Comparison and Evaluation of Three Methods for Estimating Forest above Ground Biomass Using TM and GLAS Data
Abstract
:1. Introduction
2. Materials
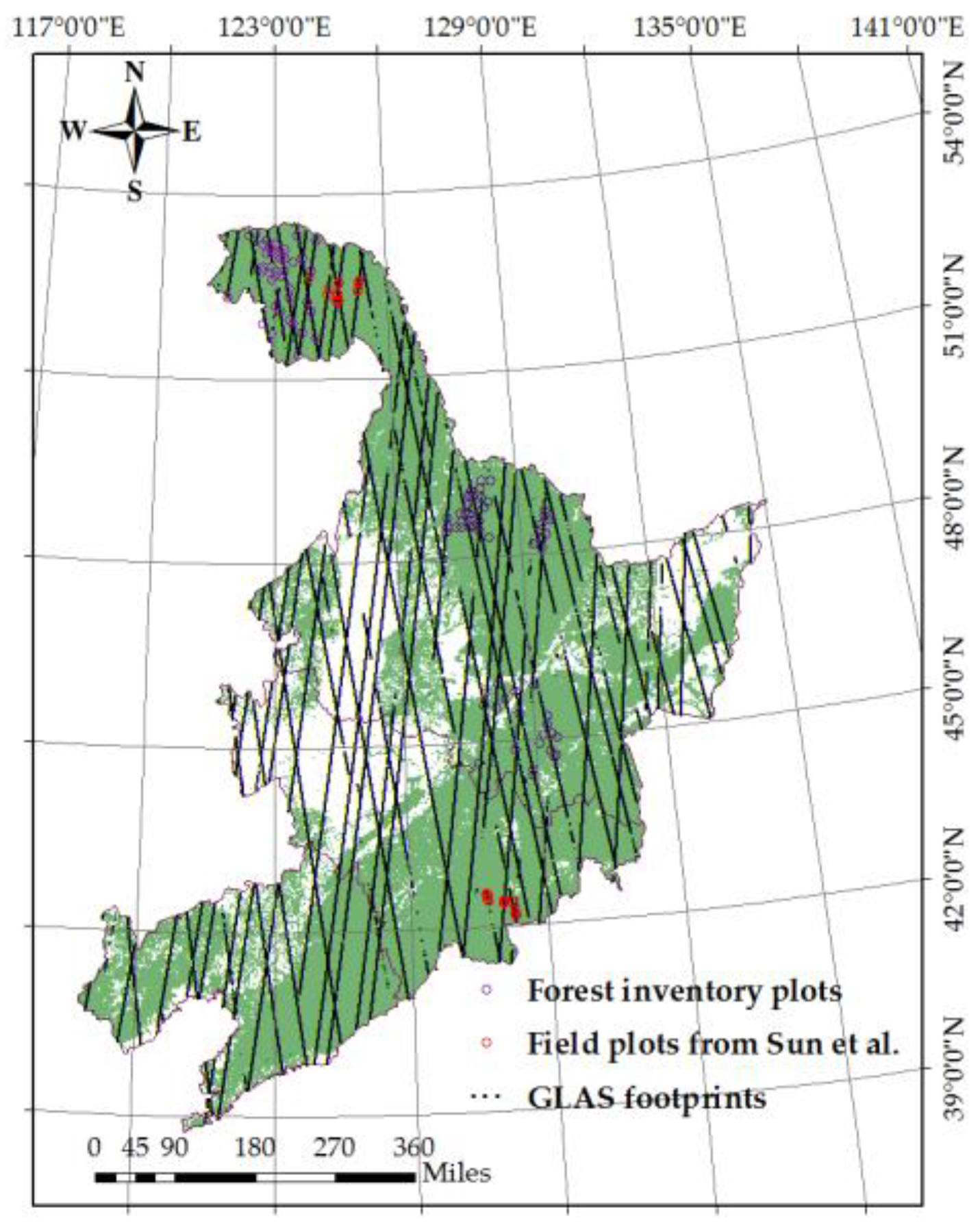
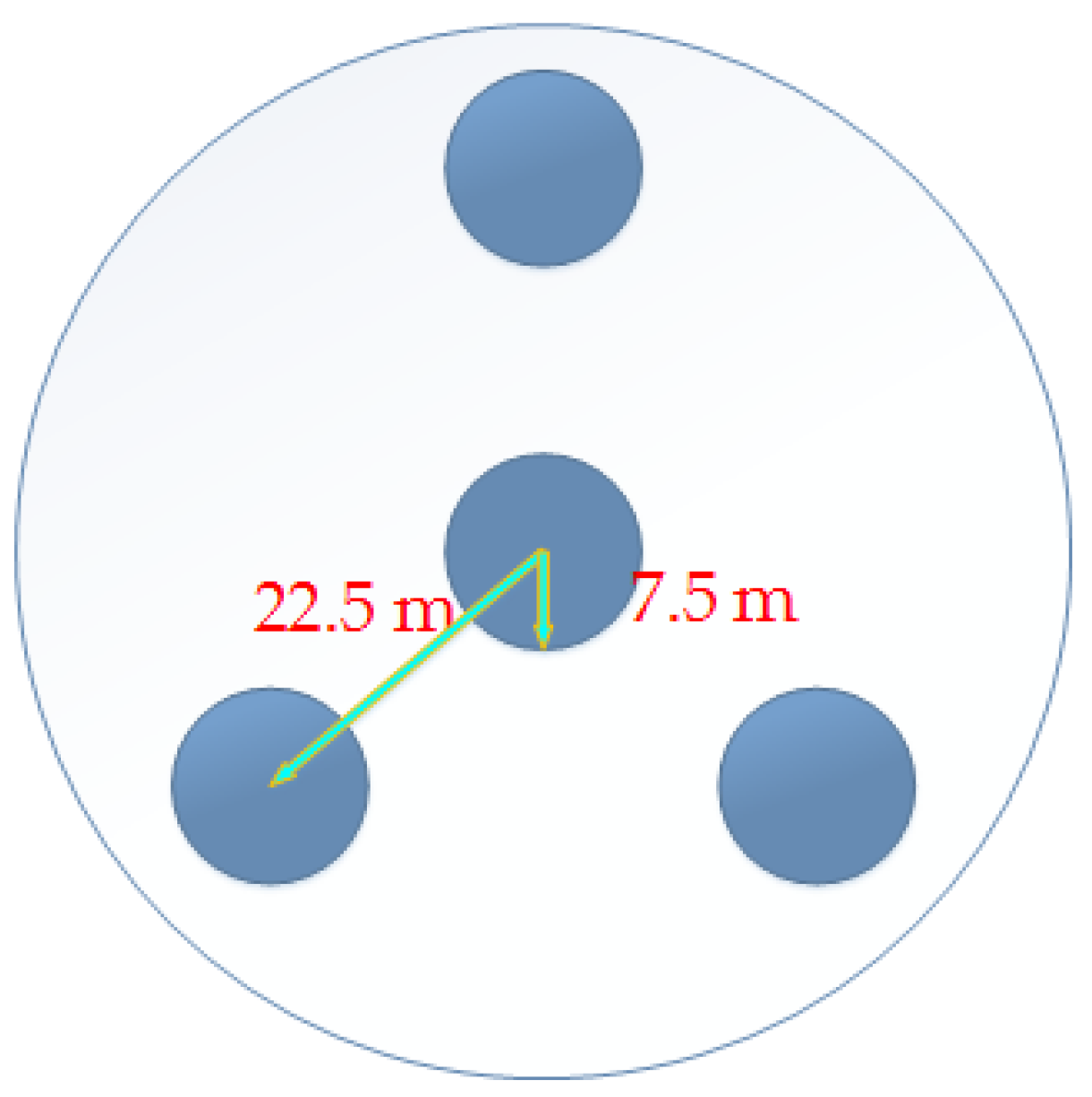
2.1. Field Data
2.2. ICESat/GLAS Data
2.3. Landsat5/TM Data
- Surface reflectance: bands 1, 2, 3, 4, 5 and 7;
- Spectral indices: normalized difference vegetation index (NDVI) [42], Enhanced Vegetation index (EVI) [43] perpendicular vegetation index (PVI) [44], soil-adjusted vegetation index (SAVI) [45], normalized difference infrared vegetation index (NDIIB6) [46], normalized difference infrared vegetation index (NDIIB7) [46], [47], [47], and [47];
