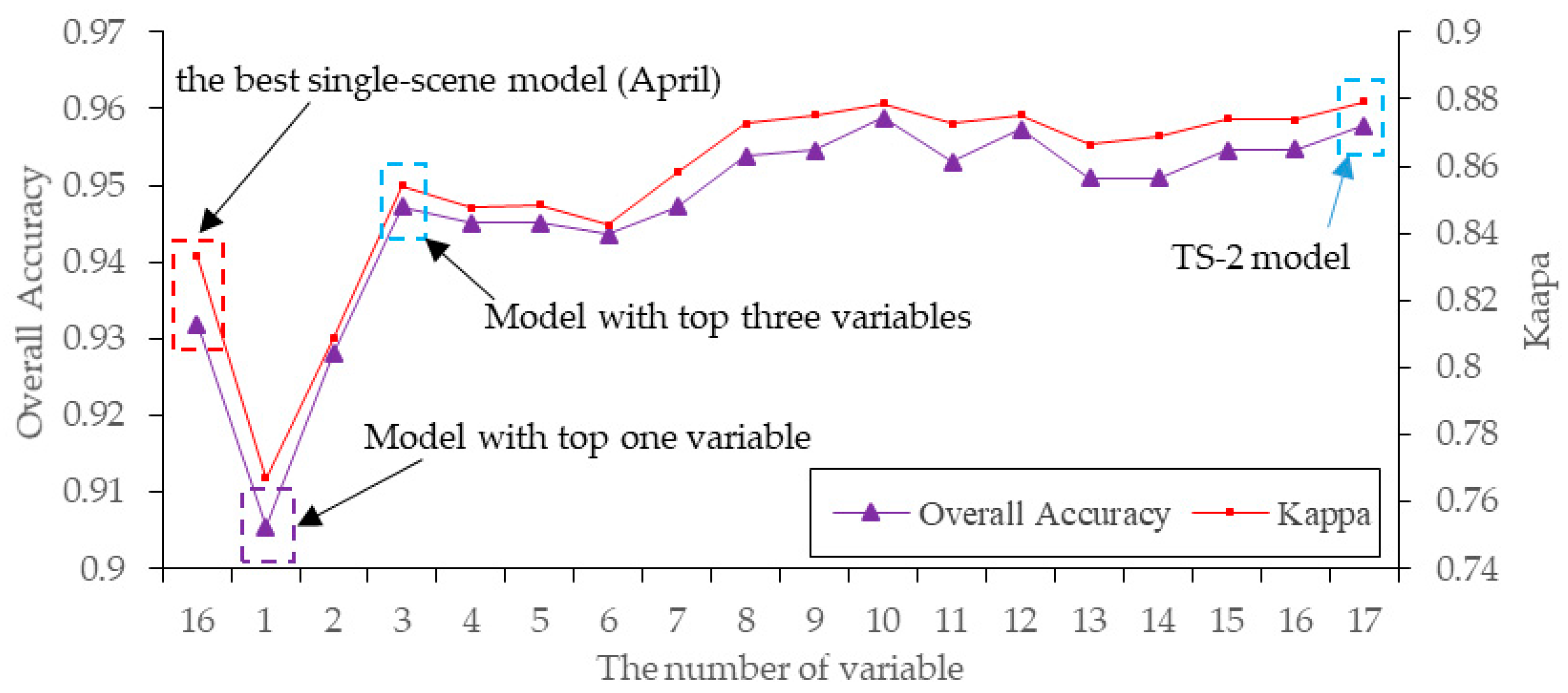
4.2. Model Complexity Analysis
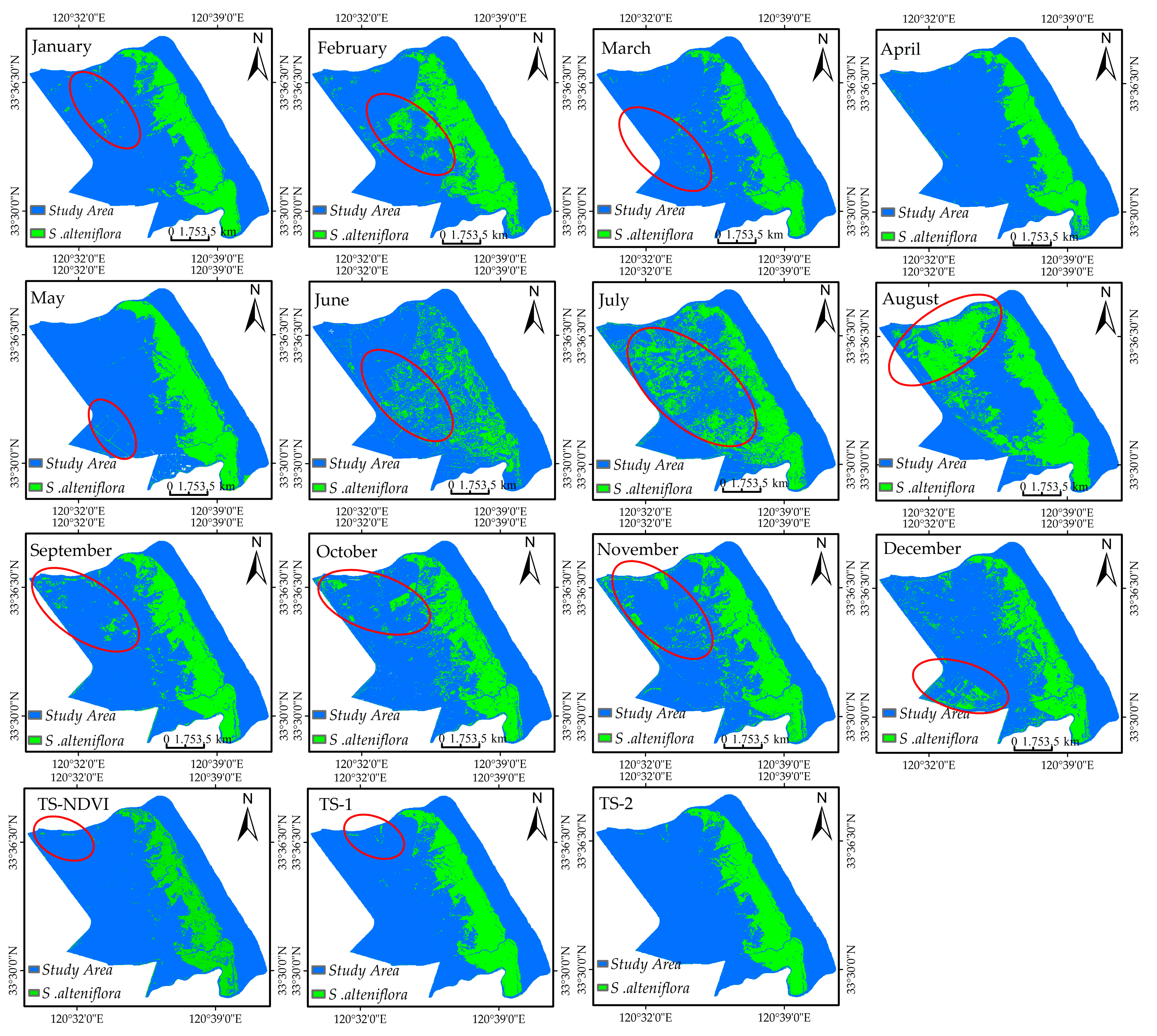
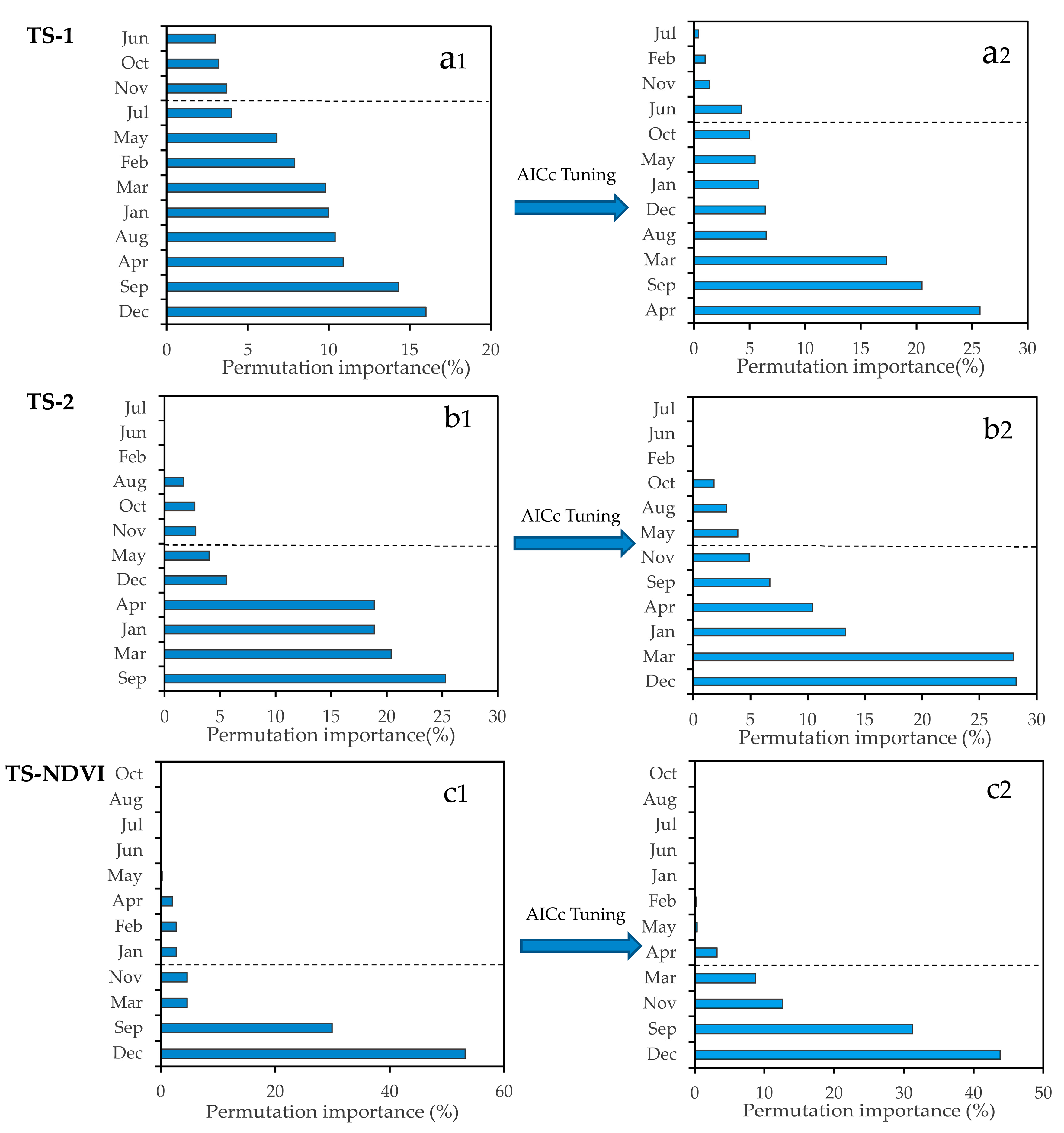
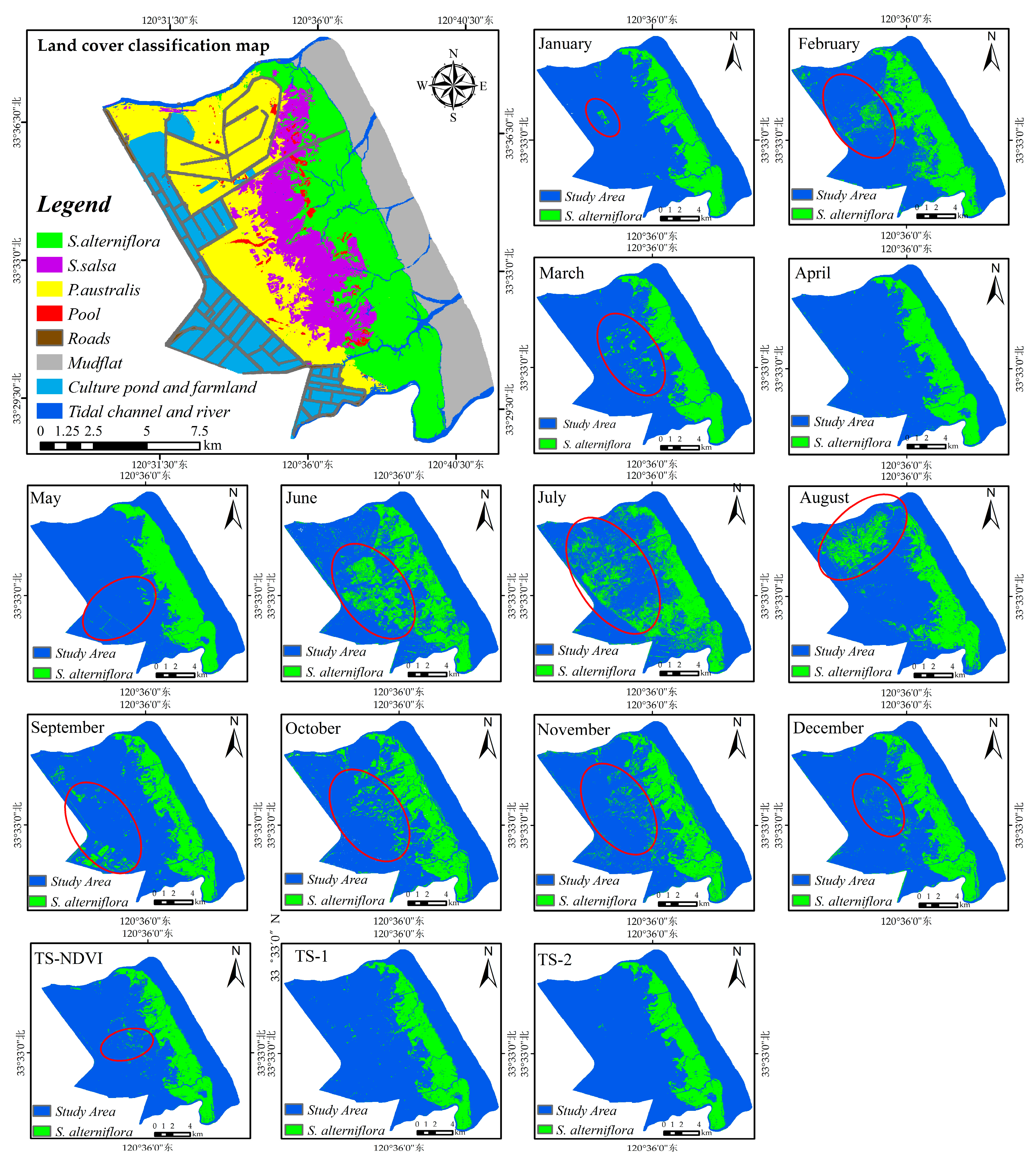
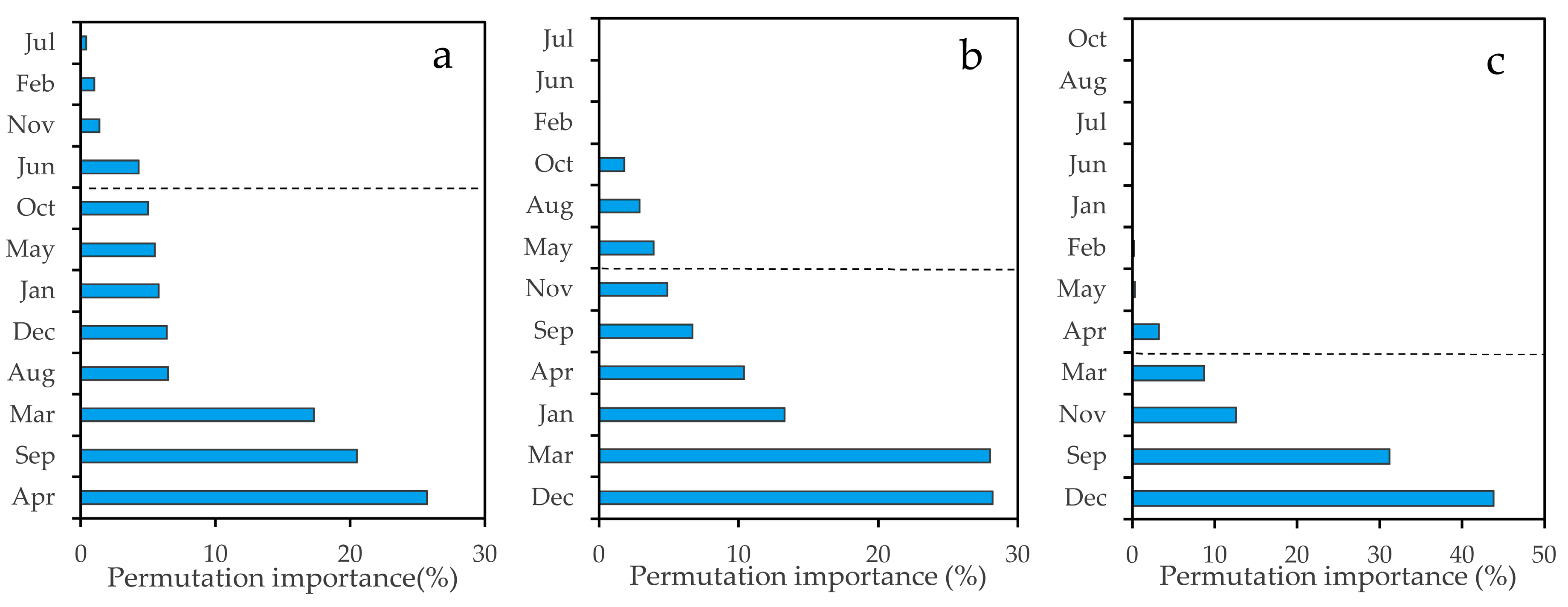
4.3. Important Months and Seasons in S. alterniflora Detection
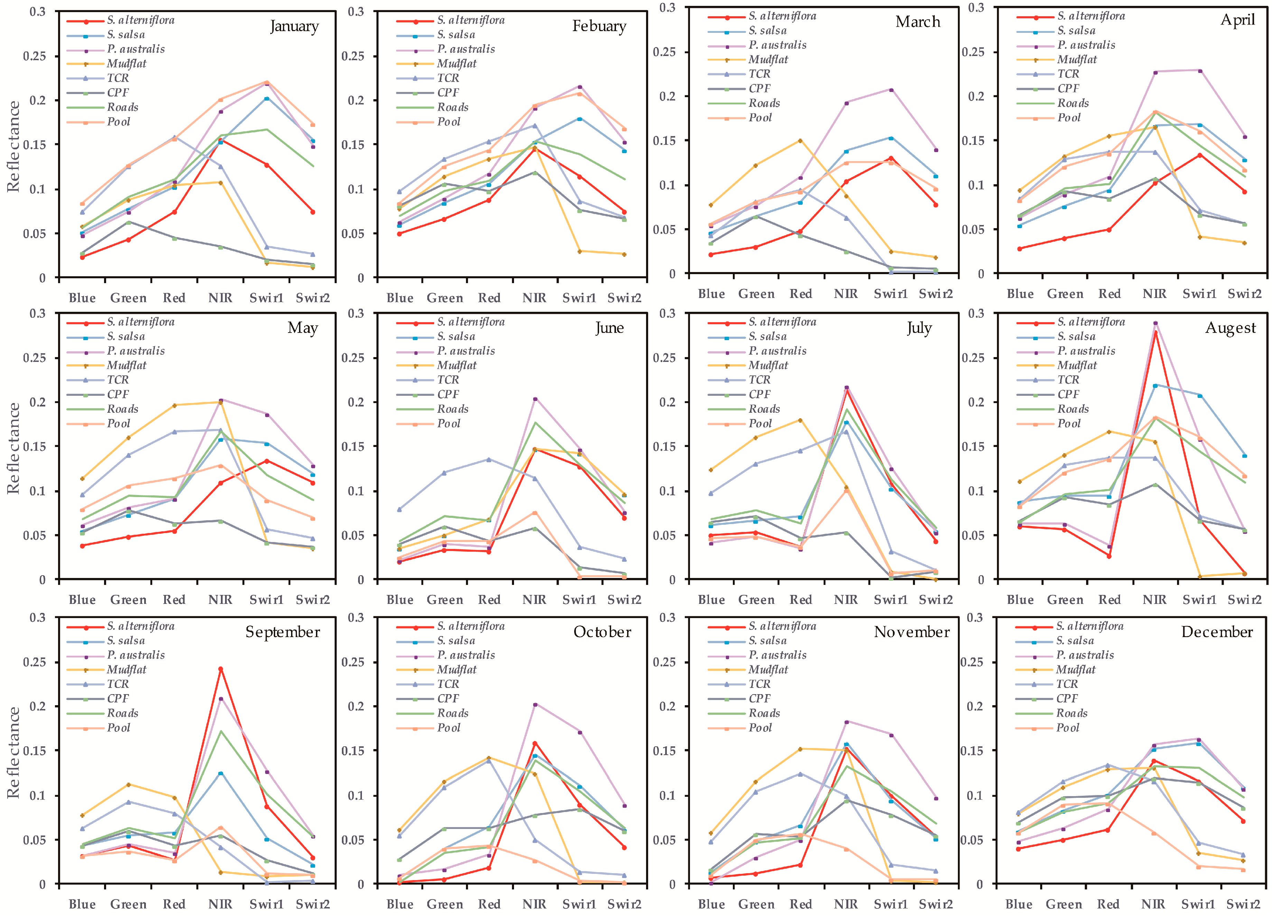
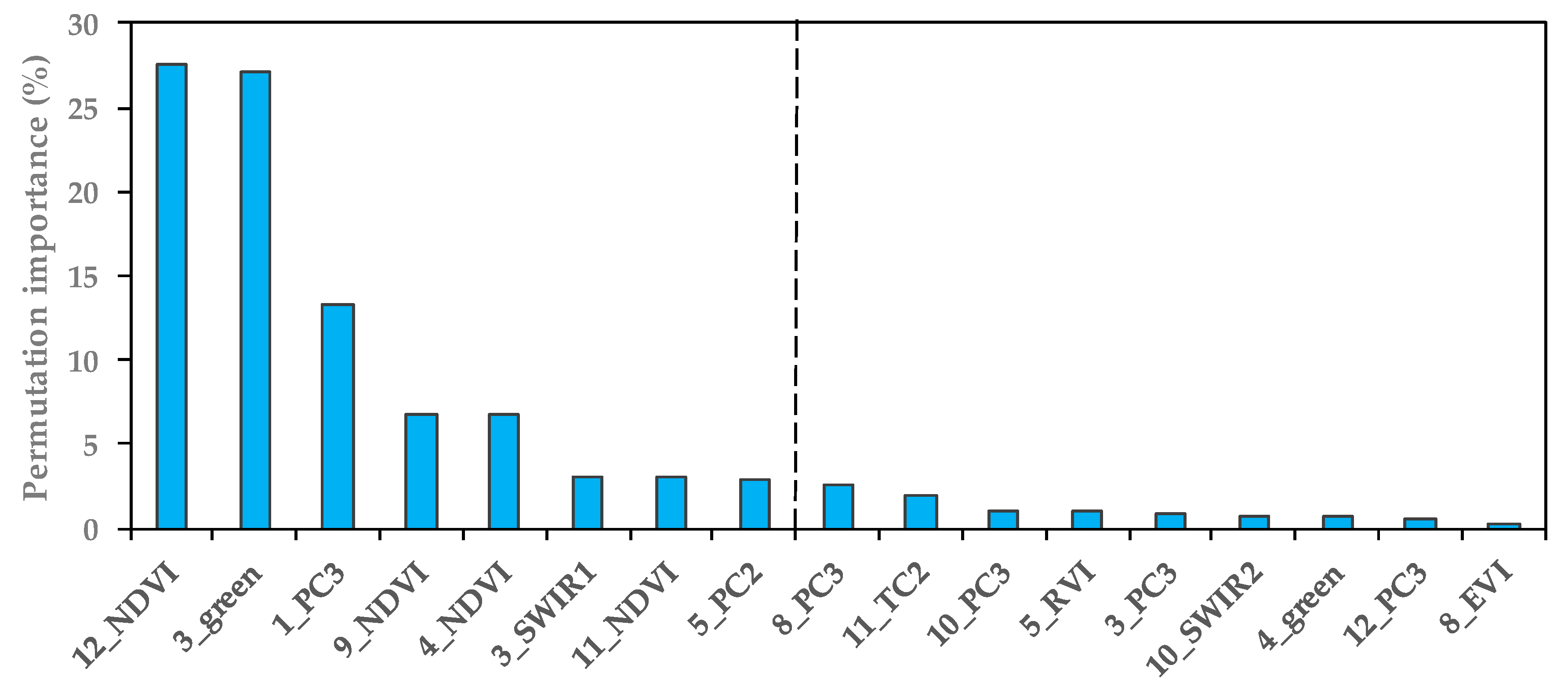
4.4. Important Band Variables and Their Roles in S. alterniflora Detection
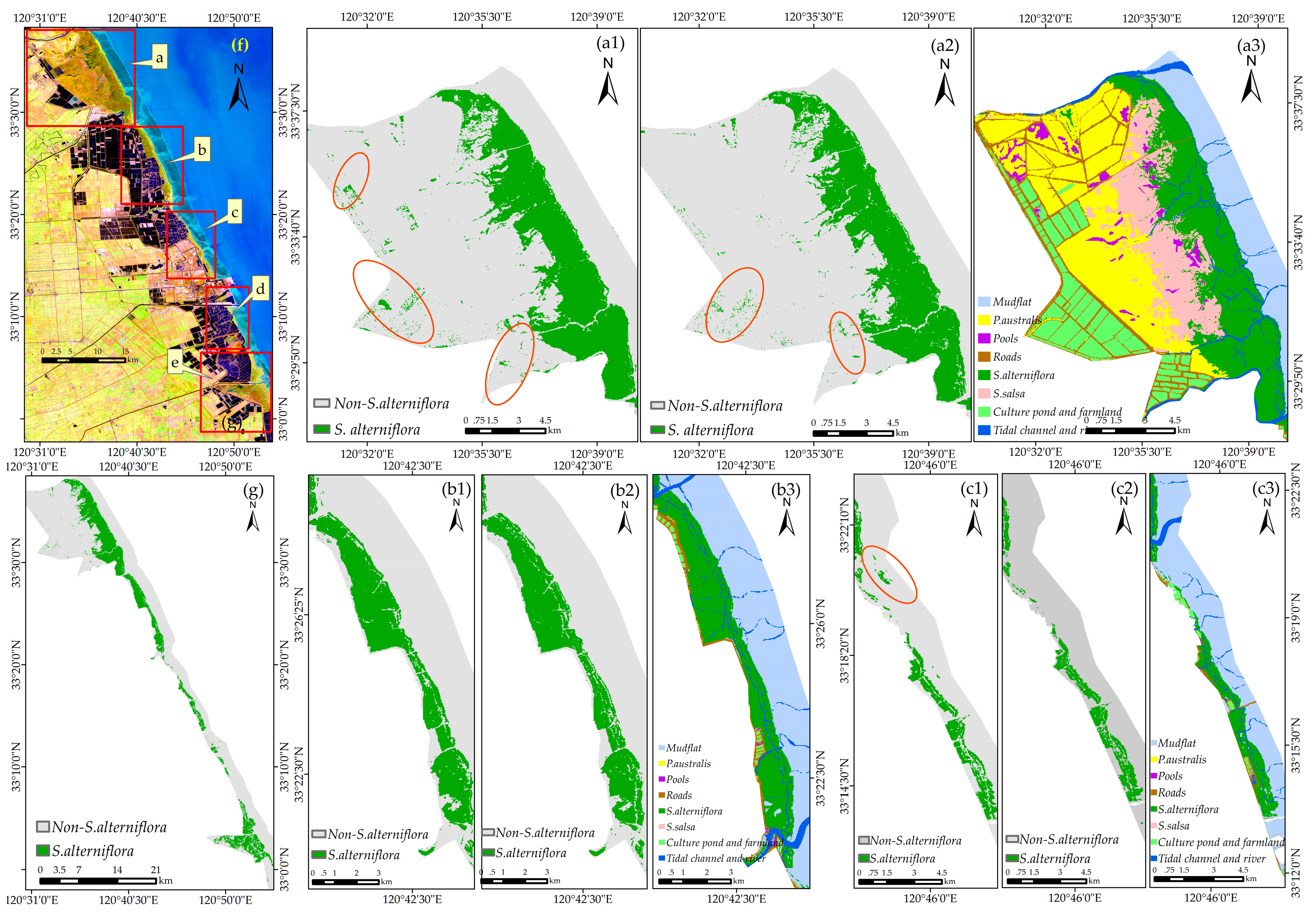
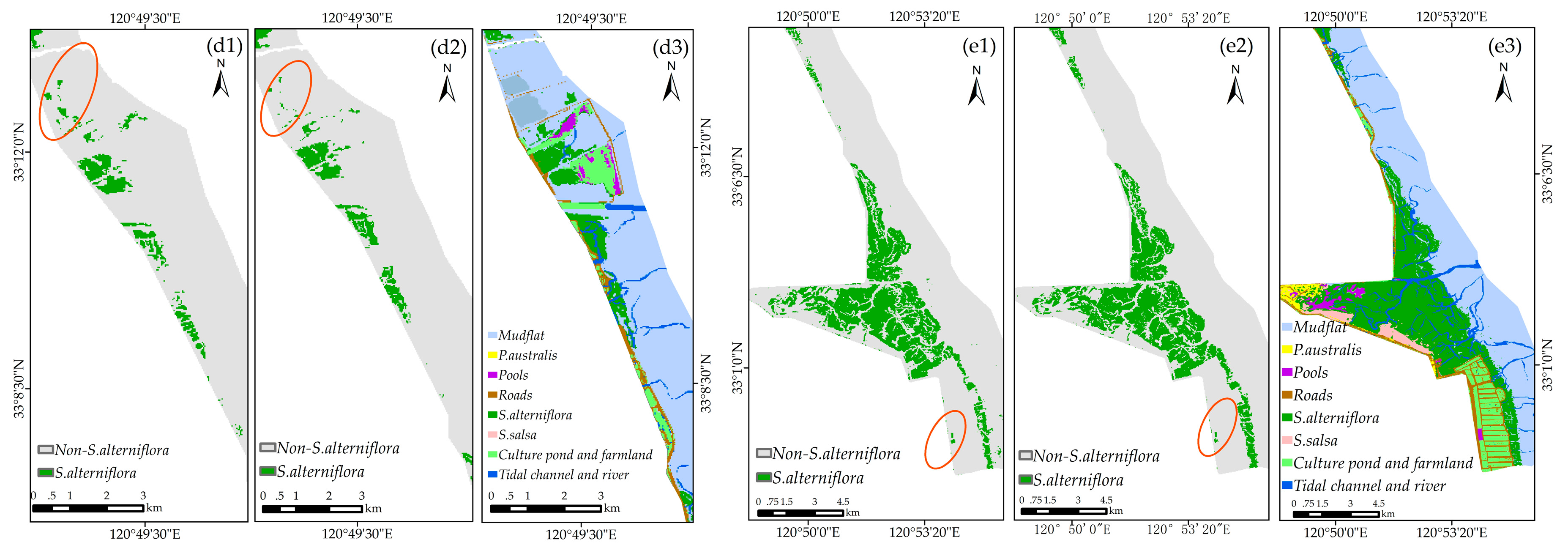
4.5. Applicability of the Method for S. alterniflora Detection in Large Regions
5. Discussion
5.1. Performance of OCC Methods for S. alterniflora Detection and Model Complexity Analysis
5.2. Important Phenological Windows and the Role of NDVI for S. alterniflora Detection
5.3. Opportunities and Challenges for S. alterniflora Detection Using the Phenology-Based OCC Approach
6. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix A
Appendix B
| Prediction | CPF | Mudflat | P. australis | Pools | Roads | TCR | S. salsa | S. alterniflora | |
|---|---|---|---|---|---|---|---|---|---|
| Reference | |||||||||
| CPF | 192 | 0 | 3 | 3 | 11 | 1 | 0 | 7 | |
| Mudflat | 1 | 84 | 0 | 0 | 0 | 0 | 0 | 0 | |
| P. australis | 4 | 0 | 239 | 2 | 6 | 0 | 5 | 1 | |
| pools | 1 | 0 | 11 | 32 | 0 | 0 | 3 | 2 | |
| Roads | 16 | 1 | 11 | 1 | 48 | 1 | 5 | 8 | |
| TCR | 3 | 9 | 4 | 0 | 7 | 20 | 6 | 45 | |
| S. salsa | 0 | 0 | 10 | 0 | 1 | 1 | 108 | 5 | |
| S. alterniflora | 1 | 4 | 4 | 1 | 8 | 10 | 4 | 389 | |
| Overall accuracy: 83.05%; Kappa: 0.7895 | |||||||||
References
- Perrings, C.; Dehnenschmutz, K.; Touza, J.; Williamson, M. How to manage biological invasions under globalization. Trends Ecol. Evol. 2005, 20, 212–215. [Google Scholar] [CrossRef] [PubMed]
- Callaway, R.M.; Maron, J.L. What have exotic plant invasions taught us over the past 20 years? Trends Ecol. Evol. 2006, 21, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Sax, D.F.; Stachowicz, J.J.; Brown, J.H.; Bruno, J.F.; Dawson, M.N.; Gaines, S.D.; Grosberg, R.K.; Hastings, A.; Holt, R.D.; Mayfield, M.M. Ecological and evolutionary insights from species invasions. Trends Ecol. Evol. 2007, 22, 465–471. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Pei, X.; Yue, S.; Wen, Y. The response of Spartina alterniflora biomass to soil factors in Yancheng, Jiangsu Province, P.R. China. Wetlands 2016, 36, 229–235. [Google Scholar] [CrossRef]
- Wang, A.; Chen, J.; Jing, C.; Ye, G.; Wu, J.; Huang, Z.; Zhou, C. Monitoring the invasion of Spartina alterniflora from 1993 to 2014 with Landsat TM and SPOT 6 Satellite Data in Yueqing Bay, China. PLoS ONE 2015, 10, e135538. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Wang, Q.; Jiang, D.; Fu, J.; Yang, Y.; Liu, X. Monitoring the invasion of Spartina alterniflora using very high resolution unmanned aerial vehicle imagery in Beihai, Guangxi (China). Sci. World J. 2014, 2014, 638296. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Jeelani, N.; Xin, L.; Cheng, X.; An, S. Spartina alterniflora invasion alters soil microbial community composition and microbial respiration following invasion chronosequence in a coastal wetland of China. Sci. Rep. 2016, 6, 26880. [Google Scholar] [CrossRef]
