Optimized Kernel Minimum Noise Fraction Transformation for Hyperspectral Image Classification
Abstract
:1. Introduction
2. Proposed OKMNF Method
2.1. Noise Estimation
| Algorithm 1. Noise Estimation. |
| Input: hyperspectral image , sub-block width . Step 1: compute the coefficients , , , and of the multiple linear regression models for each sub-block using Equation (11) or Equation (17); then: , or Step 2: estimate noise: Output: noise data . |
2.2. Kernelization and Regularization
2.3. OKMNF Transformation
| Algorithm 2. The Proposed OKMNF. |
| Input: hyperspectral image , and m training samples. Step 1: compute the residuals (noises) of training samples: . Step 2: dual transformation, kernelization and regularization of using Equation (22). Step 3: compute the eigenvectors of . Step 4: mapping all pixels onto the primal eigenvectors. Output: feature extraction result . |
3. Experiments and Results
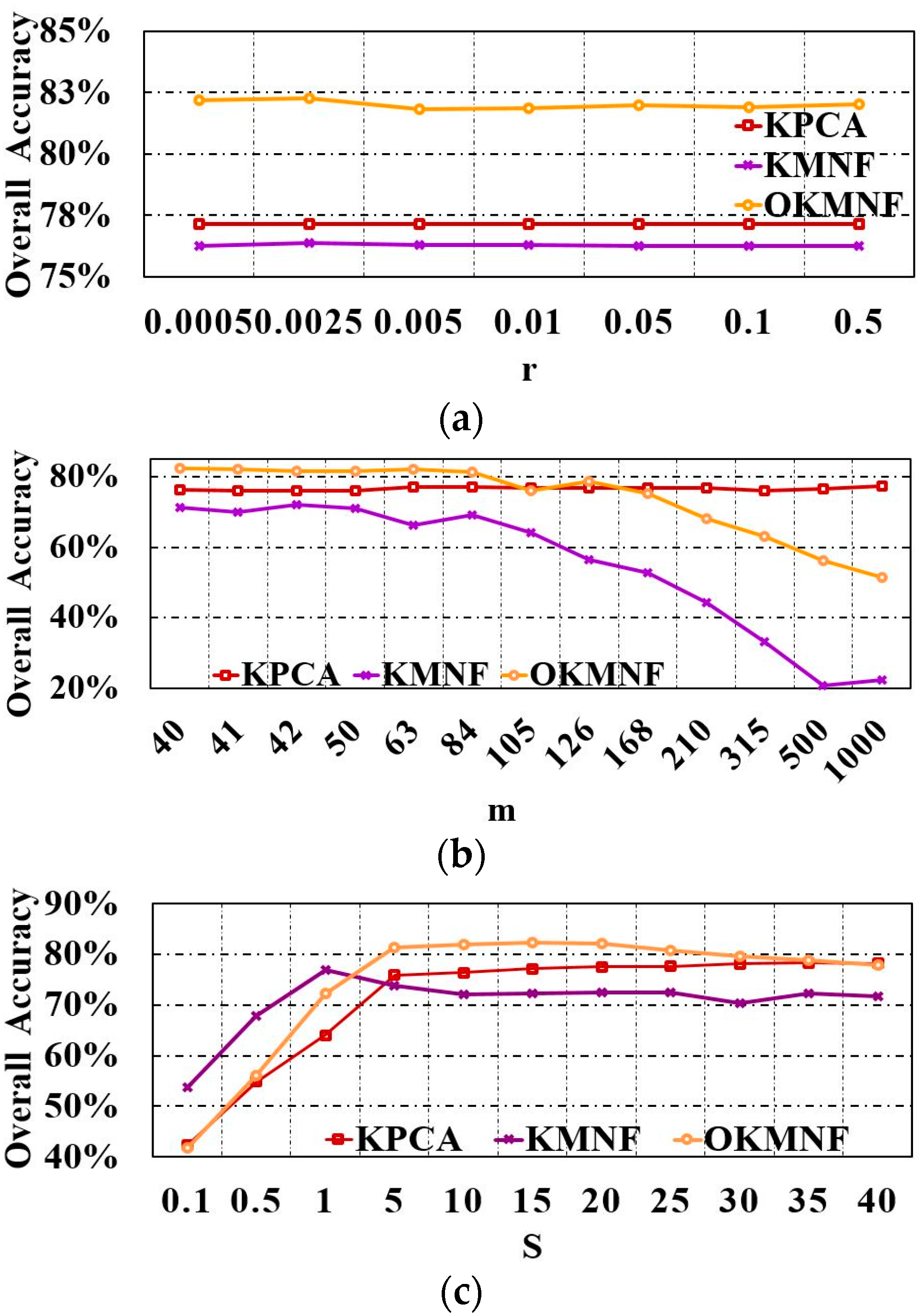
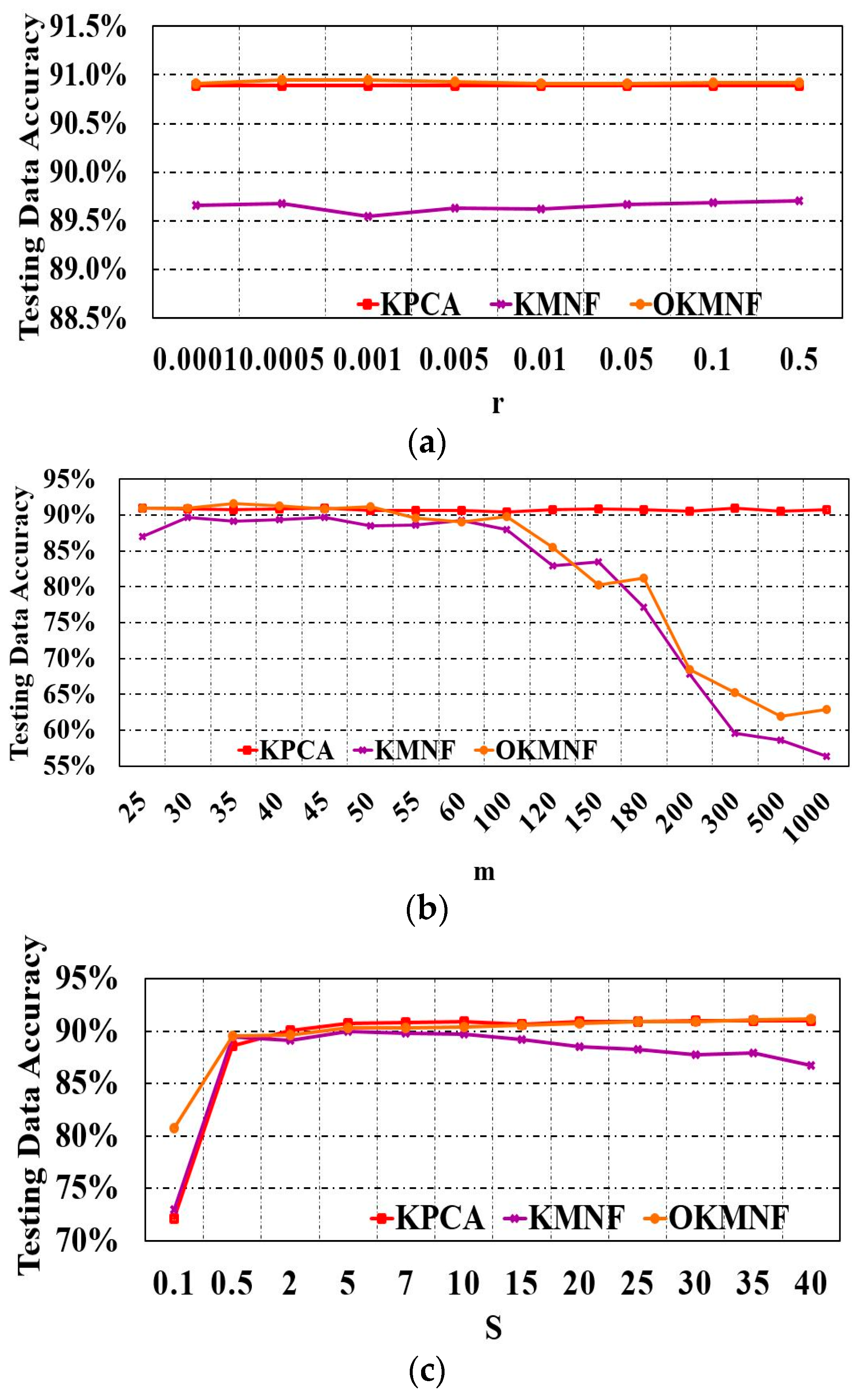
3.1. Parameter Tuning
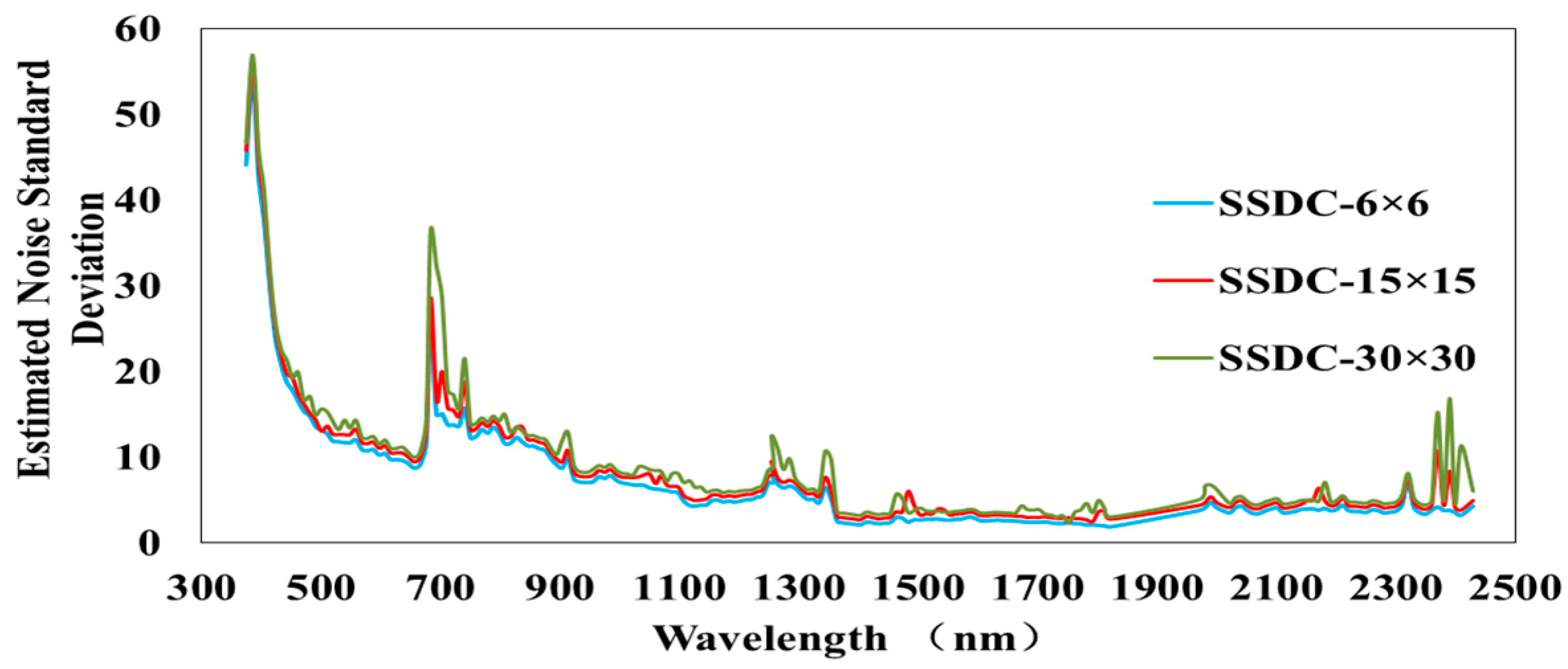
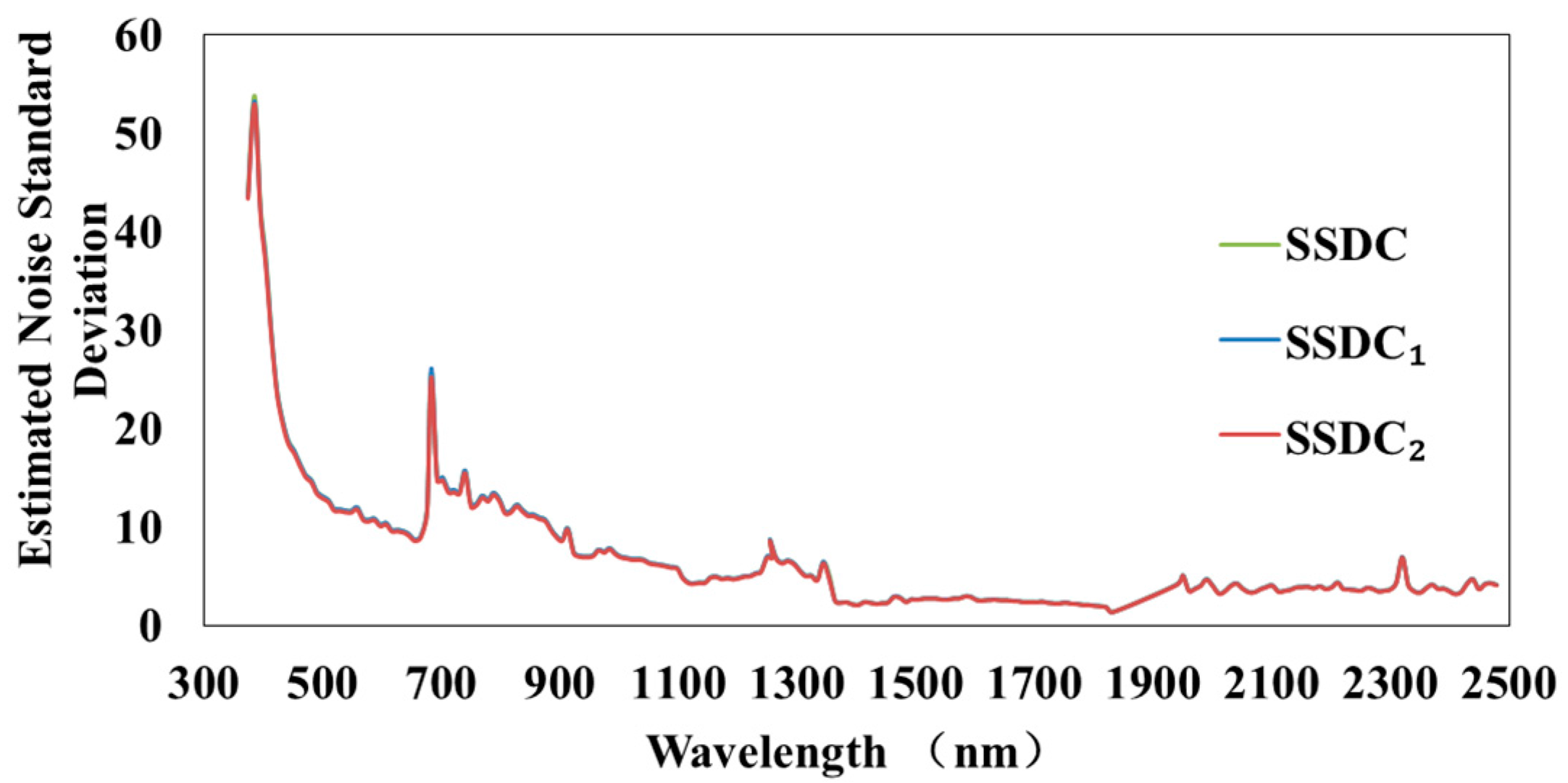
3.2. Experiments on Noise Estimation Algorithms in KMNF and OKMNF
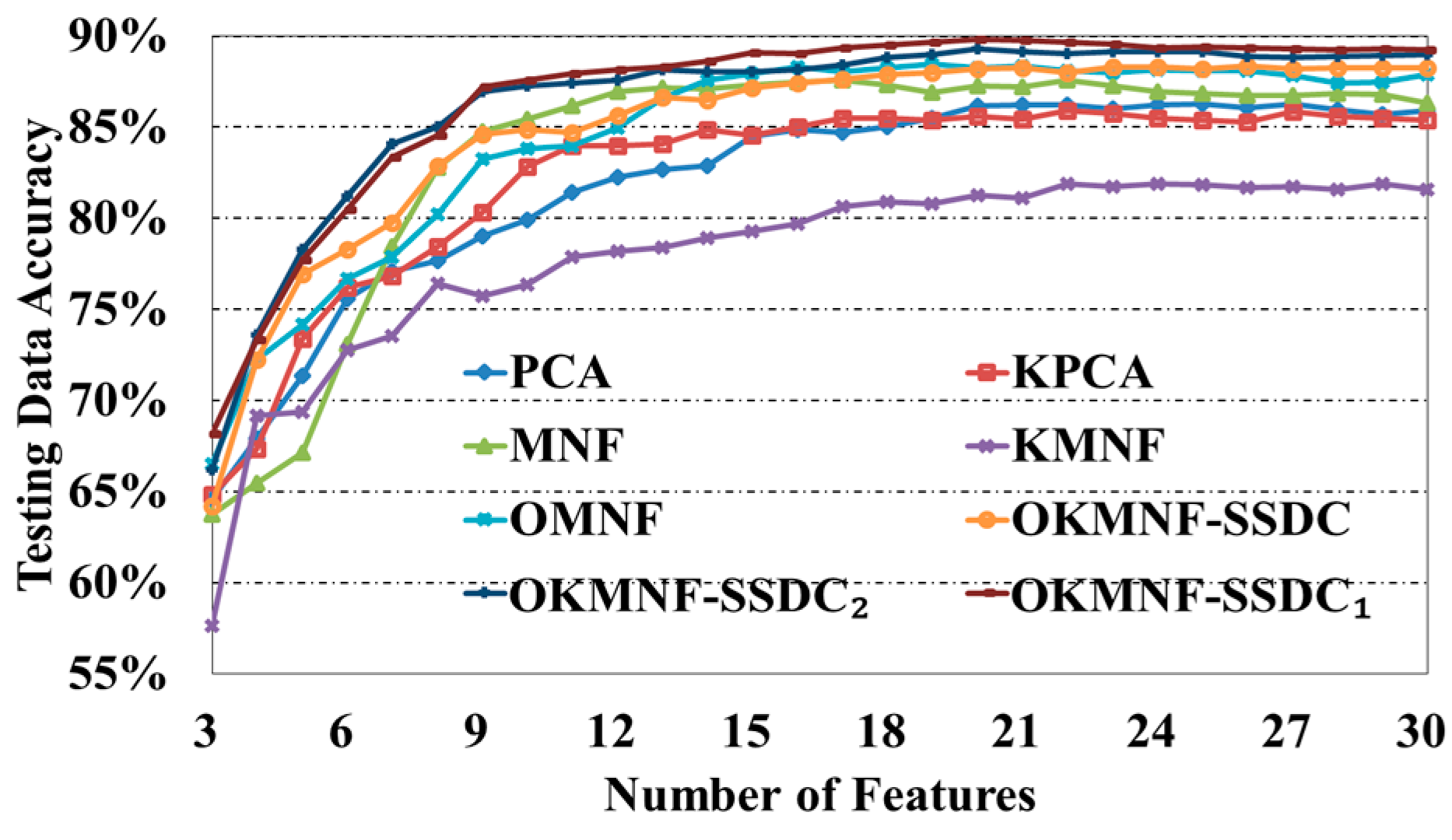
3.3. Experiments on Dimensionality Reduction Methods
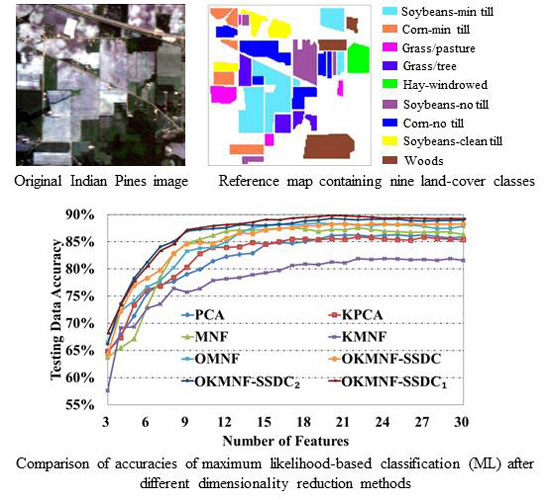
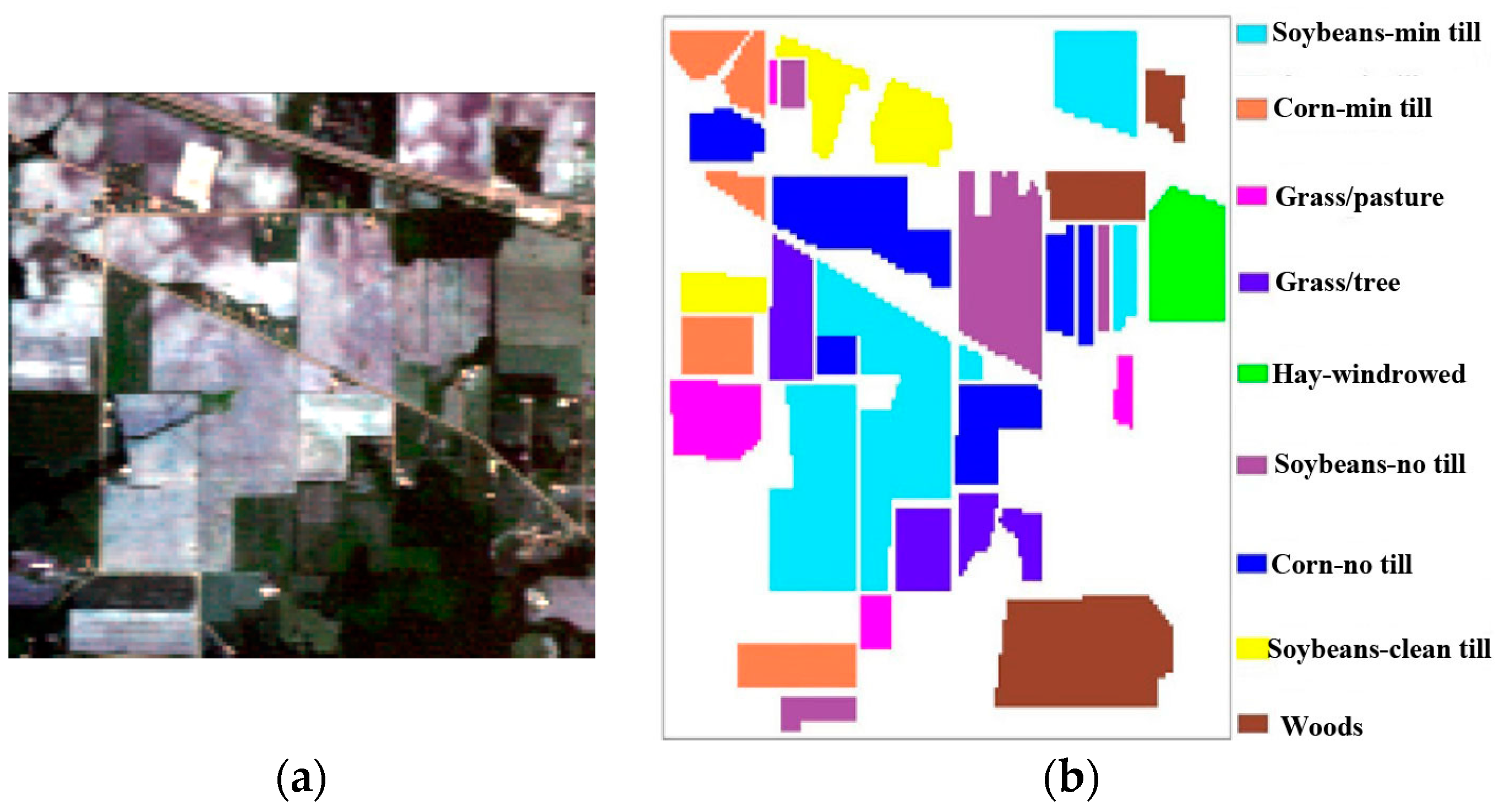
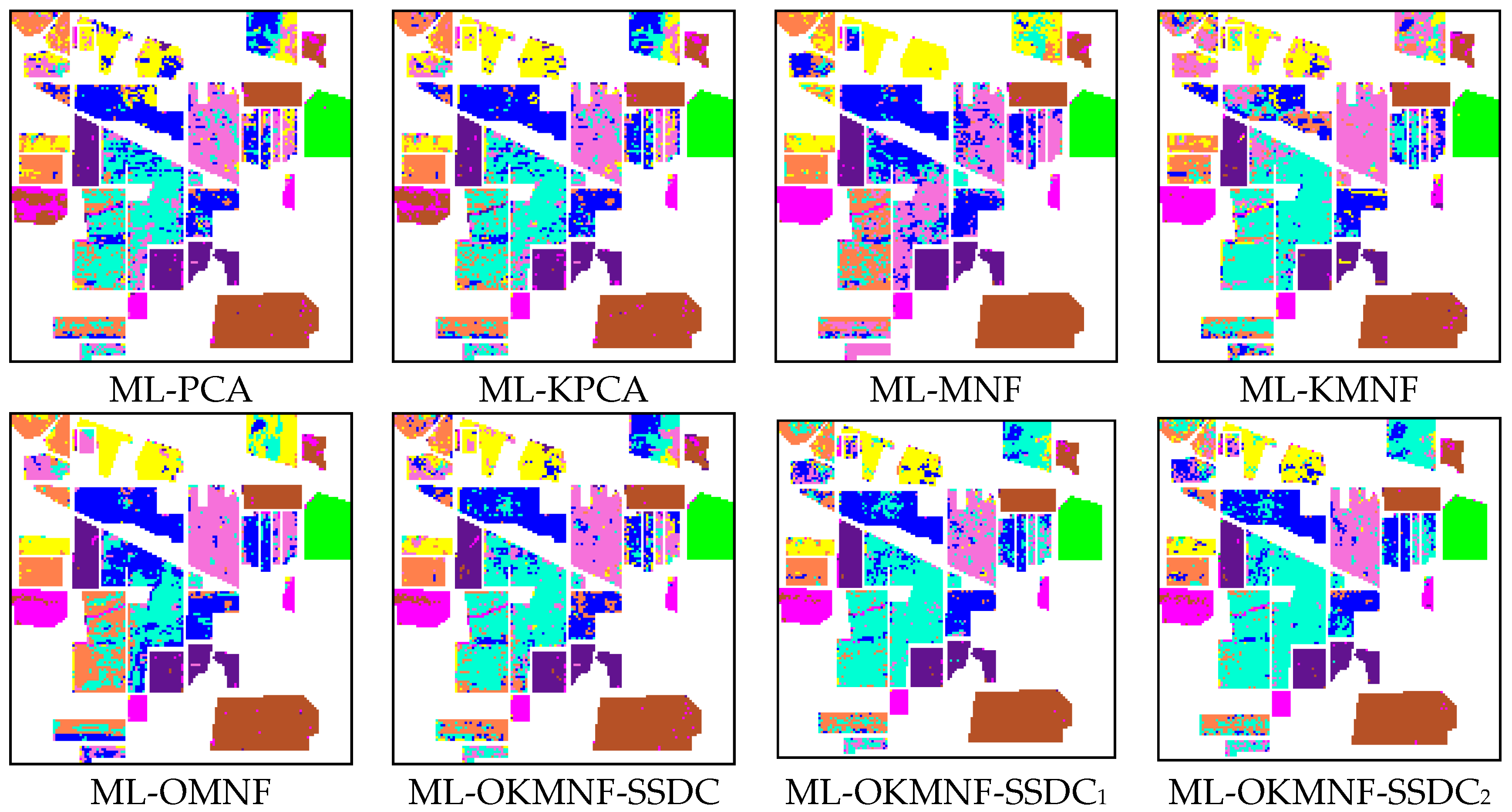
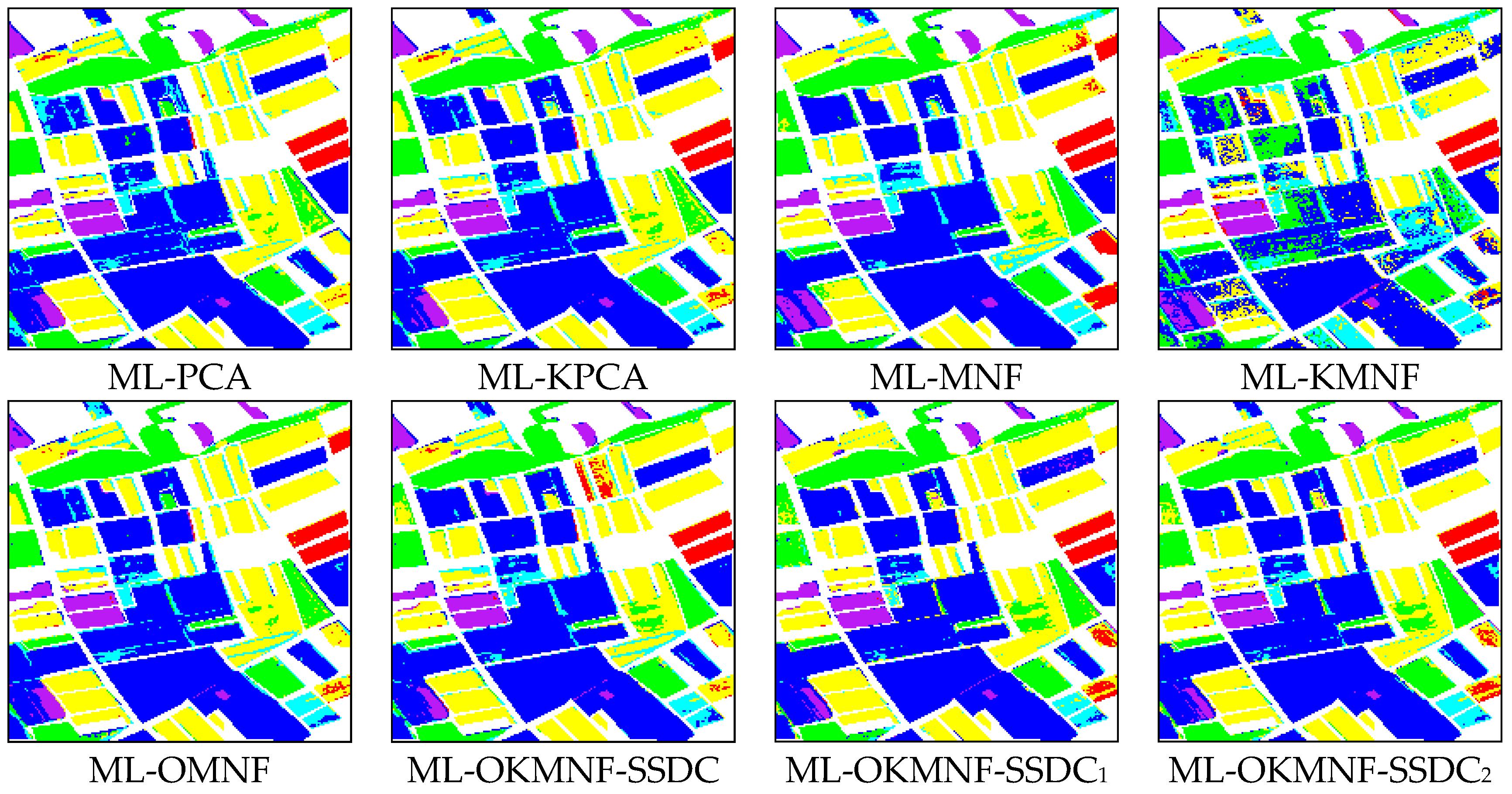
3.3.1. Experiments on the Indian Pines Image
3.3.2. Experiments on the Minamimaki Scene
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Goetz, A.F.H. Three decades of hyperspectral remote sensing of the Earth: A personal view. Remote Sens. Environ. 2009, 113, S5–S16. [Google Scholar] [CrossRef]
- Plaza, A.; Benediktsson, J.A.; Boardman, J.W.; Brazile, J.; Bruzzone, L.; Camps-Valls, G.; Chanussot, J.; Fauvel, M.; Gamba, P.; Gualtieri, A.; et al. Recent advances in techniques for hyperspectral image processing. Remote Sens. Environ. 2009, 113, S110–S122. [Google Scholar] [CrossRef]
- Hughes, G. On the mean accuracy of statistical pattern recognizers. IEEE Trans. Inf. Theory 1968, 14, 55–63. [Google Scholar] [CrossRef]
- Liu, C.H.; Zhou, J.; Liang, J.; Qian, Y.T.; Li, H.X.; Gao, Y.S. Exploring structural consistency in graph regularized joint spectral-spatial sparse coding for hyperspectral image classification. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2016, 99, 1–14. [Google Scholar] [CrossRef]