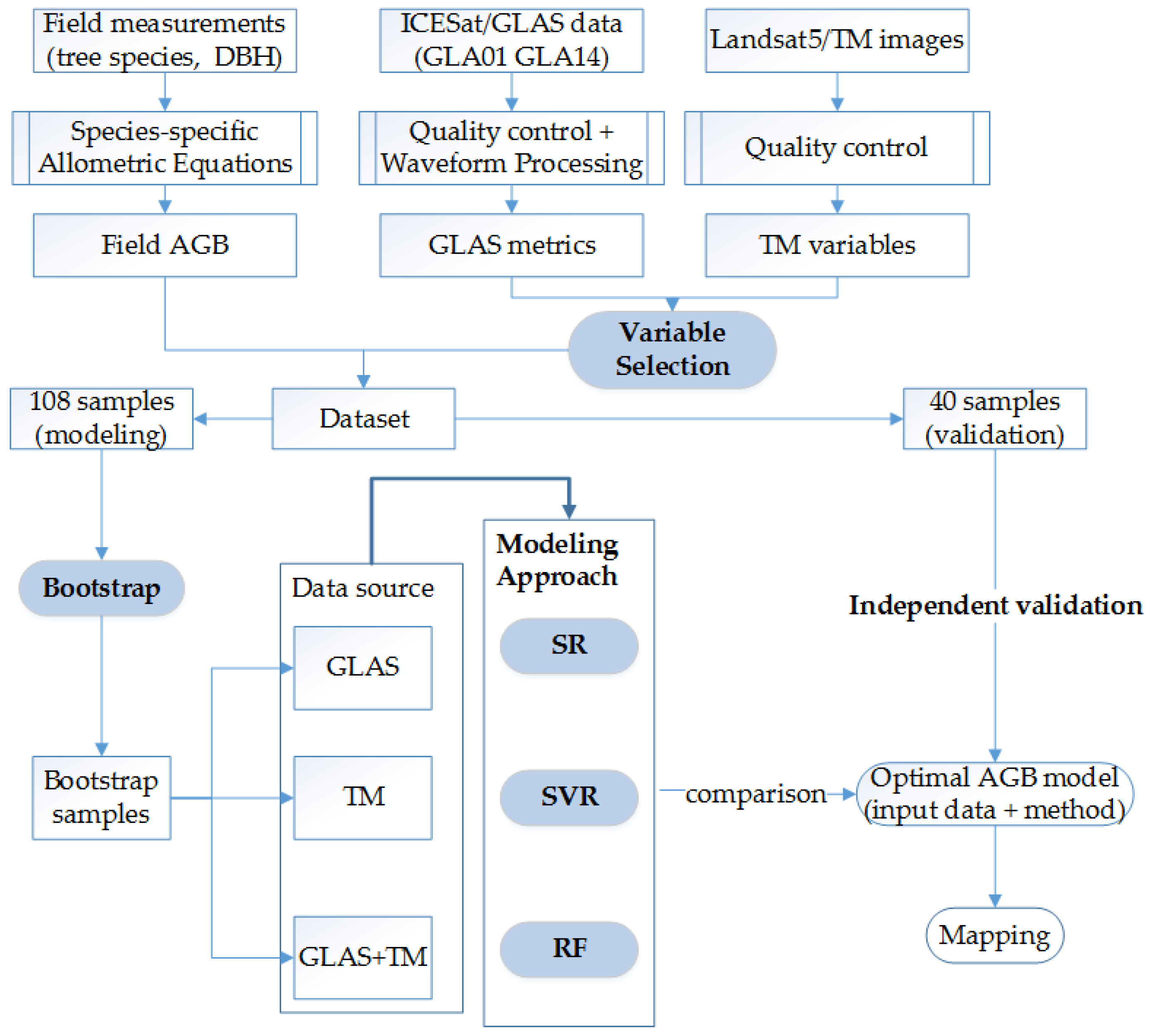
3. Methods
3.1. Variable Selection
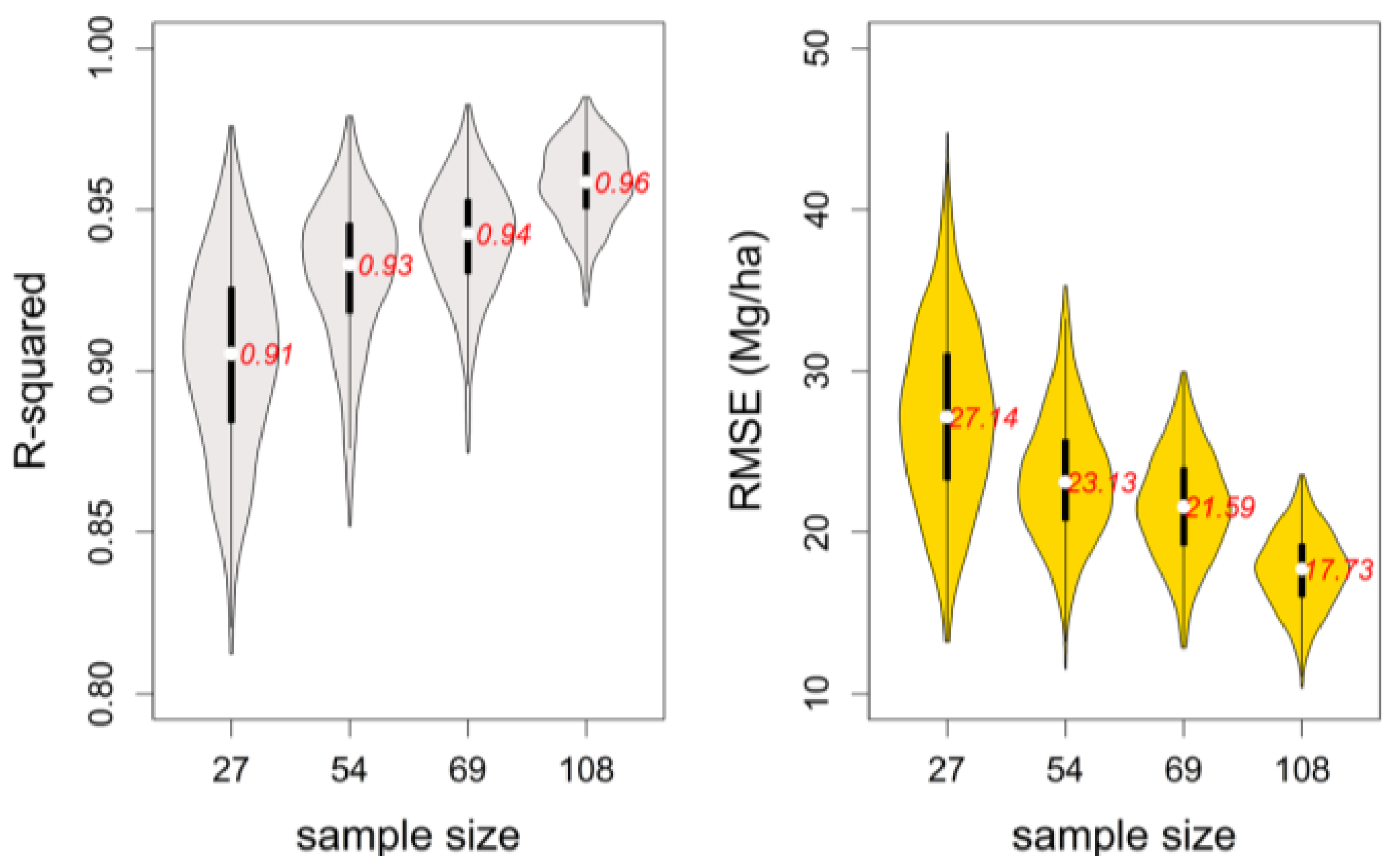
3.2. Bootstrapping
- (1)
- The original data (AGB field data and corresponding predictor variables) of size 108 was sorted by ascending AGB values.
- (2)
- After that, we divided the dataset into four equal-sized subgroups (size = 27).
- (3)
- For each subgroup, random sampling with replacement was performed and repeated 27 times. Therefore, there were 27 data for each subgroup and a total of 108 data were obtained, which was our first bootstrap sample.
- (4)
- The process (3) was repeated 300 times to obtain 300 bootstrap samples.