- Lin, W.P.; Chen, G.S.; Guo, P.P.; Zhu, W.Q.; Zhang, D.H. Remote-sensed monitoring of dominant plant species distribution and dynamics at Jiuduansha Wetland in Shanghai, China. Remote Sens. 2015, 7, 10227–10241. [Google Scholar] [CrossRef]
- Wang, C.; Liu, H.; Zhang, Y.; Li, Y. Classification of land-cover types in muddy tidal flat wetlands using remote sensing data. J. Appl. Remote Sens. 2014, 7. [Google Scholar] [CrossRef]
- Ai, J.; Gao, W.; Gao, Z.; Shi, R.; Zhang, C.; Liu, C. Integrating pan-sharpening and classifier ensemble techniques to map an invasive plant (Spartina alterniflora) in an estuarine wetland using Landsat 8 imagery. J. Appl. Remote Sens. 2016, 10. [Google Scholar] [CrossRef]
- Hladik, C.; Alber, M. Classification of salt marsh vegetation using edaphic and remote sensing-derived variables. Estuar. Coast. Shelf Sci. 2014, 141, 47–57. [Google Scholar] [CrossRef]
- Lu, J.; Zhang, Y. Spatial distribution of an invasive plant Spartina alterniflora and its potential as biofuels in China. Ecol. Eng. 2013, 52, 175–181. [Google Scholar] [CrossRef]
- Liu, X.; Liu, H.Y.; Lin, Z.S.; Li, L.H.; Lv, S.C. Spatial pattern changes of Spartina alterniflora with different invasion ages in the Yancheng coastal wetland. Acta Ecol. Sin. 2017, 37, 307–314. [Google Scholar] [CrossRef]
- Sun, C.; Liu, Y.; Zhao, S.; Zhou, M.; Yang, Y.; Li, F. Classification mapping and species identification of salt marshes based on a short-time interval NDVI time-series from HJ-1 optical imagery. Int. J. Appl. Earth Obs. 2016, 45, 27–41. [Google Scholar] [CrossRef]
- Zuo, P.; Zhao, S.; Liu, C.; Wang, C.; Liang, Y. Distribution of Spartina spp. along China’s coast. Ecol. Eng. 2012, 40, 160–166. [Google Scholar] [CrossRef]
- Liu, C.; Jiang, H.; Zhang, S.; Li, C.; Pan, X.; Lu, J.; Hou, Y. Expansion and management implications of invasive alien Spartina alterniflora in Yancheng salt marshes, China. Open J. Ecol. 2016, 6, 113–128. [Google Scholar] [CrossRef]
- Diao, C.; Wang, L. Development of an invasive species distribution model with fine-resolution remote sensing. Int. J. Appl. Earth Obs. 2014, 30, 65–75. [Google Scholar] [CrossRef]
- Bradley, B.A.; Olsson, A.D.; Wang, O.; Dickson, B.G.; Pelech, L.; Sesnie, S.E.; Zachmann, L.J. Species detection vs. habitat suitability: Are we biasing habitat suitability models with remotely sensed data? Ecol. Model. 2012, 244, 57–64. [Google Scholar] [CrossRef]
- Diao, C.; Wang, L. Incorporating plant phenological trajectory in exotic saltcedar detection with monthly time series of Landsat imagery. Remote Sens. Environ. 2016, 182, 60–71. [Google Scholar] [CrossRef]
- Bradley, B.A. Remote detection of invasive plants: A review of spectral, textural and phenological approaches. Biol. Invasions 2014, 16, 1411–1425. [Google Scholar] [CrossRef]
- Ai, J.; Gao, W.; Gao, Z.; Shi, R.; Zhang, C. Phenology-based Spartina alterniflora mapping in coastal wetland of the Yangtze Estuary using time series of GaoFen satellite no. 1 wide field of view imagery. J. Appl. Remote Sens. 2017, 11, 26020. [Google Scholar] [CrossRef]
- Ng, W.T.; Meroni, M.; Immitzer, M.; Böck, S.; Leonardi, U.; Rembold, F.; Gadain, H.; Atzberger, C. Mapping Prosopis spp. with Landsat 8 data in arid environments: Evaluating effectiveness of different methods and temporal imagery selection for Hargeisa, Somaliland. Int. J. Appl. Earth Obs. 2016, 53, 76–89. [Google Scholar] [CrossRef]