- Jia, X.P.; Kuo, B.; Crawford, M.M. Feature mining for hyperspectral image classification. Proc. IEEE 2013, 101, 676–697. [Google Scholar]
- Benediktsson, J.A.; Palmason, J.A.; Sveinsson, J.R. Classification of hyperspectral data from urban areas based on extended morphological profiles. IEEE Trans. Geosci. Remote Sens. 2005, 43, 480–491. [Google Scholar] [CrossRef]
- Qian, Y.T.; Yao, F.T.; Jia, S. Band selection for hyperspectral imagery using affinity propagation. IET Comput. Vis. 2010, 3, 213–222. [Google Scholar] [CrossRef]
- Falco, N.; Benediktsson, J.A.; Bruzzone, L. A study on the effectiveness of different independent component analysis algorithms for hyperspectral image classification. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 2183–2199. [Google Scholar] [CrossRef]
- Zabalza, J.; Ren, J.C.; Wang, Z.; Zhao, H.M.; Marshall, S. Fast implementation of singular spectrum analysis for effective feature extraction in hyperspectral imaging. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 2845–2853. [Google Scholar] [CrossRef]
- Xie, J.Y.; Hone, K.; Xie, W.X.; Gao, X.B.; Shi, Y.; Liu, X.H. Extending twin support vector machine classifier for multi-category classification problems. Intell. Data Anal. 2013, 17, 649–664. [Google Scholar]
- Chen, W.S.; Huang, J.; Zou, J.; Fang, B. Wavelet-face based subspace LDA method to solve small sample size problem in face recognition. Int. J. Wavelets Multiresolut. Inf. Process. 2009, 7, 199–214. [Google Scholar] [CrossRef]
- Gu, Y.F.; Feng, K. Optimized laplacian SVM with distance metric learning for hyperspectral image classification. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2013, 6, 1109–1117. [Google Scholar] [CrossRef]
- Ma, A.L.; Zhong, Y.F.; Zhao, B.; Jiao, H.Z.; Zhang, L.P. Semisupervised subspace-based DNA encoding and matching classifier for hyperspectral remote sensing imagery. IEEE Trans. Geosci. Remote Sens. 2016, 54, 4402–4418. [Google Scholar] [CrossRef]
- Zhang, L.F.; Zhang, L.P.; Tao, D.C.; Huang, X. On combining multiple features for hyperspectral remote sensing image classification. IEEE Trans. Geosci. Remote Sens. 2012, 50, 879–893. [Google Scholar] [CrossRef]
- Melgani, F.; Bruzzone, L. Classification of hyperspectral remote sensing images with support vector machines. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1778–1790. [Google Scholar] [CrossRef]