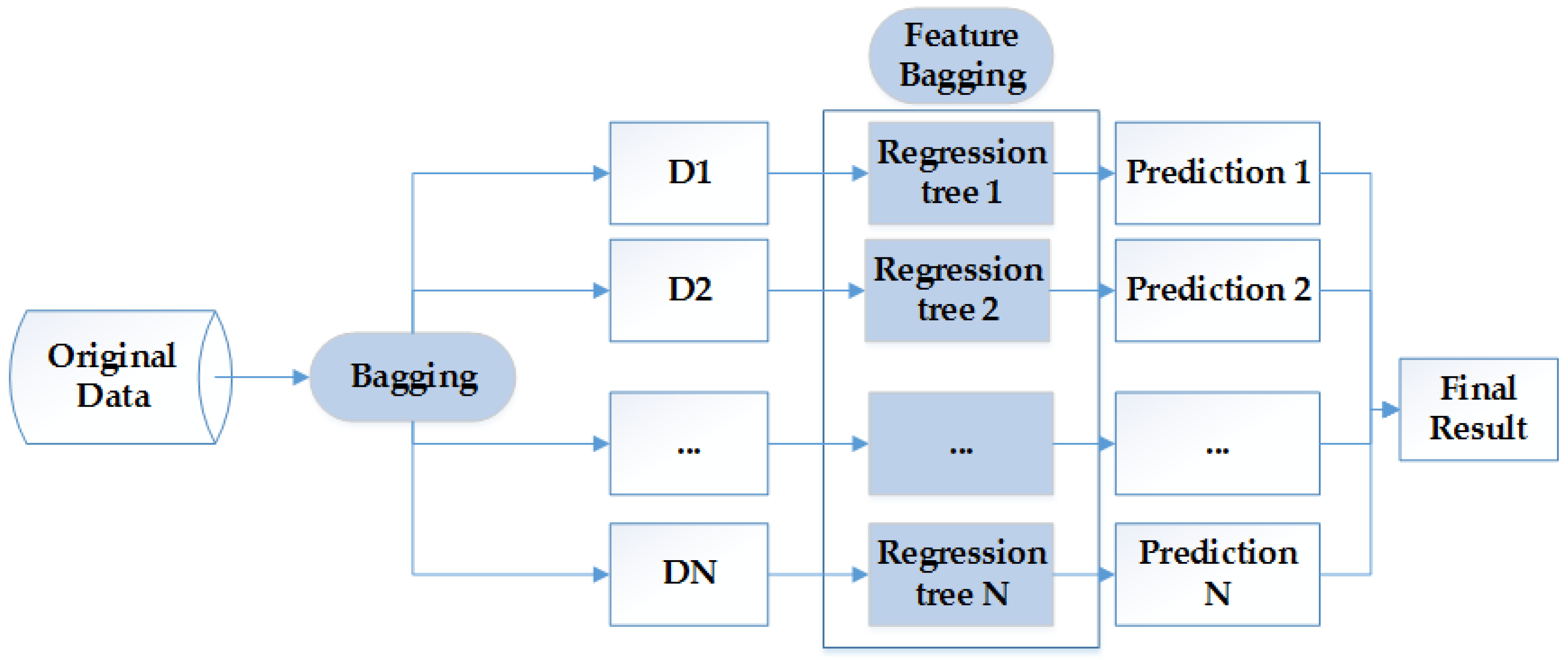
3.3. Modeling Approach
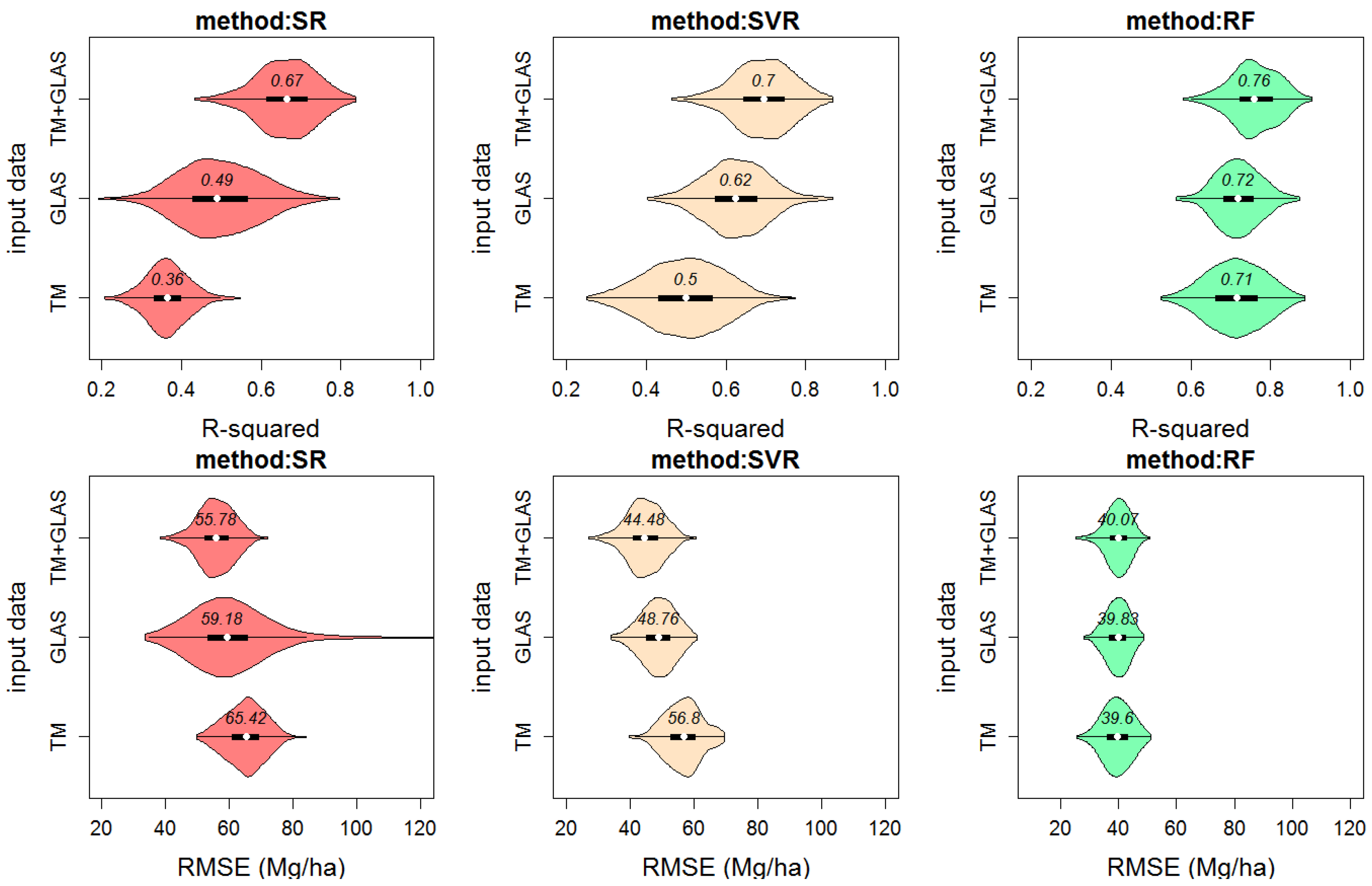
3.4. Independent Validation
4. Results
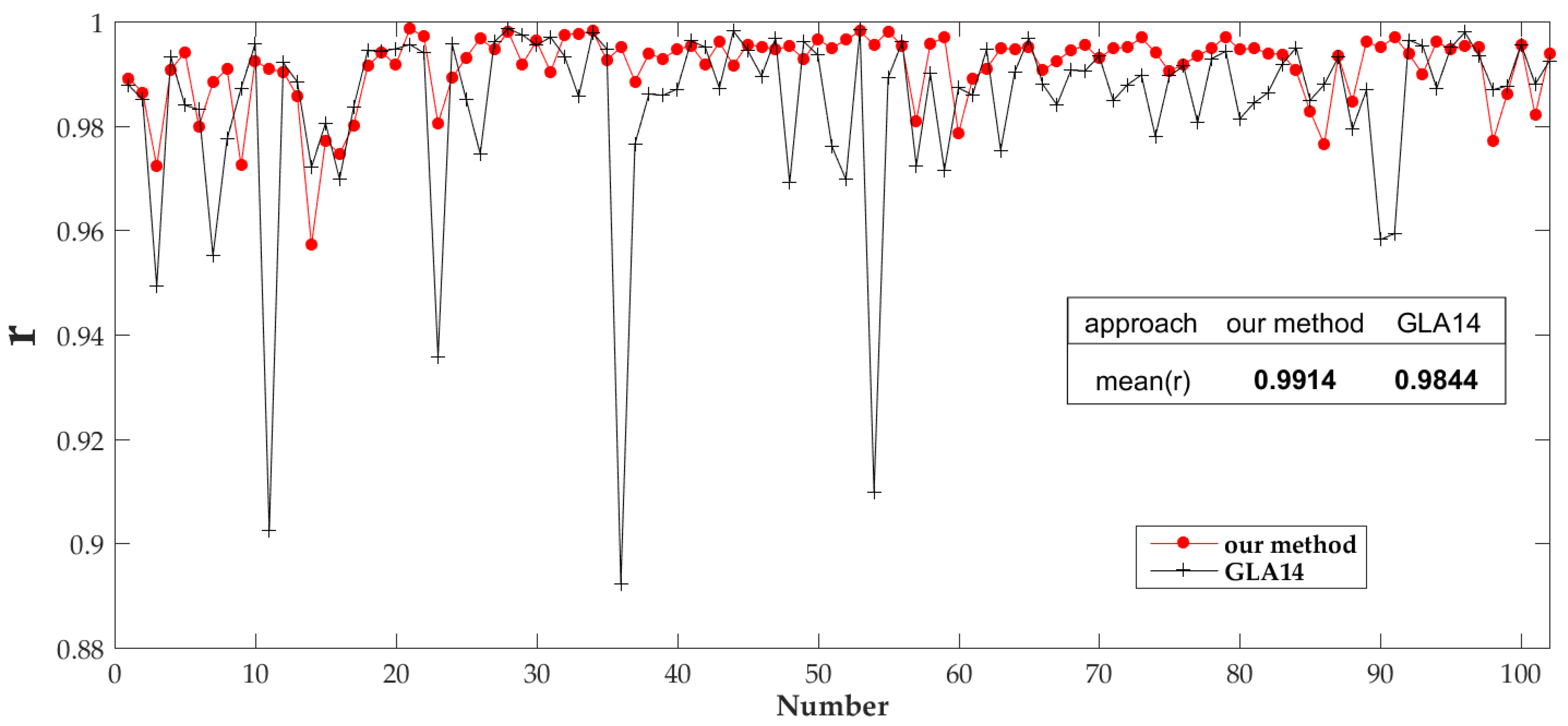
4.1. ICESat/GLAS Data Processing Results
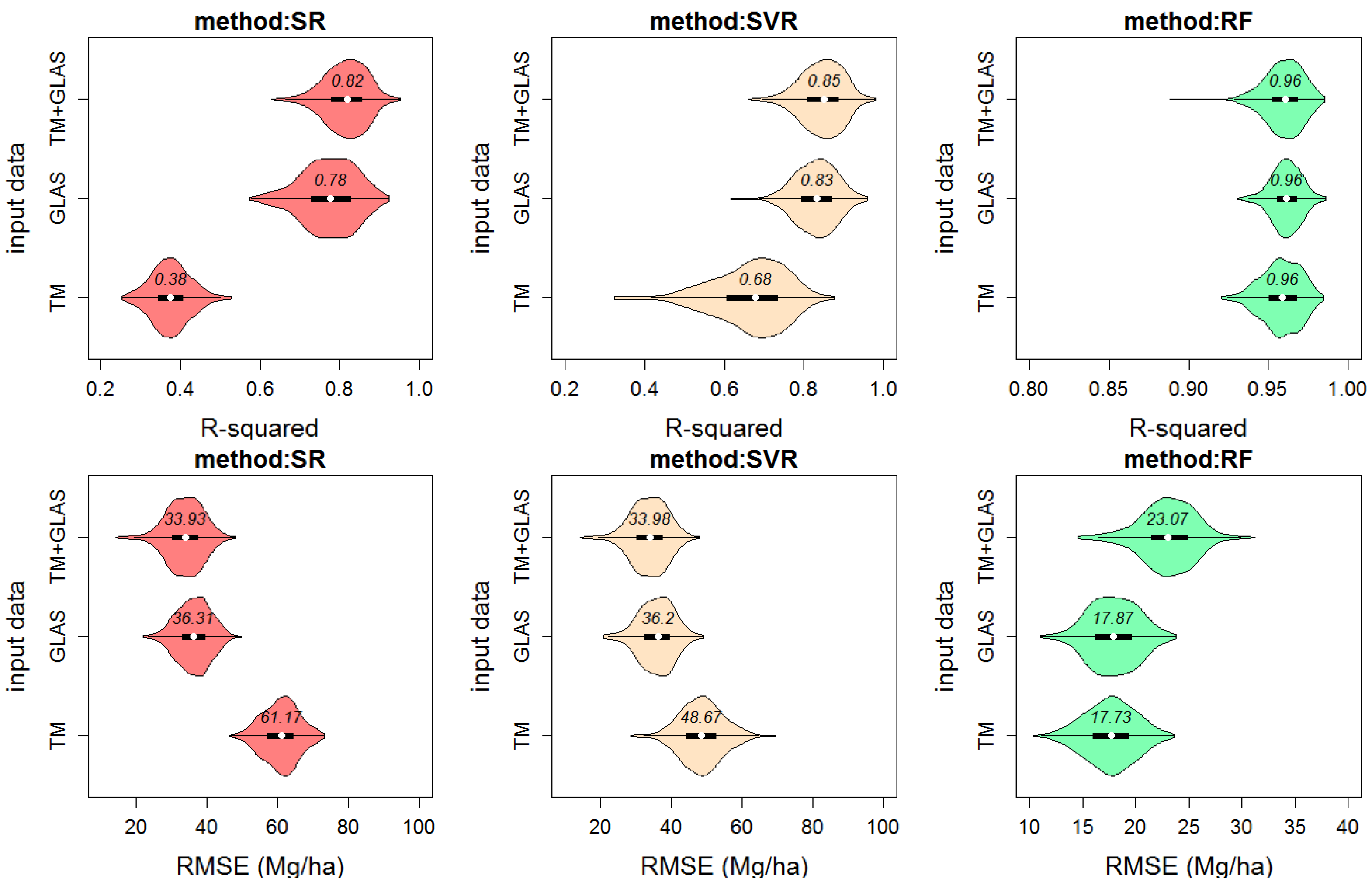
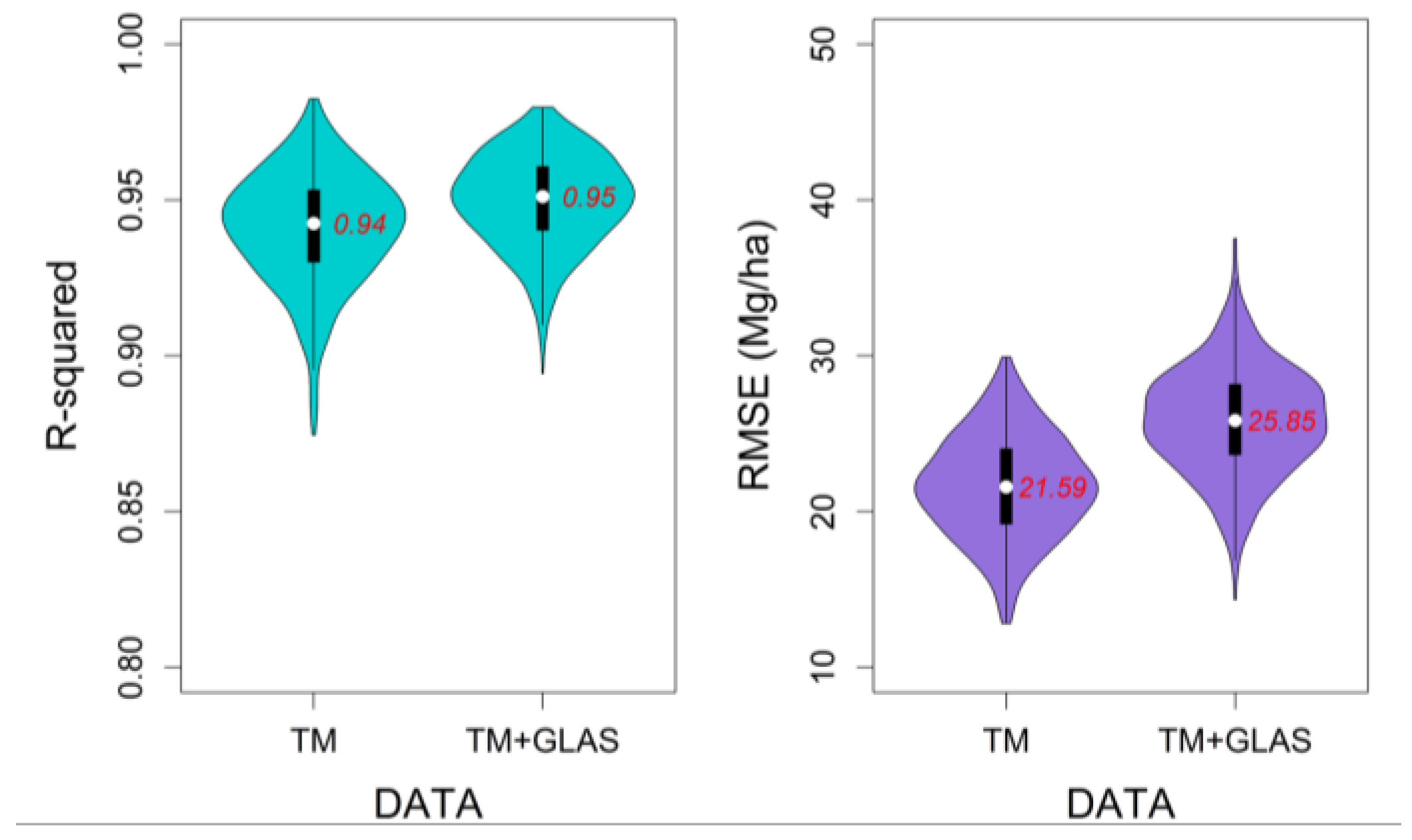
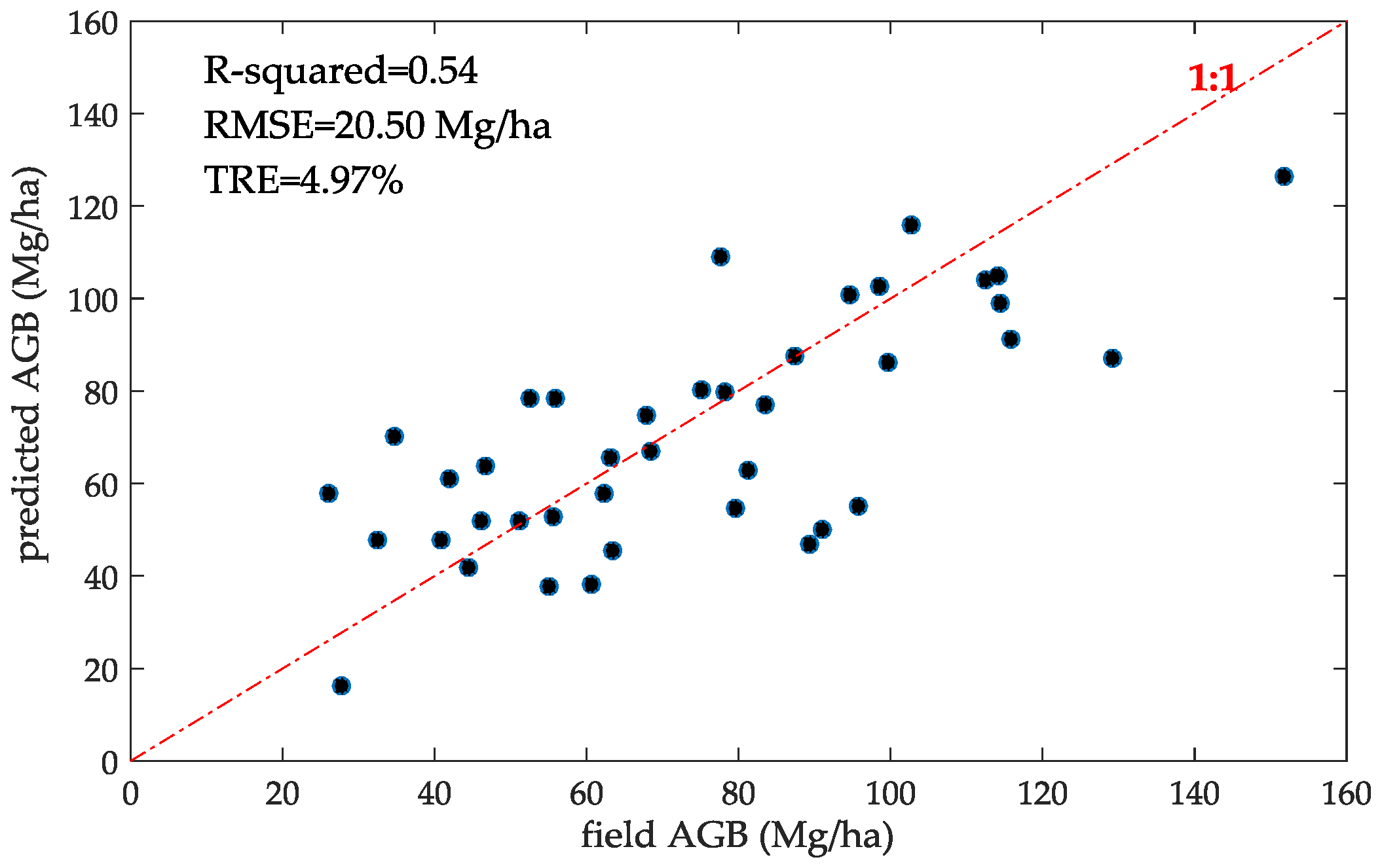