- Ng, W.; Rima, P.; Einzmann, K.; Immitzer, M.; Atzberger, C.; Eckert, S. Assessing the Potential of Sentinel-2 and Pléiades Data for the Detection of Prosopis and Vachellia spp. in Kenya. Remote Sens. 2017, 9, 74. [Google Scholar] [CrossRef]
- Meroni, M.; Ng, W.; Rembold, F.; Leonardi, U.; Atzberger, C. Mapping Prosopis juliflora in West Somaliland with Landsat 8 Satellite Imagery and Ground Information. Land Degrad. Dev. 2017, 28, 494–506. [Google Scholar] [CrossRef]
- Ouyang, Z.; Gao, Y.; Xie, X.; Guo, H.; Zhang, T.; Zhao, B. Spectral discrimination of the invasive plant Spartina alterniflora at multiple phenological stages in a saltmarsh wetland. PLoS ONE 2013, 8, e67315. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Li, H.; Li, L.; Man, W.; Jia, M.; Wang, Z.; Lu, C. Monitoring the Invasion of Spartina alterniflora Using Multi-source High-resolution Imagery in the Zhangjiang Estuary, China. Remote Sens. 2017, 9, 539. [Google Scholar] [CrossRef]
- Wang, J.; Liu, Z.; Yu, H.; Li, F.F. Mapping Spartina alterniflora Biomass Using LiDAR and Hyperspectral Data. Remote Sens. 2017, 9, 589. [Google Scholar] [CrossRef]
- Robinson, T.P.; Klinken, R.D.V.; Metternicht, G. Spatial and temporal rates and patterns of mesquite (Prosopis species) invasion in Western Australia. J. Arid Environ. 2008, 72, 175–188. [Google Scholar] [CrossRef]
- Evangelista, P.H.; Stohlgren, T.J.; Morisette, J.T.; Kumar, S. Mapping Invasive Tamarisk (Tamarix): A Comparison of Single-Scene and Time-Series Analyses of Remotely Sensed Data. Remote Sens. 2009, 1, 519–533. [Google Scholar] [CrossRef]
- Pavri, F.; Aber, J.S. Characterizing wetland landscapes: A spatiotemporal analysis of remotely sensed data at Cheyenne Bottoms, Kansas. Phys. Geogr. 2004, 25, 86–104. [Google Scholar] [CrossRef]
- Anderson, G.L.; Carruthers, R.I.; Ge, S.K.; Gong, P. Cover: Monitoring of invasive Tamarix distribution and effects of biological control with airborne hyperspectral remote sensing. Int. J. Remote Sens. 2005, 26, 2487–2489. [Google Scholar] [CrossRef]
- Peterson, E.B. Estimating cover of an invasive grass (Bromus tectorum) using tobit regression and phenology derived from two dates of Landsat ETM+ data. Int. J. Remote Sens. 2005, 26, 2491–2507. [Google Scholar] [CrossRef]
- Ji, W.; Wang, L. Phenology-guided saltcedar (Tamarix spp.) mapping using Landsat TM images in western U.S. Remote Sens. Environ. 2016, 173, 29–38. [Google Scholar] [CrossRef]
- Gao, Z.G.; Zhang, L.Q. Multi-seasonal spectral characteristics analysis of coastal salt marsh vegetation in Shanghai, China. Estuar. Coast. Shelf Sci. 2006, 69, 217–224. [Google Scholar] [CrossRef]
- Amboka, A.A.; Ngigi, T.G. Mapping and monitoring spatial-temporal cover change of Prosopis species colonization in Baringo Central, Kenya. Int. J. Eng. Sci. Invent. 2015, 4, 2319–6734. [Google Scholar]
- Wakie, T.T.; Evangelista, P.H.; Jarnevich, C.S.; Laituri, M. Mapping current and potential distribution of non-native prosopis juliflora in the afar region of Ethiopia. PLos ONE 2014, 9, e112854. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Li, A.; Dhakal, S.; Glenn, N.F.; Spaete, L.P.; Shinneman, D.J.; Pilliod, D.S.; Arkle, R.S.; McIlroy, S.K. Lidar aboveground vegetation biomass estimates in shrublands: prediction, uncertainties and application to coarser scales. Remote Sens. 2017, 9, 903. [Google Scholar] [CrossRef]