- Harsanyi, J.; Chang, C. Hyperspectral image classification and dimensionality reduction: An orthogonal subspace projection approach. IEEE Trans. Geosci. Remote Sens. 1994, 32, 779–785. [Google Scholar] [CrossRef]
- Wang, Q.; Lin, J.; Yuan, Y. Salient band selection for hyperspectral image classification via manifold ranking. IEEE Trans. Neural Netw. Learn. Syst. 2016, 27, 1279–1289. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Lin, J.; Wang, Q. Dual clustering based hyperspectral band selection by contextual analysis. IEEE Trans. Geosci. Remote Sens. 2016, 54, 1431–1445. [Google Scholar] [CrossRef]
- Yuan, Y.; Lin, J.; Wang, Q. Hyperspectral image classification via multi-task joint sparse representation and stepwise MRF optimization. IEEE Trans. Cybern. 2016, 46, 2966–2977. [Google Scholar]
- Fukunaga, K. Introduction to Statistical Pattern Recognition; Academic Press: Cambridge, MA, USA, 1990. [Google Scholar]
- Kuo, B.C.; Landgrebe, D.A. Nonparametric weighted feature extractionfor classification. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1096–1105. [Google Scholar]
- Ly, N.; Du, Q.; Fowler, J.E. Sparse graph-based discriminant analysis for hyperspectral imagery. IEEE Trans. Geosci. Remote Sens. 2014, 52, 3872–3884. [Google Scholar]
- Ly, N.; Du, Q.; Fowler, J.E. Collaborative graph-based discriminant analysis for hyperspectral imagery. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 2688–2696. [Google Scholar] [CrossRef]
- Xue, Z.H.; Du, P.J.; Li, J.; Su, H.J. Simultaneous sparse graph embedding for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2015, 53, 6114–6133. [Google Scholar] [CrossRef]
- Roger, E.R. Principal components transform with simple, automatic noise adjustment. Int. J. Remote Sens. 1996, 17, 2719–2727. [Google Scholar] [CrossRef]
- Green, A.A.; Berman, M.; Switzer, P.; Craig, M.D. A transformation for ordering multispectral data in terms of image quality with implications for noise removal. IEEE Trans. Geosci. Remote Sens. 1998, 26, 65–74. [Google Scholar] [CrossRef]
- Chen, P.H.; Jiao, L.C.; Liu, F.; Gou, S.P.; Zhao, J.Q.; Zhao, Z.Q. Dimensionality reduction of hyperspectral imagery using sparse graph learning. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2016, 99, 1–17. [Google Scholar] [CrossRef]
- Lee, J.B.; Woodyatt, S.; Berman, M. Enhancement of high spectral resolution remote-sensing data by a noise-adjusted principal components transform. IEEE Trans. Geosci. Remote Sens. 1990, 28, 295–304. [Google Scholar] [CrossRef]
