Mapping Spartina alterniflora Biomass Using LiDAR and Hyperspectral Data
Abstract
:1. Introduction
2. Materials and Methods
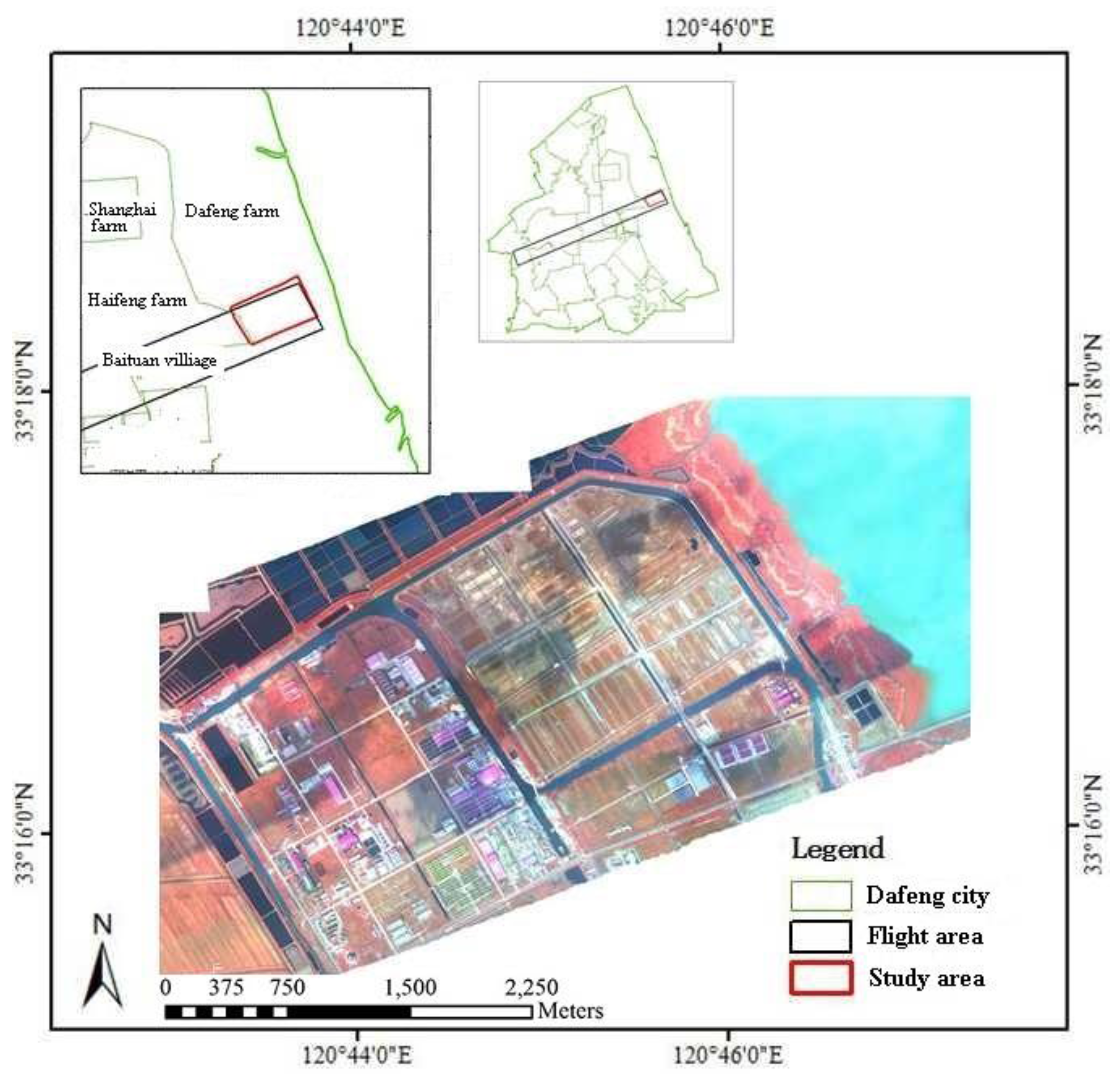
2.1. Study Area
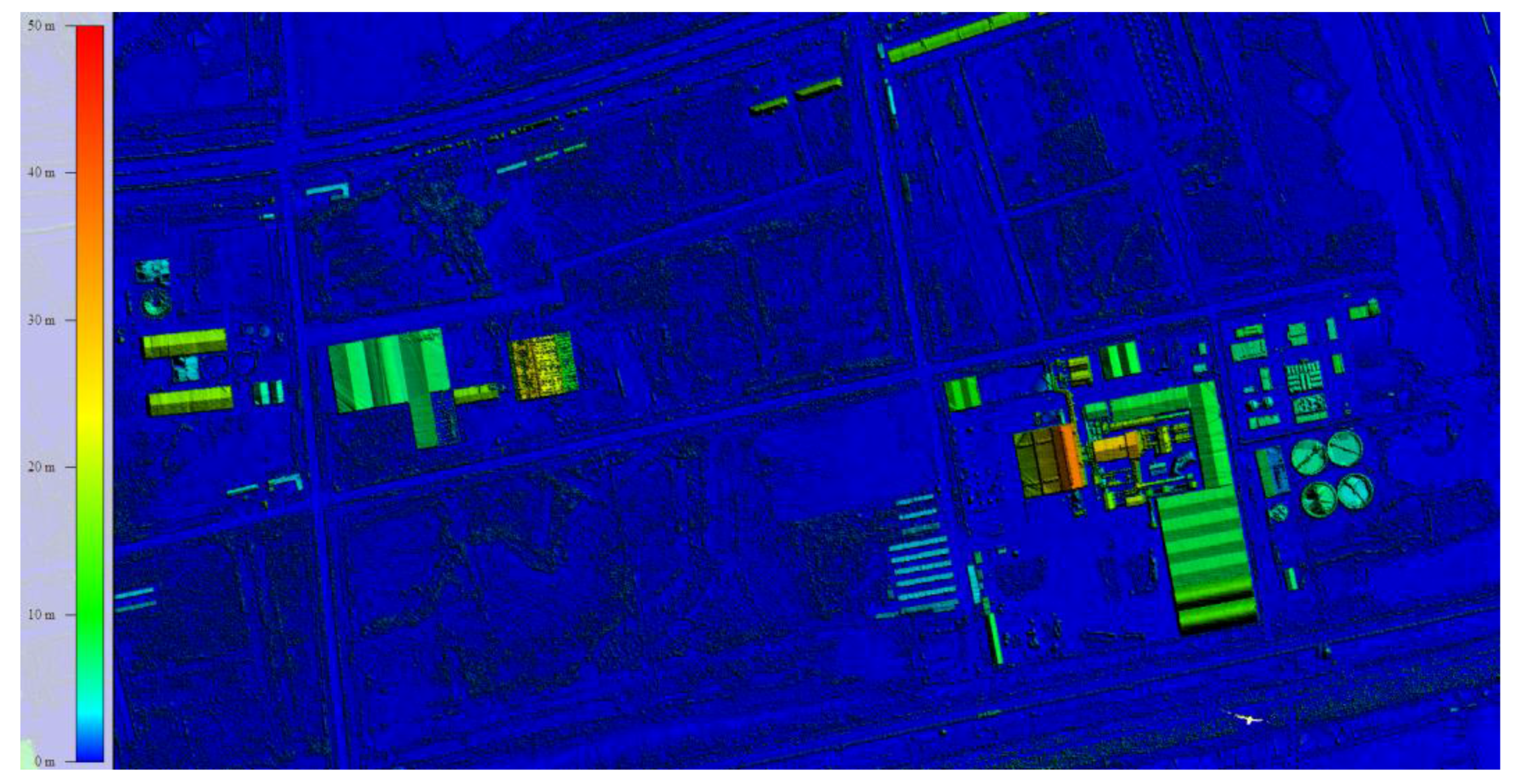
2.2. Data Collection
2.3. Collection of Vegetation Samples and Spectra
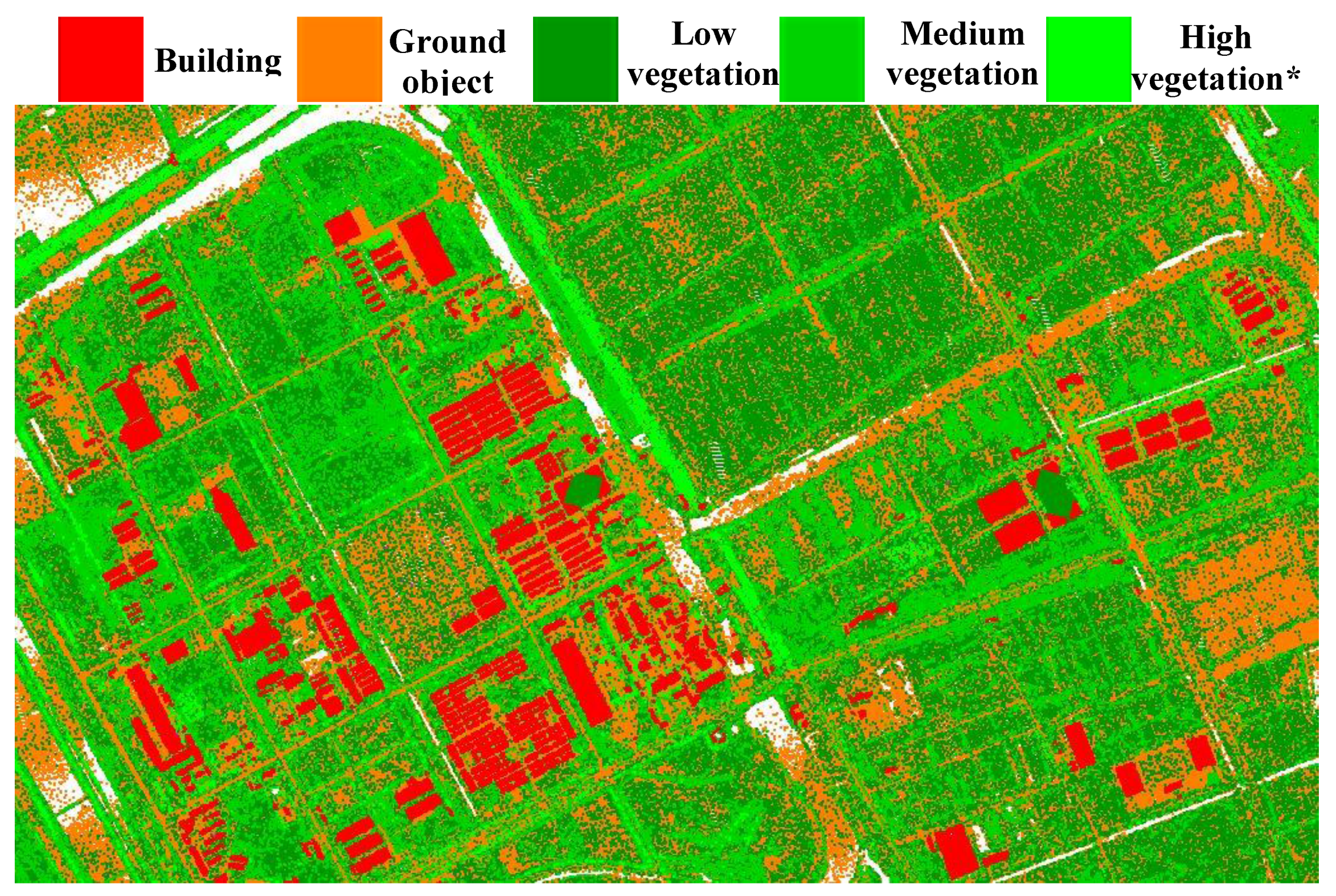
2.4. Data Pre-Processing
2.5. Biomass Estimation Model
3. Results and Discussion
3.1. Biomass Estimation Model of Wetland Vegetation
3.2. Biomass Estimation Model of Spartian Alterniflora
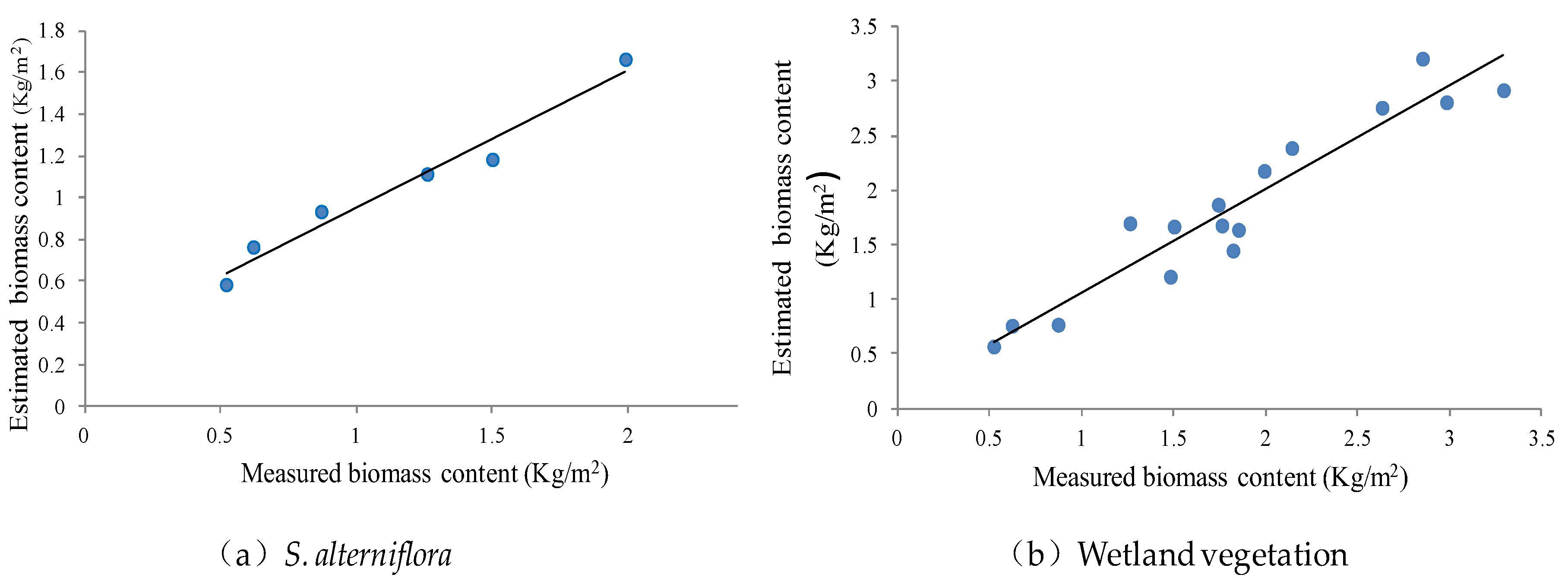
3.3. Model Accuracy Verification
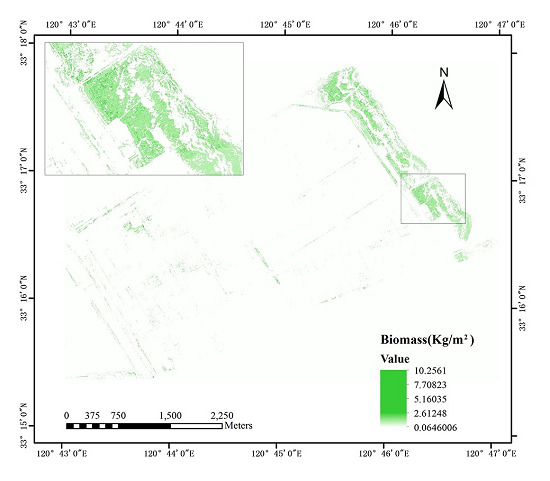
3.4. Mapping the S. alternniflora Biomass Distribution
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Shen, Y.M.; Yang, J.S.; Zeng, H.; Zhou, Q. Review of studies on alien species Spartina alterniflora in China. Mar. Environ. Sci. 2008, 27, 391–396. [Google Scholar]
- Brusati, E.D.; Grosholza, E.D. Effect of native and invasive cordgrass on Macoma petalum density, growth, and isotopic signatures. Estuar. Coast. Shelf Sci. 2007, 71, 517–522. [Google Scholar] [CrossRef]
- Rosso, P.H.; Ustin, S.L.; Hastings, A. Use of lidar to study changes associated with Spartina invasion in San Francisco Bay marshes. Remote Sens. Environ. 2006, 100, 295–306. [Google Scholar] [CrossRef]
- Zhu, D.; Gao, S. The expansion of Spartina alterniflora marsh in response to tidal flat reclamation, central Jiangsu coast, eastern China. Geogr. Res. 2014, 33, 2382–2392. (In Chinese) [Google Scholar]