- Ao, Z.; Su, Y.; Li, W.; Guo, Q.; Zhang, J. One-Class Classification of Airborne LiDAR Data in Urban Areas Using a Presence and Background Learning Algorithm. Remote Sens. 2017, 9, 1001. [Google Scholar] [CrossRef]
- Song, B.; Li, P.; Li, J.; Plaza, A. One-class classification of remote sensing images using kernel sparse representation. IEEE J. STARS 2016, 9, 1613–1623. [Google Scholar] [CrossRef]
- Schölkopf, B.; Platt, J.C. Estimating the support of a high-dimensional Distribution. Neural Comput. 2001, 13, 1443–1471. [Google Scholar] [CrossRef] [PubMed]
- Tax, D.M.J.; Duin, R.P.W. Support Vector Data Description. Mach. Learn. 2004, 54, 45–66. [Google Scholar] [CrossRef]
- Elkan, C.; Noto, K. Learning classifiers from only positive and unlabeled data. In Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Las Vegas, NV, USA, 24–27 August 2008; pp. 213–220. [Google Scholar]
- Liu, B.; Dai, Y.; Li, X.; Lee, W.S. Building text classifiers using positive and unlabeled examples. In Proceedings of the IEEE International Conference on Data Mining, Melbourne, FL, USA, 22 November 2003; pp. 179–188. [Google Scholar]
- Amici, V.; Marcantonio, M.; La Porta, N.; Rocchini, D. A multi-temporal approach in MaxEnt modelling: A new frontier for land use/land cover change detection. Ecol. Inform. 2017, 40–49. [Google Scholar] [CrossRef]
- Wan, B.; Guo, Q.; Fang, F.; Su, Y.; Wang, R. Mapping US Urban Extents from MODIS Data Using One-Class Classification Method. Remote Sens. 2015, 7, 10143–10163. [Google Scholar] [CrossRef]
- Sanchez-Hernandez, C.; Boyd, D.S.; Foody, G.M. One-class classification for mapping a specific land-cover class: SVDD classification of Fenland. IEEE Trans. Geosci. Remote Sens. 2007, 45, 1061–1073. [Google Scholar] [CrossRef]
- Sánchezazofeifa, A.; Rivard, B.; Wright, J.; Feng, J.L.; Li, P.J.; Chong, M.M.; Bohlman, S.A. Estimation of the Distribution of Tabebuia guayacan (Bignoniaceae) using high-resolution remote sensing imagery. Sensors 2011, 11, 3831–3851. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Xu, H.; Guo, J. Urban building damage detection from very high resolution imagery using OCSVM and spatial features. Int. J. Remote Sens. 2010, 31, 3393–3409. [Google Scholar] [CrossRef]
- Li, P.; Song, B.; Xu, H. Urban building damage detection from very high resolution imagery by One-Class SVM and shadow information. In Proceedings of the 2011 IEEE International Geoscience and Remote Sensing Symposium, Vancouver, BC, Canada, 24–29 July 2011; pp. 1409–1412. [Google Scholar] [CrossRef]
- Mack, B.; Roscher, R.; Stenzel, S.; Feilhauer, H.; Schmidtlein, S.; Waske, B. Mapping raised bogs with an iterative one-class classification approach. ISPRS J. Photogramm. 2016, 120, 53–64. [Google Scholar] [CrossRef]
- Mack, B.; Roscher, R.; Waske, B. Can I Trust My One-Class Classification? Remote Sens. 2014, 6, 8779–8802. [Google Scholar] [CrossRef]
- Mack, B.; Waske, B. In-depth comparisons of MaxEnt, biased SVM and one-class SVM for one-class classification of remote sensing data. Remote Sens. Lett. 2017, 8, 290–299. [Google Scholar] [CrossRef]