- Bachmann, C.M.; Ainsworth, T.L.; Fusina, R.A. Exploiting manifold geometry in hyperspectral imagery. IEEE Trans. Geosci. Remote Sens. 2005, 43, 441–454. [Google Scholar]
- Mohan, A.; Sapiro, G.; Bosch, E. Spatially coherent nonlinear dimensionality reduction and segmentation of hyperspectral images. IEEE Geosci. Remote Sens. Lett. 2007, 4, 206–210. [Google Scholar] [CrossRef]
- Nielsen, A.A. Kernel maximum autocorrelation factor and minimum noise fraction transformations. IEEE Trans. Image Process. 2011, 20, 612–624. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Chova, L.; Nielsen, A.A.; Camps-Valls, G. Explicit signal to noise ratio in reproducing kernel hilbert spaces. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Vancouver, BC, Canada, 24–29 July 2011; pp. 3570–3573. [Google Scholar]
- Nielsen, A.A.; Vestergaard, J.S. Parameter optimization in the regularized kernel minimum noise fraction transformation. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Munich, Germany, 22–27 July 2012; pp. 370–373. [Google Scholar]
- Shawe-Taylor, J.; Cristianini, N. Kernel Methods for Pattern Analysis; Cambridge University Press: New York, NY, USA, 2004. [Google Scholar]
- Li, W.; Prasad, S.; Fowler, J.E. Decision fusion in kernel-induced spaces for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2014, 52, 3399–3411. [Google Scholar] [CrossRef]
- Li, W.; Prasad, S.; Fowler, J.E.; Bruce, L.M. Locality preserving dimensionality reduction and classification for hyperspectral image analysis. IEEE Trans. Geosci. Remote Sens. 2012, 50, 1185–1198. [Google Scholar] [CrossRef]
- Li, W.; Prasad, S.; Fowler, J.E.; Bruce, L.M. Locality-preserving discriminant analysis in kernel-induced feature spaces for hyperspectral image classification. IEEE Geosci. Remote Sens. Lett. 2011, 8, 894–898. [Google Scholar] [CrossRef]
- Zhang, Y.H.; Prasad, S. Locality preserving composite kernel feature extraction for multi-source geospatial image analysis. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 1385–1392. [Google Scholar] [CrossRef]
- Kuo, B.C.; Ho, H.H.; Li, C.H.; Huang, C.C.; Taur, J.S. A kernel-based feature selection method for SVM with RBF kernel for hyperspectral image classification. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 317–326. [Google Scholar]
- Schölkopf, B.; Smola, A.; Müller, K.-R. Nonlinear component analysis as a kernel eigenvalue problem. Neural Comput. 1998, 10, 1299–1319. [Google Scholar] [CrossRef]