- Thomas, M.B.; Reid, A.M. Are exotic natural enemies an effective way of controlling invasive plants? Trends Ecol. Evol. 2007, 22, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.S.; Shen, Y.M.; Lu, L.Y.; Yan, S.G.; Wang, Y.H.; Li, J.L.; Zhang, Z.L. Formation of Spartina alterniflora salt marshes on the coast of Jiangsu Province, China. Ecol. Eng. 2004, 23, 95–105. (In Chinese) [Google Scholar] [CrossRef]
- Liu, C.Y.; Zhang, S.Q.; Jiang, H.X.; Wang, H. Spatiotemporal dynamics and landscape pattern of alien species Spartina alterniflora in Yancheng coastal wetlands of Jiangsu Province, China. Chin. J. Appl. Ecol. 2009, 20, 901–908. (In Chinese) [Google Scholar]
- Tong, Q.X.; Zheng, L.F.; Wang, J.N. Study on Imaging Spectrometer Remote Sensing Information for Wetland Vegetation. J. Remote Sens. 1997, 1, 50–57. (In Chinese) [Google Scholar]
- Fu, X.; Liu, G.H.; Huang, C.; Liu, Q.S. Remote sensing estimation models of Suaeda salsa biomass in the coastal wetland. Acta Ecol. Sin. 2012, 32, 5355–5362. [Google Scholar]
- Tang, X.G. Estimation of Forest Aboveground Biomass by Integrating ICESat/GLAS Waveform and TM Data. Doctoral Dissertation, University of Chinese Academy of Sciences, Beijing, China, 2013. [Google Scholar]
- Kasischke, E.S.; Bourgeau-Chavez, L.L. Monitoring South Florida wetlands using ERS-1 SAR imagery. Photogramm. Eng. Remote Sens. 1997, 63, 281–291. [Google Scholar]
- Temilola, E.F.; Amanda, H. Armstrong Remote Characterization of Biomass Measurements: Case Study of Mangrove Forests; INTECH Open Access Publisher: Rijeka, Croatia, 2010; p. 202. ISBN 978-953-307-113-8.2010. [Google Scholar]