- Baldeck, C.A.; Asner, G.P. Single-species detection with airborne imaging spectroscopy data: A comparison of support vector techniques. IEEE J. STARS 2015, 8, 2501–2512. [Google Scholar] [CrossRef]
- Lin, J.; Liu, X.; Li, K.; Li, X. A maximum entropy method to extract urban land by combining MODIS reflectance, MODIS NDVI, and DMSP-OLS data. Int. J. Remote Sens. 2014, 35, 6708–6727. [Google Scholar] [CrossRef]
- Phillips, S.J.; Schapire, R.E. A maximum entropy approach to species distribution modeling. In Proceedings of the 21st International Conference on Machine Learning, Banff, AB, Canada, 4–8 July 2004; ACM Press: New York, NY, USA, 2004; pp. 655–662. [Google Scholar]
- Phillips, S.J.; Dudík, M. Modeling of species distributions with Maxent: New extensions and a comprehensive evaluation. Ecography 2008, 31, 161–175. [Google Scholar] [CrossRef]
- Li, W.; Guo, Q. A maximum entropy approach to one-class classification of remote sensing imagery. Int. J. Remote Sens. 2010, 31, 2227–2235. [Google Scholar] [CrossRef]
- Ortiz, S.M.; Breidenbach, J.; Kaendler, G. Early detection of bark beetle green attack using TerraSAR-X and RapidEye data. Remote Sens. 2013, 5, 1912–1931. [Google Scholar] [CrossRef]
- Skowronek, S.; Asner, G.P.; Feilhauer, H. Performance of one-class classifiers for invasive species mapping using airborne imaging spectroscopy. Ecol. Inform. 2017, 37, 66–76. [Google Scholar] [CrossRef]
- Morales, N.S.; Fernandez, I.C.; Baca-Gonzalez, V. MaxEnt’s parameter configuration and small samples: Are we paying attention to recommendations? A systematic review. PeerJ 2017, 5, e3093. [Google Scholar] [CrossRef] [PubMed]
- Stenzel, S.; Feilhauer, H.; Mack, B.; Metz, A.; Schmidtlein, S. Remote sensing of scattered Natura 2000 habitats using a one-class classifier. Int. J. Appl. Earth Obs. 2014, 33, 211–217. [Google Scholar] [CrossRef]
- Warren, D.L.; Wright, A.N.; Seifert, S.N.; Shaffer, H.B. Incorporating model complexity and spatial sampling bias into ecological niche models of climate change risks faced by 90 California vertebrate species of concern. Divers Distrib, 2014, 20, 334–343. [Google Scholar] [CrossRef]
- Radosavljevic, A.; Anderson, R.P. Making better Maxent models of species distributions: Complexity, overfitting and evaluation. J. Biogeogr. 2014, 41, 629–643. [Google Scholar] [CrossRef]
- Syfert, M.M.; Smith, M.J.; Coomes, D.A. The effects of sampling bias and model complexity on the predictive performance of MaxEnt species distribution models. PLoS ONE 2013, 8, e551582. [Google Scholar] [CrossRef]
- Muscarella, R.; Galante, P.J.; Soley-Guardia, M.; Boria, R.A.; Kass, J.M.; Uriarte, M.A.; Anderson, R.P. ENMeval: An R package for conducting spatially independent evaluations and estimating optimal model complexity for Maxent ecological niche models. Methods Ecol. Evol. 2014, 5, 1198–1205. [Google Scholar] [CrossRef]
- Warren, D.L.; Seifert, S.N. Ecological niche modeling in Maxent: The importance of model complexity and the performance of model selection criteria. Ecol. Appl. 2011, 21, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Callejas, D.; Araujo, M.B. The effects of model and data complexity on predictions from species distributions models. Ecol. Model. 2016, 326, 4–12. [Google Scholar] [CrossRef]
- EarthExplorer. U.S. Geological Survey Earth Resources Observation and Science (EROS) Center. Available online: http://earthexplorer.usgs.gov/ (accessed on 8 August 2016).