- Gao, L.R.; Zhang, B.; Chen, Z.C.; Lei, L.P. Study on the issue of noise estimation in dimension reduction of hyperspectral images. In Proceedings of the IEEE Workshop on Hyperspectral Image and Signal Processing: Evolution in Remote Sensing (WHISPERS), Lisbon, Portugal, 6–9 June 2011; pp. 1–4. [Google Scholar]
- Zhao, B.; Gao, L.R.; Zhang, B. An optimized method of kernel minimum noise fraction for dimensionality reduction of hyperspectral imagery. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Beijing, China, 10–15 July 2016; pp. 48–51. [Google Scholar]
- Zhao, B.; Gao, L.R.; Liao, W.Z.; Zhang, B. A new kernel method for hyperspectral image feature extraction. Geo-Spat. Inf. Sci. 2017, 99, 1–11. [Google Scholar]
- Bioucas-Dias, J.M.; Plaza, A.; Dobigeon, N.; Parente, M.; Du, Q.; Gader, P.; Chanussot, J. Hyperspectral unmixing overview: Geometrical, statistical, and sparse regression-based approaches. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2012, 5, 354–379. [Google Scholar] [CrossRef]
- Jia, S.; Xie, Y.; Tang, G.H.; Zhu, J.S. Spatial-spectral-combined sparse representation-based classification for hyperspectral imagery. Soft Comput. 2014, 1–10. [Google Scholar] [CrossRef]
- Chen, C.; Li, W.; Tramel, E.W.; Cui, M.S.; Prasad, S.; Fowler, J.E. Spectral–spatial preprocessing using multihypothesis prediction for noise-robust hyperspectral image classification. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 1047–1059. [Google Scholar] [CrossRef]
- Ghamisi, P.; Benediktsson, J.A.; Jon, A.; Cavallaro, G.; Plaza, A. automatic framework for spectral–spatial classification based on supervised feature extraction and morphological attribute profiles. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 2147–2160. [Google Scholar] [CrossRef]
- Gao, L.R.; Du, Q.; Yang, W.; Zhang, B. A comparative study on noise estimation for hyperspectral imagery. In Proceedings of the IEEE Workshop on Hyperspectral Image and Signal Processing: Evolution in Remote Sensing (WHISPERS), Shanghai, China, 4–7 June2012; pp. 1–4. [Google Scholar]
- Roger, E.R.; Arnold, F.J. Reliably estimating the noise in AVIRIS hyperspectral images. Int. J. Remote Sens. 1996, 17, 1951–1962. [Google Scholar] [CrossRef]
- Gao, L.R.; Du, Q.; Zhang, B.; Yang, W.; Wu, Y.F. A comparative study on linear regression-based noise estimation for hyperspectral imagery. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2013, 6, 488–498. [Google Scholar] [CrossRef]