- Rajab Pourrahmati, M.; Baghdadi, N.; Asghar Darvishsefat, A.; Namiranian, M.; Gond, V.; Moeinazad Tehrani, S.M.; Fayad, I.; Bailly, J.S. Capability of GLAS/ICESat data to estimate forest canopy height and volume in mountainous forests of Iran. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2016, 8, 5246–5261. [Google Scholar] [CrossRef]
- Fayad, I.; Baghdadi, N.; Guitet, S.; Bailly, J.S.; Hérault, B.; Gond, V.; El Hajj, M.; Ho Tong Minh, D. Aboveground biomass mapping in French Guiana by combining remote sensing, forest inventories and environmental data. Int. J Appl. Earth Obs. 2016, 52, 502–514. [Google Scholar] [CrossRef]
- Li, X.; Yeh, G.A.; Wang, S.G.; Liu, K.; Liu, X.P. Estimating Mangrove Wetland Biomass Using Radar Remote Sensing. J. Remote Sens. 2006, 10, 387–396. (In Chinese) [Google Scholar]
- Ceballos, A.; Hernández, J.; Corvalán, P.; Galleguillos, M. Comparison of Airborne LiDAR and Satellite Hyperspectral Remote Sensing to Estimate Vascular Plant Richness in Deciduous Mediterranean Forests of Central Chile. Remote Sens. 2015, 7, 2692–2714. [Google Scholar] [CrossRef]
- Boudreau, J.; Nelson, R.F.; Margolis, H.A.; Beaudoin, A.; Guindon, L.; Kimes, D.S. Regional aboveground forest biomass using airborne and spaceborne LiDAR in QuéBec. Remote Sens. Environ. 2008, 112, 3876–3890. [Google Scholar] [CrossRef]
- Maselli, F.; Chiesi, M.; Montaghi, A.; Pranzini, E. Use of ETM+ images to extend stem volume estimates obtained from LiDAR data. ISPRS J. Photogramm. 2011, 66, 662–671. [Google Scholar] [CrossRef]
- Li, J.Q. Method for Extracting Lower Vegetation Structure of Forest Canopy Using LiDAR Data. Doctoral Dissertation, Tsinghua University, Beijing, China, 2011. (In Chinese). [Google Scholar]