- Chang, C.I.; Du, Q. Interference and noise-adjusted principal components analysis. IEEE Trans. Geosci. Remote Sens. 1999, 37, 2387–2396. [Google Scholar] [CrossRef]
- Munyati, C. Use of principal component analysis (PCA) of remote sensing images in wetland change detection on the Kafue flats, Zambia. Geocarto Int. 2004, 19, 11–22. [Google Scholar] [CrossRef]
- West, A.M.; Evangelista, P.H.; Jarnevich, C.S.; Kumar, S.; Swallow, A.; Luizza, M.W.; Chignell, S.M. Using multi-date satellite imagery to monitor invasive grass species distribution in post-wildfire landscapes: An iterative, adaptable approach that employs open-source data and software. Int. J. Appl. Earth Obs. 2017, 59, 135–146. [Google Scholar] [CrossRef]
- Google Earth, Version 6.2.0. 2016. Available online: https://www.google.com/earth/ (accessed on 18 October 2016).
- LocaSpace Viewer (LSV), Version 3.1.8. 2016. Available online: http://www.locaspace.cn/ (accessed on 11 September 2016).
- Phillips, S.J.; Elith, J. On estimating probability of presence from use-availability or presence-background data. Ecology 2013, 94, 1409–1419. [Google Scholar] [CrossRef] [PubMed]
- Phillips, S.J.; Dudík, M.; Schapire, R.E. Maxent software for modeling species niches and distributions. Version 3.4.1. 2017. Available online: http://biodiversityinformatics.amnh.org/open_source/maxent/. (accessed on 28 October 2017).
- Jarnevich, C.S.; Stohlgren, T.J.; Kumar, S.; Morisette, J.T.; Holcombe, T.R. Caveats for correlative species distribution modeling. Ecol. Inform. 2015, 29, 6–15. [Google Scholar] [CrossRef]
- Dan, L.W.; Glor, R.E.; Turelli, M. ENMTools: A toolbox for comparative studies of environmental niche models. Ecography 2010, 33, 607–611. [Google Scholar] [CrossRef]
- VanDerWal, J.; Falconi, L.; Januchowski, S.; Shoo, L.; Storlie, C. SDMTools: Species Distribution Modelling Tools: Tools for Processing Data Associated with Species Distribution Modelling Exercises, R Package Version 1.1-221; 2014. Available online: http://CRAN.R-project.org/package=SDMTools (accessed on 5 August 2014).
- Foody, G.M.; Mathur, A.; Sanchez-Hernandez, C.; Boyd, D.S. Training set size requirements for the classification of a specific class. Remote Sens. Environ. 2006, 104, 1–14. [Google Scholar] [CrossRef]
- West, A.M.; Kumar, S.; Brown, C.; Stohlgren, T.; Bromberg, J. Field validation of an invasive species Maxent model. Ecol. Inform. 2016, 36, 126–134. [Google Scholar] [CrossRef]
- Rhoden, C.M.; Peterman, W.E.; Taylor, C.A. Maxent-directed field surveys identify new populations of narrowly endemic habitat specialists. PeerJ 2017, 5, e3632. [Google Scholar] [CrossRef] [PubMed]
- Raynolds, M.K.; Walker, D.A.; Raynolds, M.K.; Walker, D.A. Increased wetness confounds Landsat-derived NDVI trends in the central Alaska North Slope region, 1985–2011. Environ. Res. Lett. 2016, 11, 85004. [Google Scholar] [CrossRef]
- Esau, I.; Miles, V.V.; Davy, R.; Miles, M.W.; Kurchatova, A. Trends in normalized difference vegetation index (NDVI) associated with urban development in northern West Siberia. Atmos. Chem. Phys. 2016, 16, 1–29. [Google Scholar] [CrossRef]
- Couvillion, B.R.; Beck, H. Marsh collapse thresholds for coastal louisiana estimated using elevation and vegetation index data. J. Coast. Res. 2013, 31, 58–67. [Google Scholar] [CrossRef]