- Gao, L.R.; Zhang, B.; Sun, X.; Li, S.S.; Du, Q.; Wu, C.S. Optimized maximum noise fraction for dimensionality reduction of Chinese HJ-1A hyperspectral data. EURASIP J. Adv. Signal Process. 2013, 1, 1–12. [Google Scholar] [CrossRef]
- Gao, L.R.; Zhang, B.; Zhang, X.; Zhang, W.J.; Tong, Q.X. A new operational method for estimating noise in hyperspectral images. IEEE Geosci. Remote Sens. Lett. 2008, 5, 83–87. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, B.; Gao, L.R.; Chen, D.M. A maximum noise fraction transform with improved noise estimation for hyperspectral images. Sci. China Ser. F Inf. Sci. 2009, 52, 1578–1587. [Google Scholar] [CrossRef]
- Landgrebe, D.A.; Malaret, E. Noise in remote-sensing systems: The effect on classification error. IEEE Trans. Geosci. Remote Sens. 1986, 24, 294–299. [Google Scholar] [CrossRef]
- Corner, B.R.; Narayanan, R.M.; Reichenbach, S.E. Noise estimation in remote sensing imagery using data masking. Int. J. Remote Sens. 2003, 24, 689–702. [Google Scholar] [CrossRef]
- Gao, B.-C. An operational method for estimating signal to noise ratios from data acquired with imaging spectrometers. Remote Sens. Environ. 1993, 43, 23–33. [Google Scholar] [CrossRef]
- Documentation for Minimum Noise Fraction Transformations. Available online: http://people.compute.dtu.dk/alan/software.html (accessed on 31 March 2017).
- Wu, Y.F.; Gao, L.R.; Zhang, B.; Zhao, H.N.; Li, J. Real-time implementation of optimized maximum noise fraction transform for feature extraction of hyperspectral images. J. Appl. Remote Sens. 2014, 8, 1–16. [Google Scholar] [CrossRef]
- Camps-Valls, G.; Bruzzone, L. Kernel-based methods for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2005, 43, 1351–1362. [Google Scholar] [CrossRef]
- Acito, N.; Diani, M.; Corsini, G. Signal-dependent noise modeling andmodel parameter estimation in hyperspectral images. IEEE Trans. Geosci. Remote Sens. 2011, 49, 2957–2971. [Google Scholar] [CrossRef]
- Zhang, X.J.; Xu, C.; Li, M.; Sun, X.L. Sparse and low-rank coupling image segmentation model via nonconvex regularization. Int. J. Pattern Recognit. Artif. Intell. 2015, 29, 1–22. [Google Scholar] [CrossRef]
- Zhu, Z.X.; Jia, S.; He, S.; Sun, Y.W.; Ji, Z.; Shen, L.L. Three-dimensional Gabor feature extraction for hyperspectral imagery classification using a memetic framework. Inf. Sci. 2015, 298, 274–287. [Google Scholar] [CrossRef]