- Wang, Q.; Liao, J.J. Estimation of Wetland Vegetation Biomass in the Poyang Lake Area Using Landsat TM and ENVISAT ASAR Data. J. Geoinf. Sci. 2010, 12, 282–291. (In Chinese) [Google Scholar] [CrossRef]
- Tan, Q.M.; Liu, Y.H.; Zhang, H.B. Classification of vegetation coverage of wetland landscape based on remote sensing in coastal area in Jiangsu province. Remote Sens. Technol. Appl. 2013, 28, 934–940. (In Chinese) [Google Scholar]
- Ghosh, S.; Mishra, D.R.; Gitelson, A.A. Long-term monitoring of biophysical characteristics of tidal wetlands in the northern Gulf of Mexico—A methodological approach using MODIS. Remote Sens. Environ. 2016, 173, 39–58. [Google Scholar] [CrossRef]
- Streutker, D.R.; Mundt, J.T.; Glenn, N.F. Mapping Sagebrush Distribution Using Fusion of Hyperspectral and LiDAR Classifications. Photogramm. Eng. Remote Sens. 2006, 72, 47–54. [Google Scholar]
- Swatantran, A.; Dubayah, R.; Roberts, D.; Hofton, M.; Blair, J.B. Mapping biomass and stress in the Sierra Nevada using LiDAR and hyperspectral data fusion. Remote Sens. Environ. 2011, 115, 2917–2930. [Google Scholar] [CrossRef]
- Vaglio, L.; Chen, Q.; Lindsell, J.; Coomes, D. Above ground biomass estimation from LiDAR and hyperspectral airbone data in West African moist forests. EGU Gen. Assem. 2013, 15, 6227. [Google Scholar]
- Du, L.; Shi, S.; Yang, J.; Sun, J.; Gong, W. Using Different Regression Methods to Estimate Leaf Nitrogen Content in Rice by Fusing Hyperspectral LiDAR Data and Laser-Induced Chlorophyll Fluorescence Data. Remote Sens. 2016, 8, 526. [Google Scholar] [CrossRef]