- Carle, M.V.; Sasser, C.E. Productivity and resilience: Long-term trends and storm-driven fluctuations in the plant community of the accreting Wax lake delta. Estuar. Coasts 2016, 39, 406–422. [Google Scholar] [CrossRef]
- Feilhauer, H.; Thonfeld, F.; Faude, U.; He, K.S.; Rocchini, D.; Schmidtlein, S. Assessing floristic composition with multispectral sensors-a comparison based on monotemporal and multiseasonal field spectra. Int. J. Appl. Earth Obs. 2013, 21, 218–229. [Google Scholar] [CrossRef]
- Immitzer, M.; Vuolo, F.; Atzberger, C. First Experience with Sentinel-2 Data for Crop and Tree Species Classifications in Central Europe. Remote Sens. 2016, 8, 166. [Google Scholar] [CrossRef]
- Meng, J.; Wu, B.; Chen, X.; Du, X.; Niu, L.; Zhang, F. Validation of HJ-1 B charge-coupled device vegetation index products with spectral reflectance of Hyperion. Int. J. Remote Sens. 2011, 32, 9051–9070. [Google Scholar] [CrossRef]
- Bian, J.; Li, A.; Wang, Q.; Huang, C. Development of dense time series 30-m image products from the Chinese HJ-1A/B constellation: A case study in Zoige Plateau, China. Remote Sens. 2015, 7, 16647–16671. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Q.Z.; Huang, H.P.; Wu, W.; Du, X.; Wang, H.Y. The combined use of remote sensing and social sensing data in fine-grained urban land use mapping: A case study in Beijing, China. Remote Sens. 2017, 9, 865. [Google Scholar] [CrossRef]
- Li, L.; Solana, C.; Canters, F.; Kervyn, M. Testing random forest classification for identifying lava flows and mapping age groups on a single Landsat 8 image. J. Volcanol. Geotherm. Res. 2017, 345, 109–124. [Google Scholar] [CrossRef]


| Description | Monthly Landsat Data | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sensor | OLI | ETM | OLI | ETM | OLI | ETM | ETM | ETM | OLI | OLI | ETM | OLI |
| Track number | 119/37 | 119/37 | 119/37 | 119/37 | 119/37 | 119/37 | 119/37 | 119/37 | 119/37 | 119/37 | 119/37 | 119/37 |
| Time | 01/27 | 02/20 | 3/16 | 04/25 | 05/03 | 06/12 | 07/14 | 08/12 | 09/08 | 10/26 | 11/03 | 12/13 |
| Vegetable Indices | Formula |
|---|---|
| Normalized Difference Vegetation Index (NDVI) | NDVI = (NIR − Red)/(NIR + Red) |
| Difference Vegetation Index (DVI) | DVI = NIR − Red |
| Ratio Vegetation Index (RVI) | RVI = NIR/Red |
| Enhanced Vegetation Index (EVI) | EVI = 6.5 × (NIR − Red)/(NIR + 7.5 × Red − 2.5 × Blue + 1) |
| Models | Maxent | BSVM | McNemar Test to Compare Maxent and BSVM (p-Value) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| OA (%) | Kappa | Sensitivity | Specificity | OA (%) | Kappa | Sensitivity | Specificity | ||
| January | 94.14 | 0.84 | 0.82 | 0.98 | 93.99 | 0.84 | 0.87 | 0.96 | 0.93 (p = 0.336) |
| February | 87.83 | 0.65 | 0.64 | 0.96 | 84.69 | 0.61 | 0.76 | 0.88 | 8.80 (p < 0.01) |
| March | 92.31 | 0.79 | 0.80 | 0.96 | 93.04 | 0.80 | 0.76 | 0.99 | 2.00 (p = 0.157) |
| April | 94.36 | 0.84 | 0.82 | 0.98 | 94.07 | 0.84 | 0.86 | 0.96 | 0.19 (p = 0.662) |
| May | 93.99 | 0.84 | 0.89 | 0.96 | 93.48 | 0.82 | 0.81 | 0.98 | 10.45 (p < 0.01) |
| June | 80.37 | 0.52 | 0.74 | 0.82 | 79.49 | 0.49 | 0.68 | 0.83 | 0.62 (p = 0.432) |
| July | 80.44 | 0.49 | 0.62 | 0.86 | 80.14 | 0.43 | 0.48 | 0.91 | 13.55 (p < 0.01) |