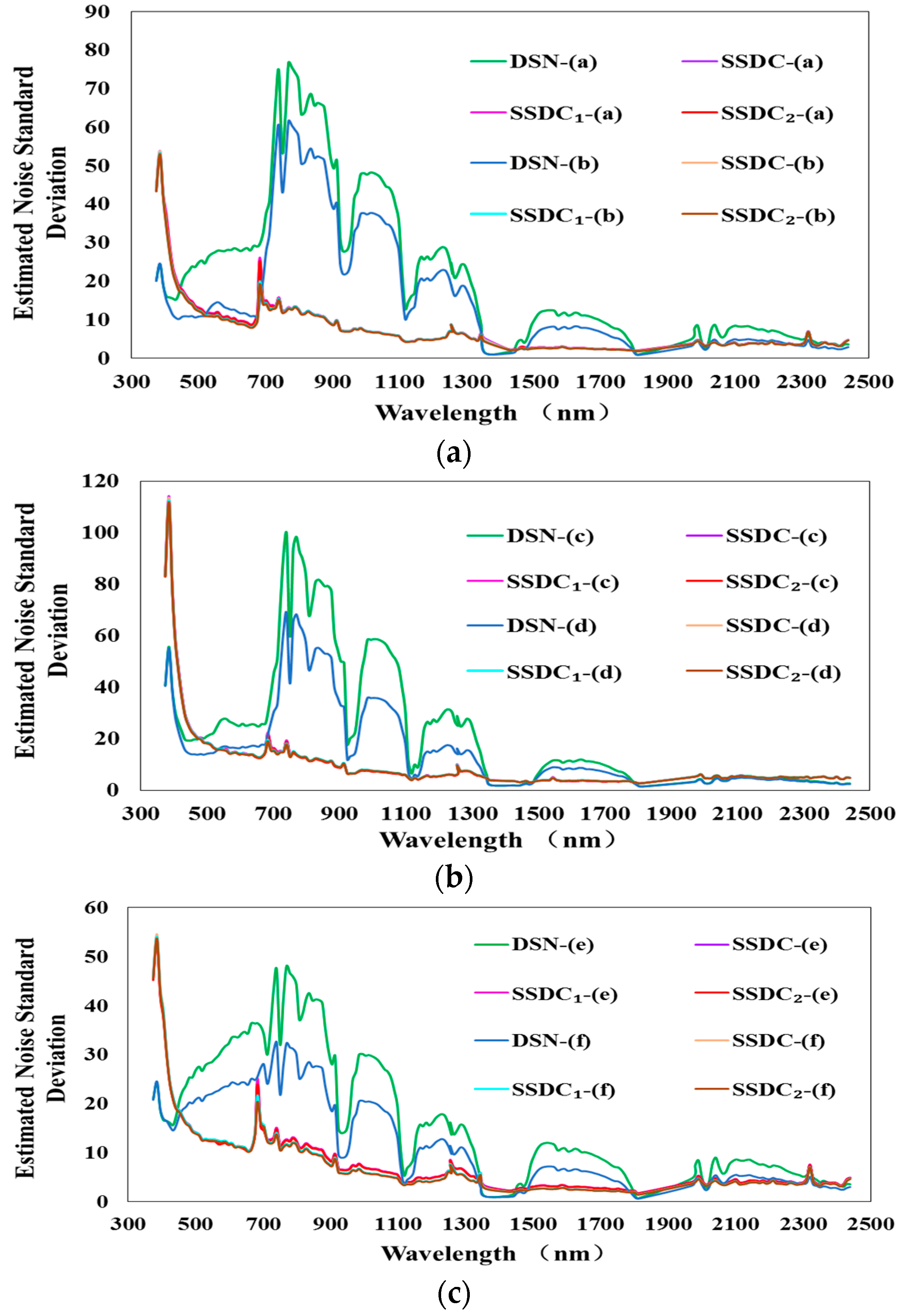



| Spatial Resolution | Acquired Site | Acquired Time | Image Description | |
|---|---|---|---|---|
| (a) | 20 m | Jasper Ridge | 3 April 1997 | Dominated by a heterogeneous city area |
| (b) | Dominated by a homogeneous vegetation area | |||
| (c) | 3.4 m | Low Altitude | 5 July 1996 | Dominated by a heterogeneous city area |
| (d) | Homogeneous farmland | |||
| (e) | 20 m | Moffett Field | 20 June 1997 | A mix of a heterogeneous city area and a homogeneous bare soil |
| (f) | Dominated by a homogeneous water |
| Classes | Training | Testing |
|---|---|---|
| Corn-no till | 359 | 1075 |
| Corn-min till | 209 | 625 |
| Grass/Pasture | 124 | 373 |
| Grass/Trees | 187 | 560 |
| Hay-windrowed | 122 | 367 |
| Soybean-no till | 242 | 726 |
| Soybean-min till | 617 | 1851 |
| Soybean-clean till | 154 | 460 |
| Woods | 324 | 970 |
| Total | 2338 | 7007 |
| Number of Features | PCA | KPCA | MNF | KMNF | OMNF | OKMNF-SSDC | OKMNF-SSDC1 | OKMNF-SSDC2 |
|---|---|---|---|---|---|---|---|---|
| 3 | 64.57% | 64.84% | 63.75% | 57.63% | 66.50% | 64.21% | 68.19% | 66.19% |
| 4 | 67.90% | 67.33% | 65.44% | 69.14% | 72.25% | 72.23% | 73.34% | 73.60% |
| 5 | 71.35% | 73.41% | 67.14% | 69.36% | 74.15% | 76.93% | 77.74% | 78.29% |
| 6 | 75.60% | 76.23% | 73.09% | 72.76% | 76.69% | 78.31% | 80.48% | 81.22% |
| 7 | 77.08% | 76.81% | 78.43% | 73.53% | 77.88% | 79.73% | 83.35% | 84.07% |
| 8 | 77.65% | 78.45% | 82.76% | 76.39% | 80.21% | 82.86% | 84.56% | 85.03% |
| 9 | 79.01% | 80.32% | 84.74% | 75.71% | 83.27% | 84.59% | 87.21% | 86.93% |
| 10 | 79.92% | 82.82% | 85.43% | 76.35% | 83.84% | 84.87% | 87.56% | 87.26% |
| 11 | 81.40% | 83.96% | 86.16% | 77.88% | 83.97% | 84.69% | 87.94% | 87.44% |
| 12 | 82.27% | 83.96% | 86.93% | 78.18% | 84.96% | 85.66% | 88.17% | 87.60% |
| 13 | 82.67% | 84.10% | 87.15% | 78.42% | 86.61% | 86.63% | 88.33% | 88.13% |
| 14 | 82.90% | 84.84% | 87.08% | 78.94% | 87.57% | 86.50% | 88.63% | 88.04% |
| 15 | 84.49% | 84.54% | 87.33% | 79.31% | 87.95% | 87.17% | 89.10% | 88.04% |
| 16 | 84.87% | 85.03% | 87.48% | 79.72% | 88.30% | 87.43% | 89.04% | 88.17% |
| 17 | 84.72% | 85.50% | 87.55% | 80.66% | 88.05% | 87.64% | 89.37% | 88.41% |
| 18 | 85.02% | 85.50% | 87.34% | 80.89% | 88.28% | 87.91% | 89.51% | 88.84% |
| 19 | 85.50% | 85.37% | 86.91% | 80.78% | 88.47% | 87.98% | 89.68% | 89.00% |