- Priem, F.; Canters, F. Synergistic Use of LiDAR and APEX Hyperspectral Data for High-Resolution Urban Land Cover Mapping. Remote Sens. 2016, 8, 787. [Google Scholar] [CrossRef]
- Clark, M.L.; Roberts, D.A.; Ewel, J.J.; Clark, D.B. Estimation of tropical rain forest aboveground biomass with small-footprint LiDAR and hyperspectral sensors. Remote Sens. Environ. 2011, 115, 2931–2942. [Google Scholar] [CrossRef]
- Wang, C.; Nie, S.; Xi, X.H.; Luo, S.Z.; Sun, X.F. Estimating the Biomass of Maize with Hyperspectral and LiDAR Data. Remote Sens. 2017, 9, 11. [Google Scholar] [CrossRef]
- Liu, X.L.; Bo, Y.C. Object-Based Crop Species Classification Based on the Combination of Airborne Hyperspectral Images and LiDAR Data. Remote Sens. 2015, 7, 922–950. [Google Scholar] [CrossRef]
- Adam, E.; Mutanga, O.; Rugege, D. Multispectral and hyperspectral remote sensing for identification and mapping of wetland vegetation: A review. Wetl. Ecol. Manag. 2010, 18, 281–296. [Google Scholar] [CrossRef]
- Wang, Y. Remote sensing of coastal environments: An overview. In Remote Sensing of Coastal Environments; Wang, J., Ed.; CRC: Boca Raton, FL, USA, 2010; pp. 1–21. [Google Scholar]
- Klemas, V. Remote sensing of coastal wetland biomass: An overview. J. Coast. Res. 2013, 29, 1016–1018. [Google Scholar] [CrossRef]
- O’Donnell, J.P.R.; Schalles, J.F. Examination of Abiotic Drivers and Their Influence on Spartina alterniflora Biomass over a Twenty-Eight Year Period Using Landsat 5 TM Satellite Imagery of the Central Georgia Coast. Remote Sens. 2016, 8, 477. [Google Scholar] [CrossRef]