| August | 88.13 | 0.68 | 0.72 | 0.93 | 86.81 | 0.62 | 0.62 | 0.95 | 13.90(p < 0.01) |
| September | 89.74 | 0.71 | 0.71 | 0.96 | 90.77 | 0.74 | 0.74 | 0.97 | 6.53 (p < 0.01) |
| October | 88.13 | 0.68 | 0.73 | 0.93 | 89.16 | 0.69 | 0.71 | 0.95 | 17.69 (p < 0.01) |
| November | 90.32 | 0.74 | 0.75 | 0.95 | 90.11 | 0.72 | 0.69 | 0.97 | 22.74 (p < 0.01) |
| December | 93.41 | 0.82 | 0.79 | 0.98 | 93.33 | 0.82 | 0.86 | 0.96 | 17.28 (p < 0.01) |
| TS-1 | 94.14 | 0.84 | 0.82 | 0.98 | 94.29 | 0.84 | 0.79 | 0.99 | 1.51 (p = 0.218) |
| TS-2 | 95.46 | 0.88 | 0.87 | 0.98 | 95.89 | 0.89 | 0.87 | 0.97 | 0.94 (p = 0.330) |
| TS-NDVI | 93.77 | 0.83 | 0.84 | 0.97 | 92.97 | 0.80 | 0.75 | 0.99 | 1.79 (p = 0.072) |
| Accuracy indices | Mean | Paired Differences 95% Confidence Interval | t | df | Significance (Two-Tailed) | |
|---|---|---|---|---|---|---|
| Lower | Upper | |||||
| Kappa: BSVM—Maxent | −0.124 | −0.451 | 0.209 | −0.800 | 14 | 0.473 |
| OA: BSVM—Maxent | −0.164 | −0.036 | 0.003 | −1.731 | 14 | 0.105 |
| Specificity: BSVM—Maxent | −0.001 | −0.175 | 0.152 | −0.148 | 14 | 0.885 |
| Sensitivity: BSVM—Maxent | −0.496 | −0.967 | −0.002 | −2.258 | 14 | 0.400 |
| Omission: BSVM—Maxent | 0.851 | −0.007 | 0.872 | 1.824 | 14 | 0.900 |
| Parameters | Experimental Setup 1 | Experimental Setup 2 | Experimental Setup 3 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Jan | Feb | Mar | Apr | May | Jun | Jul | Aug | Sep | Oct | Nov | Dec | TS-1 | TS-2 | TS-NDVI | |
| FC | LQ | LQHP | LQH | LQH | LQHP | LQ | LQ | LQHP | LQH | LQ | L | L | LQH | LQH | LQ |
| RM | 1 | 1.5 | 3.5 | 2 | 2 | 2 | 2.5 | 2 | 4 | 1.5 | 2 | 2 | 4 | 1.5 | 2 |
| Accuracy indices | Mean | Paired Differences 95% Confidence Interval | t | df | Significance (Two-Tailed) | |
|---|---|---|---|---|---|---|
| Lower | Upper | |||||
| OA: Default—AICc | 0.011 | −0.003 | 0.025 | 1.671 | 14 | 0.117 |
| Kappa: Default—AICc | 0.005 | −0.001 | 0.012 | 1.992 | 14 | 0.066 |
| Specificity: Default—AICc | 0.010 | −0.002 | 0.225 | 1.774 | 14 | 0.098 |
| Sensitivity Default—AICc | −0.007 | −0.033 | 0.019 | −0.560 | 14 | 0.585 |
| Rank | Jan | Feb | Mar | Apr | May | Jun | Jul | Aug | Sep | Oct | Nov | Dec |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | TC2 | red | TC1 | NIR | PC2 | NIR | SWIR1 | TC3 | NDVI | red | NDVI | NDVI |
| 2 | SWIR1 | EVI | green | TC3 | NIR | TC2 | TC3 | PC2 | green | green | SWIR2 | green |
| 3 | PC2 | NIR | NDVI | green | blue | EVI | SWIR2 | PC1 | red | TC1 | SWIR1 | NIR |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Liu, H.; Gong, H.; Lin, Z.; Lv, S. Appling the One-Class Classification Method of Maxent to Detect an Invasive Plant Spartina alterniflora with Time-Series Analysis. Remote Sens. 2017, 9, 1120. https://doi.org/10.3390/rs9111120
Liu X, Liu H, Gong H, Lin Z, Lv S. Appling the One-Class Classification Method of Maxent to Detect an Invasive Plant Spartina alterniflora with Time-Series Analysis. Remote Sensing. 2017; 9(11):1120. https://doi.org/10.3390/rs9111120
Chicago/Turabian StyleLiu, Xiang, Huiyu Liu, Haibo Gong, Zhenshan Lin, and Shicheng Lv. 2017. "Appling the One-Class Classification Method of Maxent to Detect an Invasive Plant Spartina alterniflora with Time-Series Analysis" Remote Sensing 9, no. 11: 1120. https://doi.org/10.3390/rs9111120