| 20 | 86.16% | 85.59% | 87.27% | 81.25% | 88.25% | 88.20% | 89.82% | 89.30% |
| 21 | 86.21% | 85.41% | 87.21% | 81.13% | 88.37% | 88.24% | 89.77% | 89.14% |
| 22 | 86.23% | 85.89% | 87.57% | 81.88% | 88.10% | 88.01% | 89.64% | 89.03% |
| 23 | 86.00% | 85.76% | 87.28% | 81.76% | 88.00% | 88.30% | 89.55% | 89.15% |
| 24 | 86.24% | 85.49% | 86.97% | 81.88% | 88.17% | 88.28% | 89.35% | 89.14% |
| 25 | 86.27% | 85.40% | 86.87% | 81.82% | 88.08% | 88.23% | 89.42% | 89.14% |
| 26 | 86.06% | 85.30% | 86.74% | 81.66% | 88.11% | 88.34% | 89.34% | 88.89% |
| 27 | 86.27% | 85.84% | 86.76% | 81.72% | 87.85% | 88.20% | 89.28% | 88.82% |
| 28 | 85.96% | 85.59% | 86.84% | 81.60% | 87.43% | 88.27% | 89.27% | 88.88% |
| 29 | 85.71% | 85.50% | 86.80% | 81.92% | 87.50% | 88.25% | 89.28% | 88.91% |
| 30 | 85.89% | 85.39% | 86.31% | 81.60% | 87.87% | 88.24% | 89.23% | 88.97% |
| Classes | Training | Testing |
|---|---|---|
| Bare soil | 1238 | 11,150 |
| plastic | 33 | 300 |
| Chinese cabbage | 29 | 245 |
| forest | 111 | 1000 |
| Japanese cabbage | 425 | 3830 |
| pasture | 20 | 153 |
| Total | 1856 | 16,678 |
| Method | Number of Features | |||||
|---|---|---|---|---|---|---|
| 3 | 4 | 5 | 6 | 7 | 8 | |
| PCA | 85.14% | 89.13% | 89.86% | 90.17% | 90.44% | 90.75% |
| KPCA | 87.43% | 88.41% | 89.46% | 90.19% | 90.22% | 90.87% |
| MNF | 86.34% | 88.73% | 89.48% | 89.69% | 90.32% | 90.59% |
| KMNF | 68.30% | 83.81% | 86.02% | 87.69% | 88.61% | 89.66% |
| OMNF | 87.82% | 88.51% | 89.32% | 90.10% | 89.88% | 90.51% |
| OKMNF-SSDC | 88.10% | 88.94% | 89.98% | 90.60% | 90.63% | 90.97% |
| OKMNF-SSDC1 | 89.46% | 90.18% | 90.44% | 91.19% | 91.39% | 91.68% |
| OKMNF-SSDC2 | 89.24% | 90.17% | 90.78% | 91.56% | 91.88% | 91.89% |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, L.; Zhao, B.; Jia, X.; Liao, W.; Zhang, B. Optimized Kernel Minimum Noise Fraction Transformation for Hyperspectral Image Classification. Remote Sens. 2017, 9, 548. https://doi.org/10.3390/rs9060548
Gao L, Zhao B, Jia X, Liao W, Zhang B. Optimized Kernel Minimum Noise Fraction Transformation for Hyperspectral Image Classification. Remote Sensing. 2017; 9(6):548. https://doi.org/10.3390/rs9060548
Chicago/Turabian StyleGao, Lianru, Bin Zhao, Xiuping Jia, Wenzhi Liao, and Bing Zhang. 2017. "Optimized Kernel Minimum Noise Fraction Transformation for Hyperspectral Image Classification" Remote Sensing 9, no. 6: 548. https://doi.org/10.3390/rs9060548