- Pearson, R.L.; Miller, L.D. Remote mapping of standing crop bioma.ss for estimation of the productivity of the shortgrassprairie. In Proceedings of the Eighth International Symposium on Remote Sensing of Environment, Ann Arbor, MI, USA, 2–6 October 1972; p. 1355. [Google Scholar]
- Rouse, J.W.J.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring vegetation systems in the Great Plains with ERTS. In Third Earth Resources Technology Satellite-1 Symposium, Volume I: Technical Presentations; NASA SP-351; NASA: Washington, DC, USA, 1974; p. 309. [Google Scholar]
- Gitelson, A.A.; Kaufman, Y.J.; Merzlyak, M.N. Use of a green channel in remote sensing of global vegetation from EOS-MODIS. Remote Sens. Environ. 1996, 58, 289–298. [Google Scholar] [CrossRef]
- Jordan, C.F. Derivation of Leaf-Area Index from Quality of Light on the Forest Floor. Ecology 1969, 4, 663–666. [Google Scholar] [CrossRef]
- Roujean, J.L.; Breon, F.M. Estimating PAR absorbed by vegetation from bidirectional reflectance measurements. Remote Sens. Environ. 1995, 51, 375–384. [Google Scholar] [CrossRef]
- Broge, N.H.; Leblanc, E. Comparing prediction power and stability of broadband and hyperspectral vegetation indices for estimation of green leaf area index and canopy chlorophyll density. Remote Sens. Environ. 2011, 76, 156–172. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Stark, R.; Rundquist, D. Novel algorithms for remote estimation of vegetation fraction. Remote Sens. Environ. 2002, 80, 76–87. [Google Scholar] [CrossRef]
- Kaufman, Y.J.; Tanre, D. Atmospherically resistant vegetation index (ARVI) for EOS-MODIS. IEEE Trans. Geosci. Remote Sens. 1992, 30, 261–270. [Google Scholar] [CrossRef]