4.2. AGB Model Results
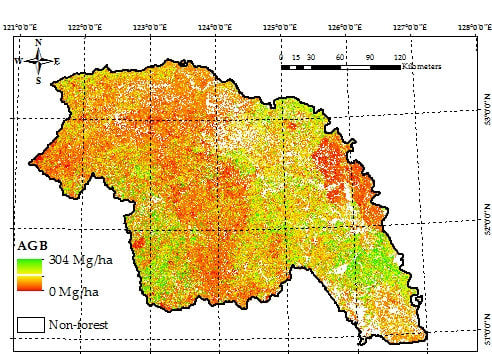
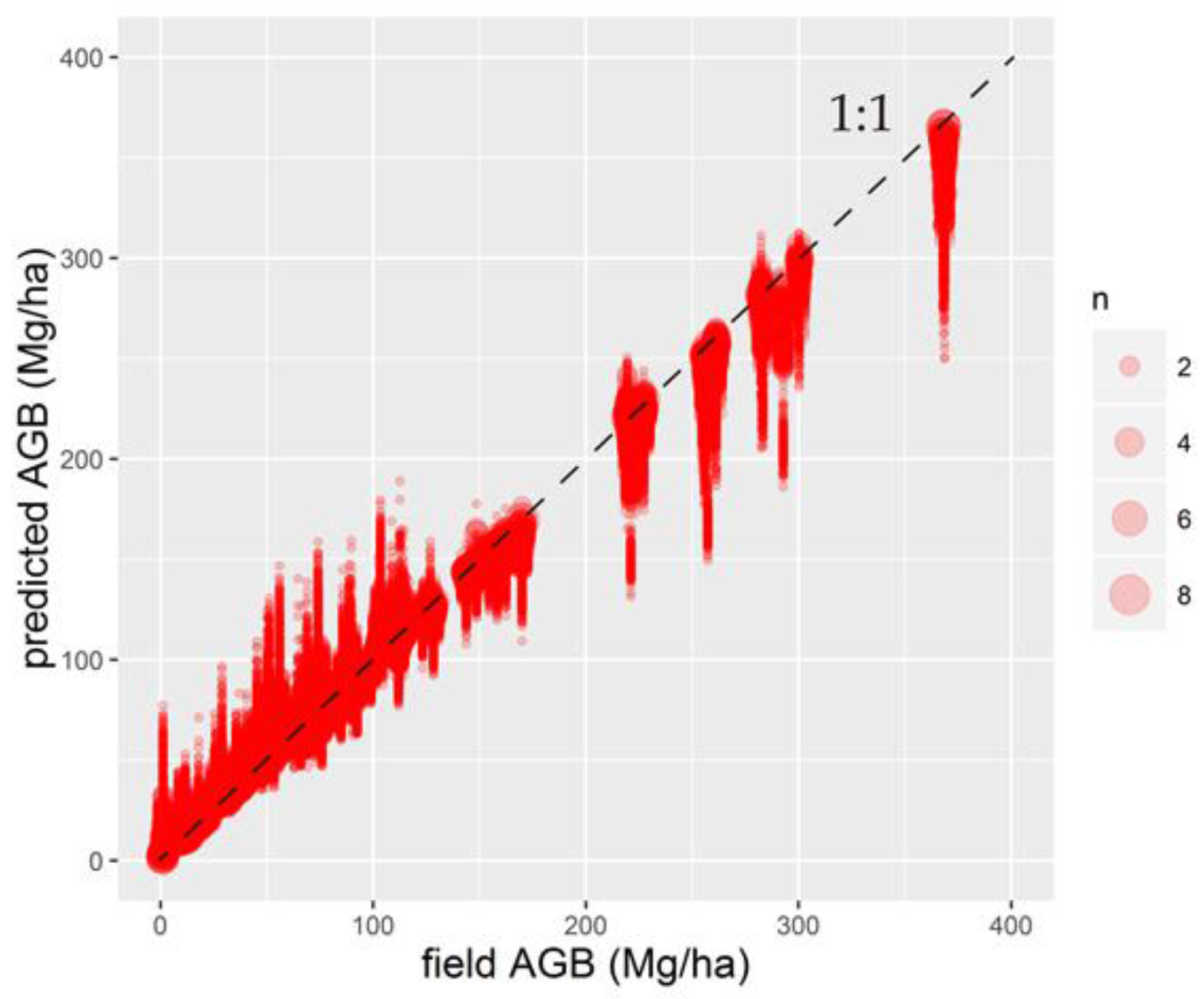
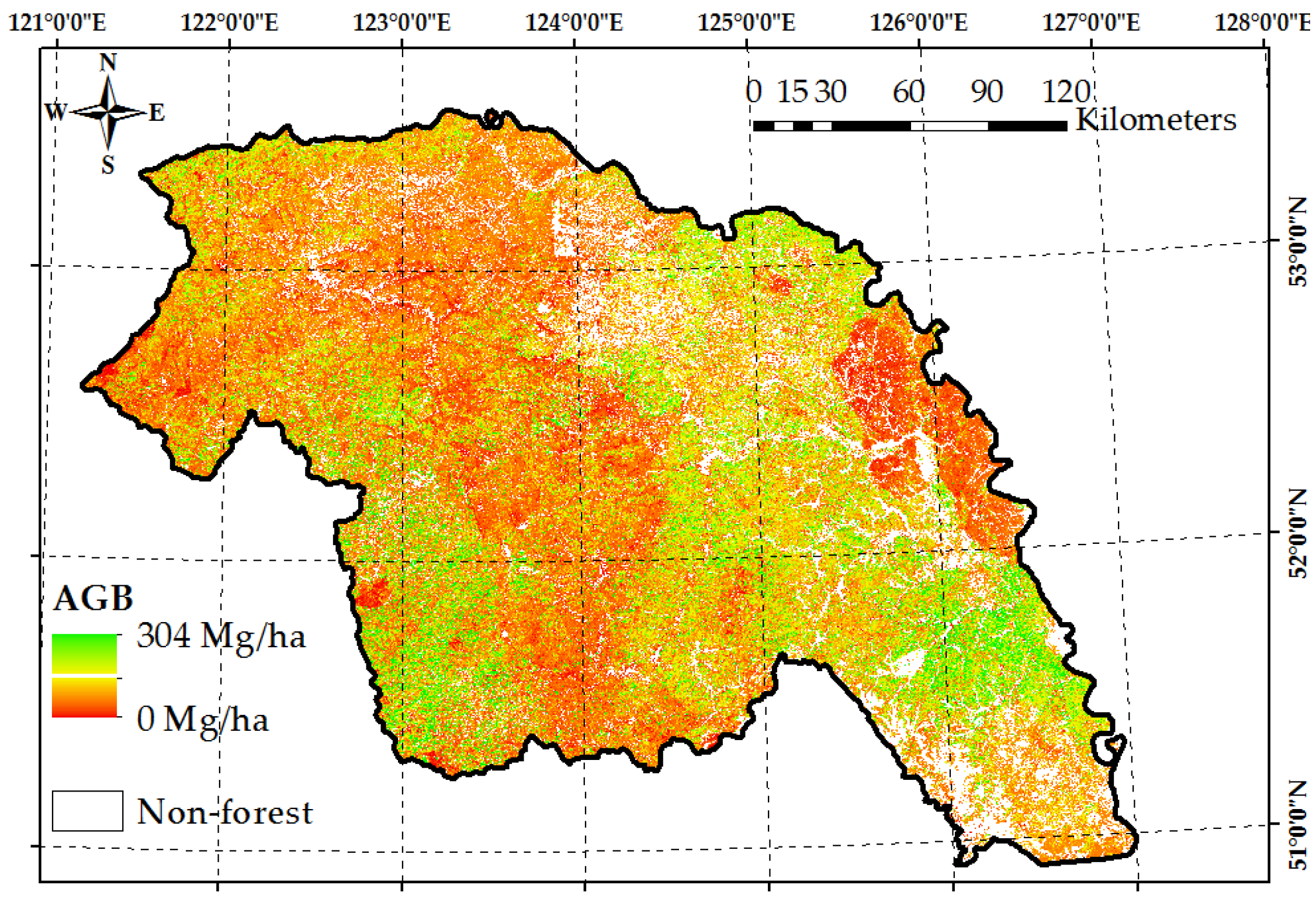
4.3. Wall-to-Wall AGB Prediction over the Daxing’anling Mountains in Heilongjiang Province
5. Discussion
5.1. Spatio-Temporal Matching between GLAS Data and Measured Data
5.2. GLAS Data and Terrain Effects
5.3. Influence of Regional Coverage Types on Estimation
5.4. Effects of TM Data on Regional Biomass Mapping
5.5. Effects of range of AGB values on validation
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Olson, J.S.; Watts, J.A.; Allison, L.J. Carbon in Live Vegetation of Major World Ecosystems; Oak Ridge National Laboratory: Oak Ridge, TN, USA, 1983.