- Pinty, B.M.; Verstraete, M. GEMI: A non-linear index to monitor global vegetation from satellites. Vegetatin 1992, 101, 15–20. [Google Scholar] [CrossRef]
- Liu, H.Q.; Huete, A.A. Feedback based modification of the NDVI to minimize canopy background and atmospheric noise. IEEE Trans. Geosci. Remote Sens. 1995, 33, 457–465. [Google Scholar]
- Chang, C.W.; Laird, D.A.; Mausbach, M.J.; Hurburgh, C.R. Near-infrared reflectance spectroscopy—Principal components regression analyses of soil properties. Soil Sci. Soc. Am. J. 2001, 65, 480–490. [Google Scholar] [CrossRef]
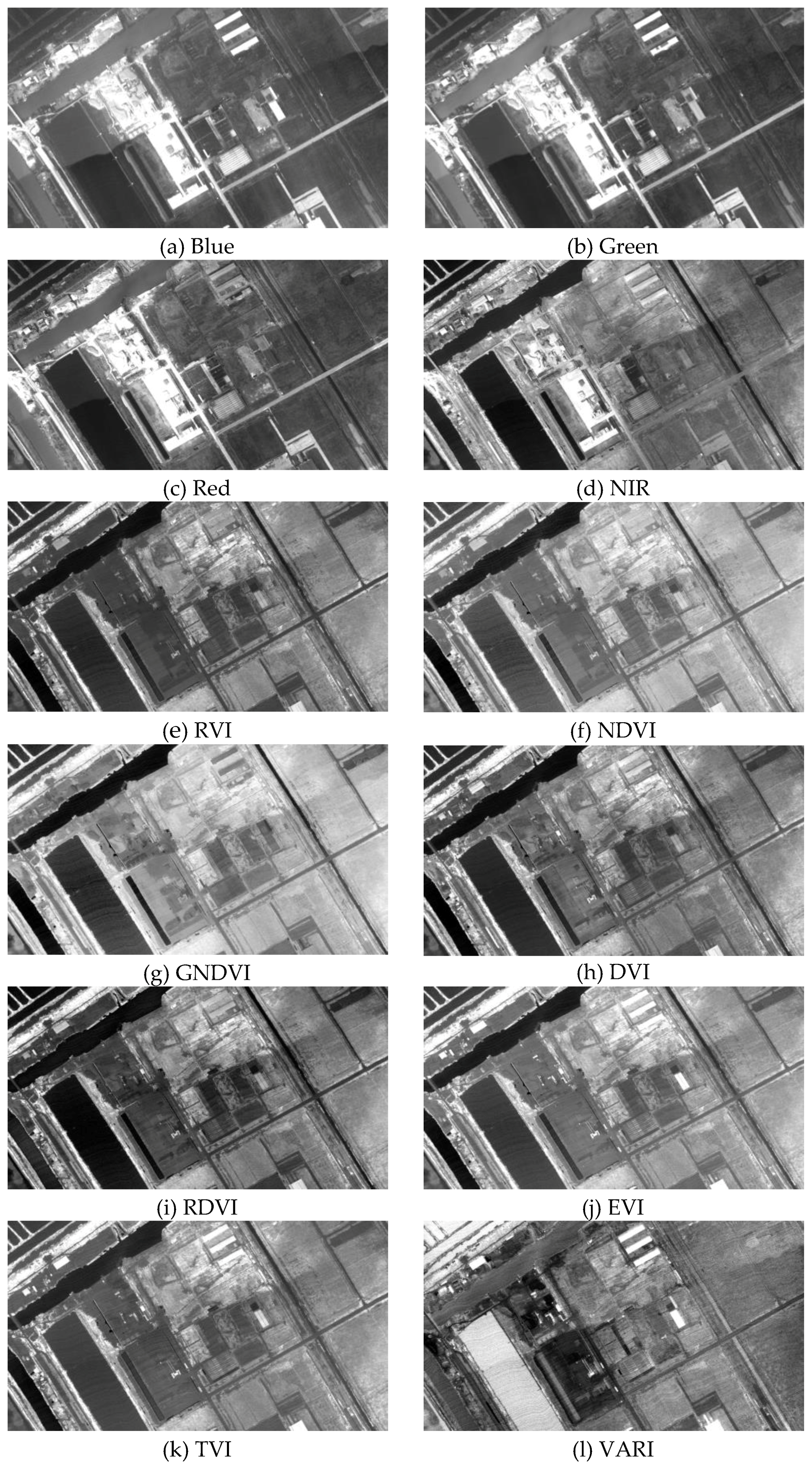

| VI | R452 | R534 | R694 | R865 | RVI | NDVI | GNDVI | DVI |
|---|---|---|---|---|---|---|---|---|
| Wetland | 0.515 ** | 0.639 ** | 0.087 | 0.546 ** | 0.007 | 0.006 | 0.071 | 0.488 ** |
| VI | RDVI | TVI | VARI | ARVI | GEMI | EVI | Height | |
| Wetland | 0.294 | 0.506 ** | 0.109 | 0.013 | 0.507 ** | 0.007 | 0.421 ** |
| Independent Variables Introduction | Multivariate Linear Estimation Models | R2 Value |
|---|---|---|
| Variables forced entry | Y = 35.684X1 − 3.531X2 + 7.582X3 − 0.078X4 + 0.18X5 − 1.061 | 0.650 |
| Variables forward entry | Y = 48.517X1 − 0.686 | 0.620 |
| Y = 35.411X1 + 0.124X5 − 1.228 | 0.704 | |
| Variable backward elimination | Y = 38.665X1 − 2.681X2 + 0.165X5 − 1.085 | 0.691 |
| Y = 35.446X1 − 3.559X2 + 7.582X3 + 0.179X5 − 1.083 | 0.672 | |
| Y = 35.684X1 − 3.531X2 + 7.582X3 − 0.078X4 + 0.18X5 − 1.061 | 0.650 | |
| Variable stepwise entry | Y = 35.411X1 + 0.124X5 − 1.228 | 0.704 |