- Brown, S.; Sathaye, J.; Cannell, M.; Cannell, M.; Kauppi, P.E. Mitigation of carbon emissions to the atmosphere by forest management. Commonw. For. Rev. 1996, 75, 80–91. [Google Scholar]
- Zhang, Y.; Liang, S.; Sun, G. Forest biomass mapping of northeastern China using GLAS and MODIS data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 140–152. [Google Scholar] [CrossRef]
- Lu, D. The potential and challenge of remote sensing–based biomass estimation. Int. J. Remote Sens. 2006, 27, 1297–1328. [Google Scholar] [CrossRef]
- McRoberts, R.E.; Næsset, E.; Gobakken, T. Inference for lidar-assisted estimation of forest growing stock volume. Remote Sens. Environ. 2013, 128, 268–275. [Google Scholar] [CrossRef]
- Ahamed, T.; Tian, L.; Zhang, Y.; Ting, K.C. A review of remote sensing methods for biomass feedstock production. Biomass Bioenergy 2011, 35, 2455–2469. [Google Scholar] [CrossRef]
- Lu, D.; Batistella, M.; Moran, E. Satellite estimation of aboveground biomass and impacts of forest stand structure. Photogramm. Eng. Remote Sens. 2005, 71, 967–974. [Google Scholar] [CrossRef]
- Dobson, M.C.; Ulaby, F.T.; LeToan, T.; Beaudoin, A. Dependence of radar backscatter on coniferous forest biomass. IEEE Trans. Geosci. Remote Sens. 1992, 30, 412–415. [Google Scholar] [CrossRef]
- Drake, J.B.; Dubayah, R.O.; Clark, D.B.; Knox, R.G.; Blair, J.B.; Hofton, M.A.; Chazdone, R.L.; Weishampelf, J.F.; Prince, S. Estimation of tropical forest structural characteristics using large-footprint lidar. Remote Sens. Environ. 2002, 79, 305–319. [Google Scholar] [CrossRef]
- Yu, Y.; Yang, X.; Fan, W. Estimates of forest structure parameters from GLAS data and multi-angle imaging spectrometer data. Int. J. Appl. Earth Obs. Geoinform. 2015, 38, 65–71. [Google Scholar] [CrossRef]
- Mougin, E.; Proisy, C.; Marty, G.; Fromard, F.; Puig, H.; Betoulle, J.L.; Rudant, J.P. Multifrequency and multipolarization radar backscattering from mangrove forests. IEEE Trans. Geosci. Remote Sens. 1999, 37, 94–102. [Google Scholar] [CrossRef]
- Sandberg, J.; Tsoukas, H. Grasping the logic of practice: Theorizing through practical rationality. Acad. Manag. Rev. 2011, 36, 338–360. [Google Scholar] [CrossRef]
- Hajj, M.E.; Baghdadi, N.; Fayad, I.; Vieilledent, G.; Bailly, J.S.; Minh, D.H.T. Interest of integrating spaceborne LiDAR data to improve the estimation of biomass in high biomass forested areas. Remote Sens. 2017, 9, 213. [Google Scholar] [CrossRef]
- Fayad, I.; Baghdadi, N.; Guitet, S.; Bailly, J.S.; Hérault, B.; Gond, V.; Hajj, M.E.; Minh, D.H.T. Aboveground biomass mapping in French Guiana by combining remote sensing, forest inventories and environmental data. Int. J. Appl. Earth Obs. Geoinform. 2016, 52, 502–514. [Google Scholar] [CrossRef]
- Lu, D.; Chen, Q.; Wang, G.; Liu, L.; Li, G.; Moran, E. A survey of remote sensing-based aboveground biomass estimation methods in forest ecosystems. Int. J. Digit. Earth 2016, 9, 63–105. [Google Scholar] [CrossRef]
- Dixon, R.K.; Brown, S.; Houghton, R.E.A.; Solomon, A.M.; Trexler, M.C.; Wisniewski, J. Carbon pools and flux of global forest ecosystems. Science 1994, 263, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Saatchi, S.; Malhi, Y.; Zutta, B.; Buermann, W.; Anderson, L.O.; Araujo, A.M.; Phillips, O.L.; Peacock, J.; Steege, H.T.; Gonzalez, G.L.; et al. Mapping landscape scale variations of forest structure, biomass, and productivity in Amazonia. Biogeosci. Discuss. 2009, 6, 5461–5505. [Google Scholar] [CrossRef]
- Pang, Y.; Lefsky, M.; Sun, G.; Miller, M.E.; Li, Z. Temperate forest height estimation performance using ICESat GLAS data from different observation periods. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2008, 37, 777–782. [Google Scholar]
- Sun, G.; Ranson, K.; Masek, J.; Guo, Z.; Pang, Y.; Fu, A.; Wang, D. Estimation of tree height and forest biomass from GLAS data. J. For. Plan. 2008, 13, 157–164. [Google Scholar]
- Chen, X.; Zeng, W.; Xiong, Z.; Zhang, M. New development of China National Forest Inventory (NFI)—On revision of NFI technical regulations. For. Resour. Manag. 2004, 5, 40–45. [Google Scholar]
- Chen, J.; Chen, J.; Liao, A.; Cao, X.; Chen, L.; Chen, X.; He, C.; Han, G.; Peng, S.; Lu, M.; et al. Global land cover mapping at 30 m resolution: A POK-based operational approach. ISPRS J. Photogramm. Remote Sens. 2014, 103, 7–27. [Google Scholar] [CrossRef]
- Zeng, W. Methodoly on Moeling of Single-Tree Biomass Equations for National Biomass Estimation in China. Ph.D. Thesis, Chinese Academy of Forestry, Beijing, China, 2011. (In Chinese). [Google Scholar]
- Zeng, W.S.; Zhang, H.R.; Tang, S.Z. Using the dummy variable model approach to construct compatible single-tree biomass equations at different scales—A case study for Masson pine (Pinus massoniana) in southern China. Can. J. For. Res. 2011, 41, 1547–1554. [Google Scholar] [CrossRef]
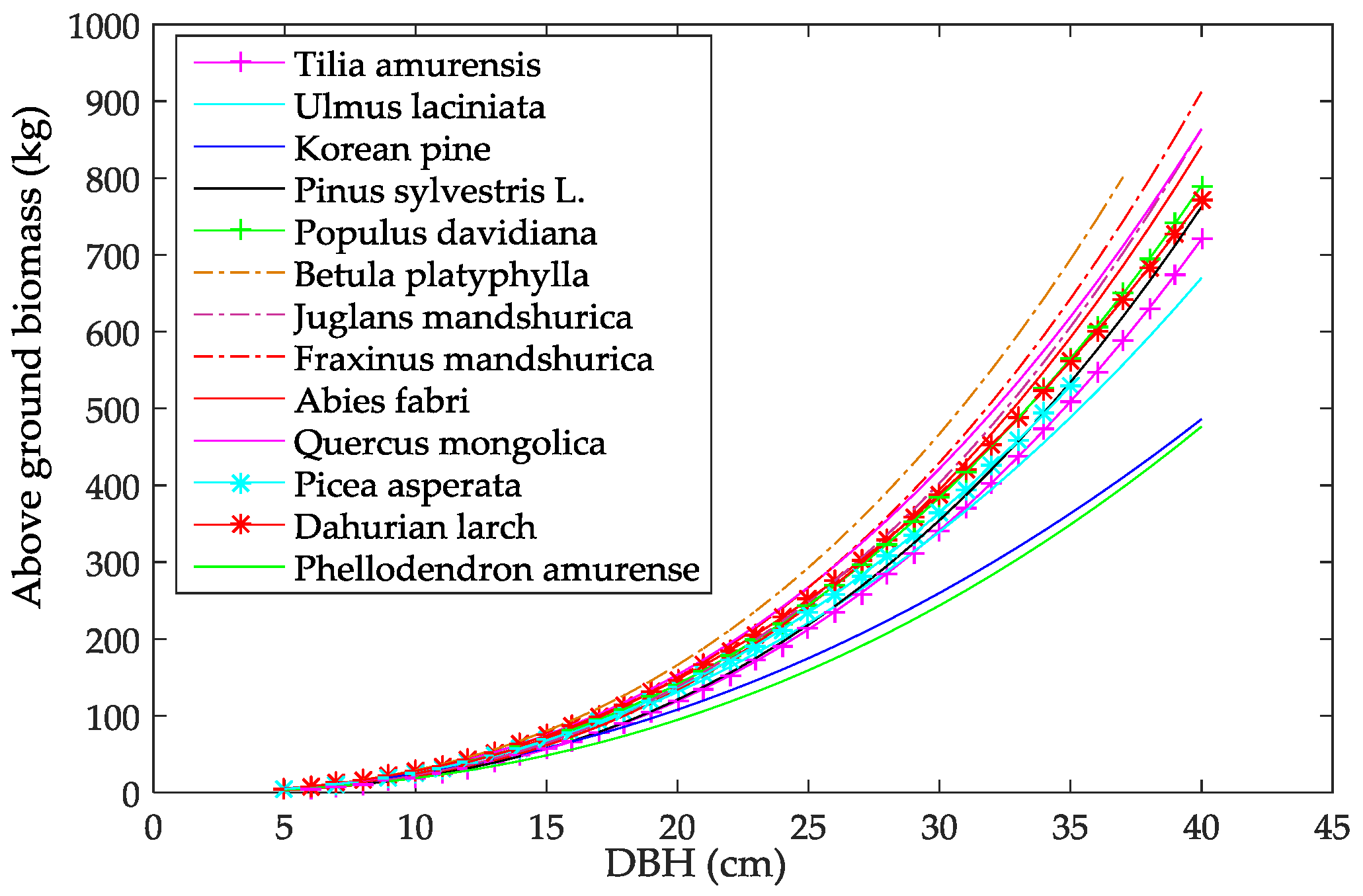
- Dong, L.; Zhang, L.; Li, F. Developing additive systems of biomass equations for nine hardwood species in Northeast China. Trees 2015, 29, 1149–1163. [Google Scholar] [CrossRef]
- Wang, H. Dynamic Simulating System for Stand Growth of Forest in Northeast China. Ph.D. Thesis, Northeast Foreast University, Harbin, China, 2012. (In Chinese). [Google Scholar]
- Wang, C. Biomass allometric equations for 10 co-occurring tree species in Chinese temperate forests. For. Ecol. Manag. 2006, 222, 9–16. [Google Scholar] [CrossRef]
- Dong, L.; Zhang, L.; Li, F. A compatible system of biomass equations for three conifer species in northeast, China. For. Ecol. Manag. 2014, 329, 306–317. [Google Scholar] [CrossRef]
- National Snow & Ice Data Center. Available online: http://nsidc.org/ (accessed on 2 April 2017).
- Chi, H.; Sun, G.; Huang, J.; Guo, Z.; Ni, W.; Fu, A. National forest aboveground biomass mapping from ICESat/GLAS data and MODIS imagery in China. Remote Sens. 2015, 7, 5534–5564. [Google Scholar] [CrossRef]
- Wu, D.; Fan, W. Synergistic use of ICESat/GLAS and MISR data for estimating forest aboveground biomass. Bull. Bot. Res. 2015, 3, 397–405. (In Chinese) [Google Scholar]
- Baghdadi, N.; El Hajj, M.; Bailly, J.S.; Fabre, F. Viability statistics of GLAS/ICESat data acquired over tropical forests. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 1658–1664. [Google Scholar] [CrossRef]
- Pourrahmati, M.R.; Baghdadi, N.N.; Darvishsefat, A.A.; Namiranian, M.; Fayad, I.; Bailly, J. S.; Gond, V. Capability of GLAS/ICESat data to estimate forest canopy height and volume in mountainous forests of Iran. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 5246–5261. [Google Scholar] [CrossRef]
- Sihag, R. Wavelet thresholding for image de-noising. In Proceedings of the International Conference on VLSI, Communication & Instrumentation (ICVCI), Kottayam, Kerala, India, 7–9 April 2011; Volume 201, pp. 20–23. [Google Scholar]
- Sun, G.; Ranson, K.; Kimes, D.; Blair, J.B.; Kovacs, K. Forest vertical structure from GLAS: An evaluation using LVIS and SRTM data. Remote Sens. Environ. 2008, 112, 107–117. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, D.; Huang, W.; Gao, Z.; Yang, X.; Li, C.; Wang, J. Application and evaluation of wavelet-based denoising method in hyperspectral imagery data. In Proceedings of the International Conference on Computer and Computing Technologies in Agriculture, Beijing, China, 29–31 October 2011; Springer: Berlin/Heidelberg, Germany, 2011; pp. 461–469. [Google Scholar]
- Yi, T.-H.; Li, H.-N.; Zhao, X.-Y. Noise smoothing for structural vibration test signals using an improved wavelet thresholding technique. Sensors 2012, 12, 11205–11220. [Google Scholar] [CrossRef] [PubMed]
- Ranson, K.J.; Kimes, D.; Sun, G.; Nelson, R.; Kharuk, V.; Montesano, P. Using MODIS and GLAS data to develop timber volume estimates in central Siberia. In Proceedings of the 2007 IEEE International Geoscience and Remote Sensing Symposium, Barcelona, Spain, 23–28 July 2007; pp. 2306–2309. [Google Scholar]
- Lefsky, M.A.; Keller, M.; Pang, Y.; De Camargo, P.B.; Hunter, M.O. Revised method for forest canopy height estimation from Geoscience Laser Altimeter System waveforms. J. Appl. Remote Sens. 2007, 1, 013537. [Google Scholar]
- Kimes, D.S.; Ranson, K.J.; Sun, G.; Blair, J.B. Predicting lidar measured forest vertical structure from multi-angle spectral data. Remote Sens. Environ. 2006, 100, 503–511. [Google Scholar] [CrossRef]
- Lefsky, M.A.; Harding, D.; Cohen, W.B.; Parker, G.; Shugart, H.H. Surface lidar remote sensing of basal area and biomass in deciduous forests of eastern Maryland, USA. Remote Sens. Environ. 1999, 67, 83–98. [Google Scholar] [CrossRef]
- Harding, D.J.; Carabajal, C.C. ICESat waveform measurements of within-footprint topographic relief and vegetation vertical structure. Geophys. Res. Lett. 2005, 32, 322. [Google Scholar] [CrossRef]
- Rouse, J.W.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring Vegetation Systems in the Great Plains with ERTS; NASA Special Publication; NASA: Washington, DC, USA, 1974; Volume 351, p. 309.
- Huete, A.; Didan, K.; Miura, T.; Rodriguez, E.P.; Gao, X.; Ferreira, L.G. Overview of the radiometric and biophysical performance of the MODIS vegetation indices. Remote Sens. Environ. 2002, 83, 195–213. [Google Scholar] [CrossRef]
- Richardson, A.J.; Wiegand, C.L. Distinguishing vegetation from soil background information. Photogramm. Eng. Remote Sens. 1978, 43, 1541–1552. [Google Scholar]
- Huete, A.R. A soil-adjusted vegetation index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Hunt, E.R.; Rock, B.N. Detection of changes in leaf water content using near-and middle-infrared reflectances. Remote Sens. Environ. 1989, 30, 43–54. [Google Scholar]
- Cai, T.; Ju, C.; Yao, Y. Quantitative estimation of vegetation coverage in Mu Us sandy land based on RS and GIS. Chin. J. Appl. Ecol. 2005, 16, 2301–2305. (In Chinese) [Google Scholar]
- Crist, E.P. A TM Tasseled Cap equivalent transformation for reflectance factor data. Remote Sens. Environ. 1985, 17, 301–306. [Google Scholar] [CrossRef]
- Duane, M.V.; Cohen, W.B.; Campbell, J.L.; Hudiburg, T.; Turner, D.P.; Weyermann, D.L. Implications of alternative field-sampling designs on landsat-based mapping of stand age and carbon stocks in oregon forests. For. Sci. 2010, 21, 77–79. [Google Scholar]
- Powell, S.L.; Cohen, W.B.; Healey, S.P.; Kennedy, R.E.; Moisen, G.G.; Pierce, K.B.; Ohmann, J.L. Quantification of live aboveground forest biomass dynamics with Landsat time-series and field inventory data: A comparison of empirical modeling approaches. Remote Sens. Environ. 2010, 114, 1053–1068. [Google Scholar] [CrossRef]
- Juan, S.; Lantz, F. Application of bootstrap techniques in econometrics: The example of cost estimation in the automotive industry. Oil Gas Sci. Technol. 2001, 56, 373–388. [Google Scholar] [CrossRef]
- Wehrens, R.; Putter, H.; Buydens, L.M.C. The bootstrap: A tutorial. Chemom. Intell. Lab. Syst. 2000, 54, 35–52. [Google Scholar] [CrossRef]