| Y = 48.517X1 − 0.686 | 0.620 |
| VI | R452 | R534 | R694 | R865 | RVI | NDVI | GNDVI | DVI |
|---|---|---|---|---|---|---|---|---|
| S. alterniflora | 0.090 | 0.135 | 0.024 | 0.196 | 0.520 ** | 0.635 ** | 0.241 | 0.238 |
| VI | RDVI | TVI | VARI | ARVI | GEMI | EVI | Height | |
| S. alterniflora | 0.320 | 0.254 | 0.599 ** | 0.631 ** | 0.284 | 0.288 | 0.817 ** |
| Independent Variable Introduction | Multivariate Linear Estimation Models | R2 Value |
|---|---|---|
| Variables forced entry | Y = 0.54X1 − 0.802X2 − 1.004X3 + 2.714X4 + 0.053X5 + 1.101 | 0.841 |
| Variables forward entry | Y = 0.514X1 + 1.11X4 + 0.388 | 0.902 |
| Y = 0.765X1 + 0.119 | 0.811 | |
| Variable backward elimination | Y = 0.514X1 + 1.11X4 + 0.388 | 0.902 |
| Y = 0.512X1 − 0.812X3 + 2.626X4 + 0.787 | 0.891 | |
| Y = 0.533X1 − 1.829X3 + 3.106X4 + 0.056X5 + 1.018 | 0.880 | |
| Y = 0.54X1 − 0.802X2 − 1.004X3 + 2.714X4 + 0.053X5 + 1.101 | 0.841 | |
| Variable stepwise entry | Y = 0.514X1 + 1.11X4 + 0.388 | 0.902 |
| Y = 0.765X1 + 0.119 | 0.811 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Liu, Z.; Yu, H.; Li, F. Mapping Spartina alterniflora Biomass Using LiDAR and Hyperspectral Data. Remote Sens. 2017, 9, 589. https://doi.org/10.3390/rs9060589
Wang J, Liu Z, Yu H, Li F. Mapping Spartina alterniflora Biomass Using LiDAR and Hyperspectral Data. Remote Sensing. 2017; 9(6):589. https://doi.org/10.3390/rs9060589
Chicago/Turabian StyleWang, Jing, Zhengjun Liu, Haiying Yu, and Fangfang Li. 2017. "Mapping Spartina alterniflora Biomass Using LiDAR and Hyperspectral Data" Remote Sensing 9, no. 6: 589. https://doi.org/10.3390/rs9060589