- Freedman, D.A. Bootstrapping regression models. Ann. Stat. 1981, 9, 1218–1228. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Hartig, F.; Latifi, H.; Berger, C.; Hernández, J.; Corvalán, P.; Koch, B. Importance of sample size, data type and prediction method for remote sensing-based estimations of aboveground forest biomass. Remote Sens. Environ. 2014, 154, 102–114. [Google Scholar] [CrossRef]
- Avitabile, V.; Baccini, A.; Friedl, M.A.; Schmullius, C. Capabilities and limitations of Landsat and land cover data for aboveground woody biomass estimation of Uganda. Remote Sens. Environ. 2012, 117, 366–380. [Google Scholar] [CrossRef]
- Zhao, K.; Popescu, S.; Meng, X.; Pang, Y.; Agca, M. Characterizing forest canopy structure with lidar composite metrics and machine learning. Remote Sens. Environ. 2011, 115, 1978–1996. [Google Scholar] [CrossRef]
- Müller, K.-R.; Mika, S.; Rätsch, G.; Tsuda, K.; Schölkopf, B. An introduction to kernel-based learning algorithms. IEEE Trans. Neural Netw. 2001, 12, 181–201. [Google Scholar] [CrossRef] [PubMed]
- Crone, S.F.; Lessmann, S.; Pietsch, S. Parameter sensitivity of support vector regression and neural networks for forecasting. In Proceedings of the IEEE International Conference on Data Mining (DMIN), Hongkong, China, 18–22 December 2006; pp. 396–402. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Ihaka, R.; Gentleman, R. R: A language for data analysis and graphics. J. Comput. Grph. Stat. 1996, 5, 299–314. [Google Scholar] [CrossRef]
- Dalgaard, P. Introductory statistics with R. J. R. Stat. Soc. Ser. A Stat. Soc. 2009, 172, 274–275. [Google Scholar]
- Zeng, W.; Tang, S. Goodness evaluation and precision of tree biomass equations. Sci. Silvae Sin. 2011, 47, 106–113. [Google Scholar]
- Potter, K.; Hagen, H.; Kerren, A.; Dannenmann, P. Methods for presenting statistical information: The box plot. Vis. Larg. Unstruct. Data Sets S 2006, 4, 97–106. [Google Scholar]
- Huang, G.; Xia, C. MODIS-based estimation of forest biomass in northeast China. For. Resour. Manag. 2005, 4, 40–44. (In Chinese) [Google Scholar]
- Pang, Y.; Li, Z.; Lefsky, M.; Sun, G.; Yu, X. Model based terrain effect analyses on ICEsat GLAS waveforms. In Proceedings of the 2006 IEEE International Symposium on Geoscience and Remote Sensing, Vancouver, BC, Canada, 28–30 August 2006; pp. 3232–3235. [Google Scholar]
- Wang, X.; Huang, H.; Gong, P.; Liu, C.; Li, C.; Li, W. Forest canopy height extraction in rugged areas with ICESAT/GLAS data. IEEE Trans. Geosci. Remote Sens. 2014, 52, 4650–4657. [Google Scholar] [CrossRef]
| Data | Acquisition Time | |
|---|---|---|
| Field data | 2005, 2006 and 2007 | |
| GLAS data | L2A | 25 September 2003–19 November 2003 |
| L2D | 25 November 2008–17 December 2008 | |
| L3A | 3 October 2004–8 November 2004 | |
| L3B | 17 February 2005–24 March 2005 | |
| L3C | 20 May 2005–23 June 2005 | |
| L3D | 21 October 2005–24 November 2005 | |
| L3F | 24 May 2006–26 June 2006 | |
| L3G | 25 October 2006–27 November 2006 | |
| Landsat5/TM data | July 2005 | |
| Types | GLAS Metric Abbreviations | Descriptions |
|---|---|---|
| The height metrics | Extent | The distance from signal beginning to signal ending [34]. |
| Treeht | The distance from signal beginning to ground peak [34,38]. | |
| Treeht2 Treeht3 | Top tree heights with corrections [34]. | |
| H25 H75 | Quartile heights calculated by subtracting the ground elevation from elevation at which 25% or 75% of the returned energy occurs [34,39]. | |
| H10 H20 H100 | Decimal heights calculated by subtracting the ground elevation from elevation at which 10% (20%...100%) of the returned energy occurs [34]. | |
| LEE | The distance from the elevation of signal beginning to the first elevation at which the signal strength of the waveform is half of the maximum signal [38]. | |
| TEE | The distance from the last elevation at which the signal strength of the waveform is half of the maximum signal to the elevation of signal ending [38]. | |
| HOME | The height of median energy (HOME) [9]. | |
| Meanh Medh | Mean canopy height, median canopy height [40]. | |
| QMCH | Quadratic mean canopy height (QMCH) calculated from the canopy height profiles [40]. | |
| The intensity metrics | Canopy cover | The ratio of the canopy echo area to the total wave area [41]. |
| AVAW | The area under the waveform from vegetation [41]. |
| Spectral Variables | Formula |
|---|---|
| NDVI | |
| EVI | |
| PVI | |
| SAVI | |
| NDIIB6 | |
| NDIIB7 | |
| TCB | |
| TCG | |
| TCW | |
| TCdistance | |
| TCangle |
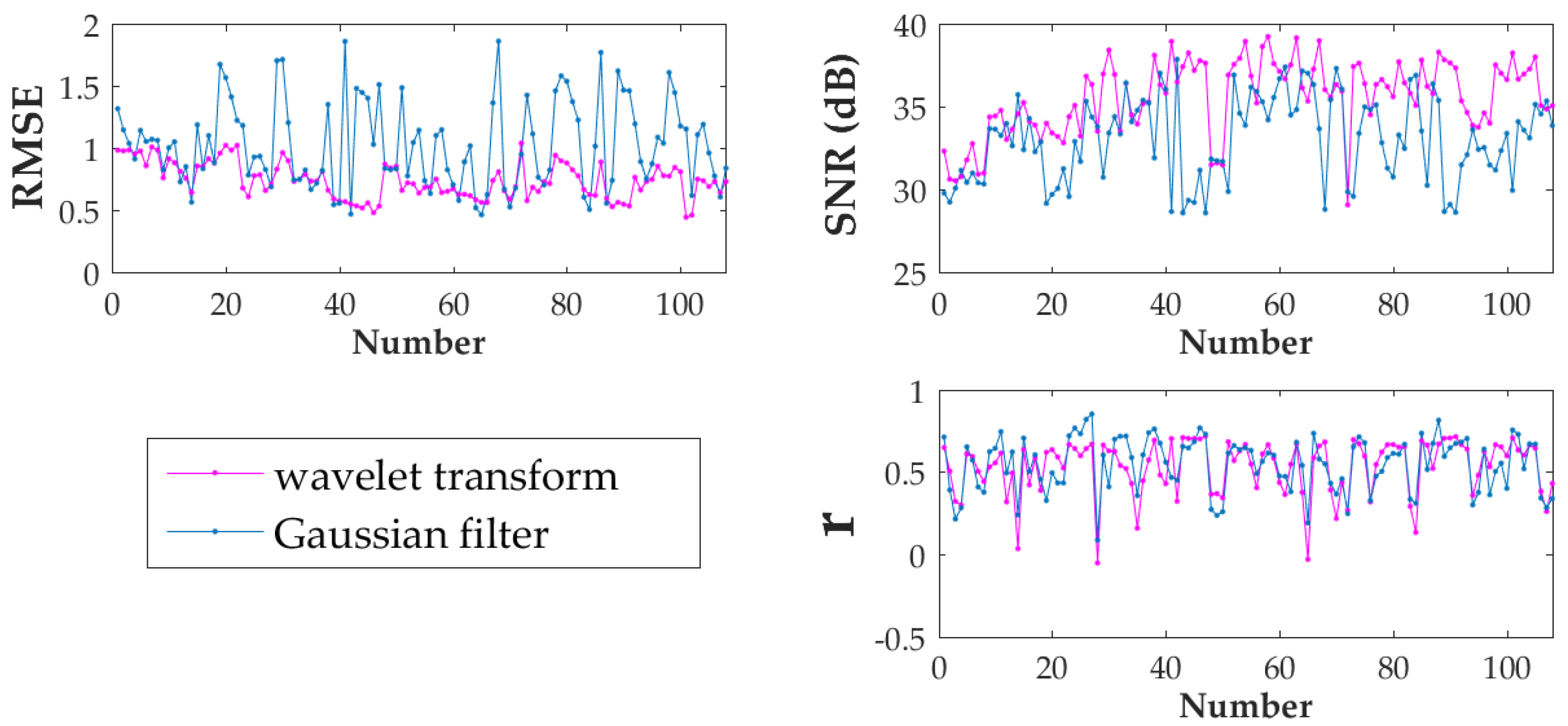
| Method | RMSE | SNR(dB) | r |
|---|---|---|---|
| wavelet transform | 0.73 | 35.67 | 0.53 |
| Gaussian filter | 1.02 | 33.15 | 0.54 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, K.; Wang, J.; Zeng, W.; Song, J. Comparison and Evaluation of Three Methods for Estimating Forest above Ground Biomass Using TM and GLAS Data. Remote Sens. 2017, 9, 341. https://doi.org/10.3390/rs9040341
Liu K, Wang J, Zeng W, Song J. Comparison and Evaluation of Three Methods for Estimating Forest above Ground Biomass Using TM and GLAS Data. Remote Sensing. 2017; 9(4):341. https://doi.org/10.3390/rs9040341
Chicago/Turabian StyleLiu, Kaili, Jindi Wang, Weisheng Zeng, and Jinling Song. 2017. "Comparison and Evaluation of Three Methods for Estimating Forest above Ground Biomass Using TM and GLAS Data" Remote Sensing 9, no. 4: 341. https://doi.org/10.3390/rs9040341