Classification of Tree Species in a Diverse African Agroforestry Landscape Using Imaging Spectroscopy and Laser Scanning
Abstract
:1. Introduction
- compare the performance of the different feature sets derived from IS and ALS data using SVM and RF classifiers;
- find species or groups of species with ecological or economical function that can be detected relatively accurately; and
- evaluate the impact of up-sampling and different approaches to group the species on the classification accuracy.
2. Material and Methods
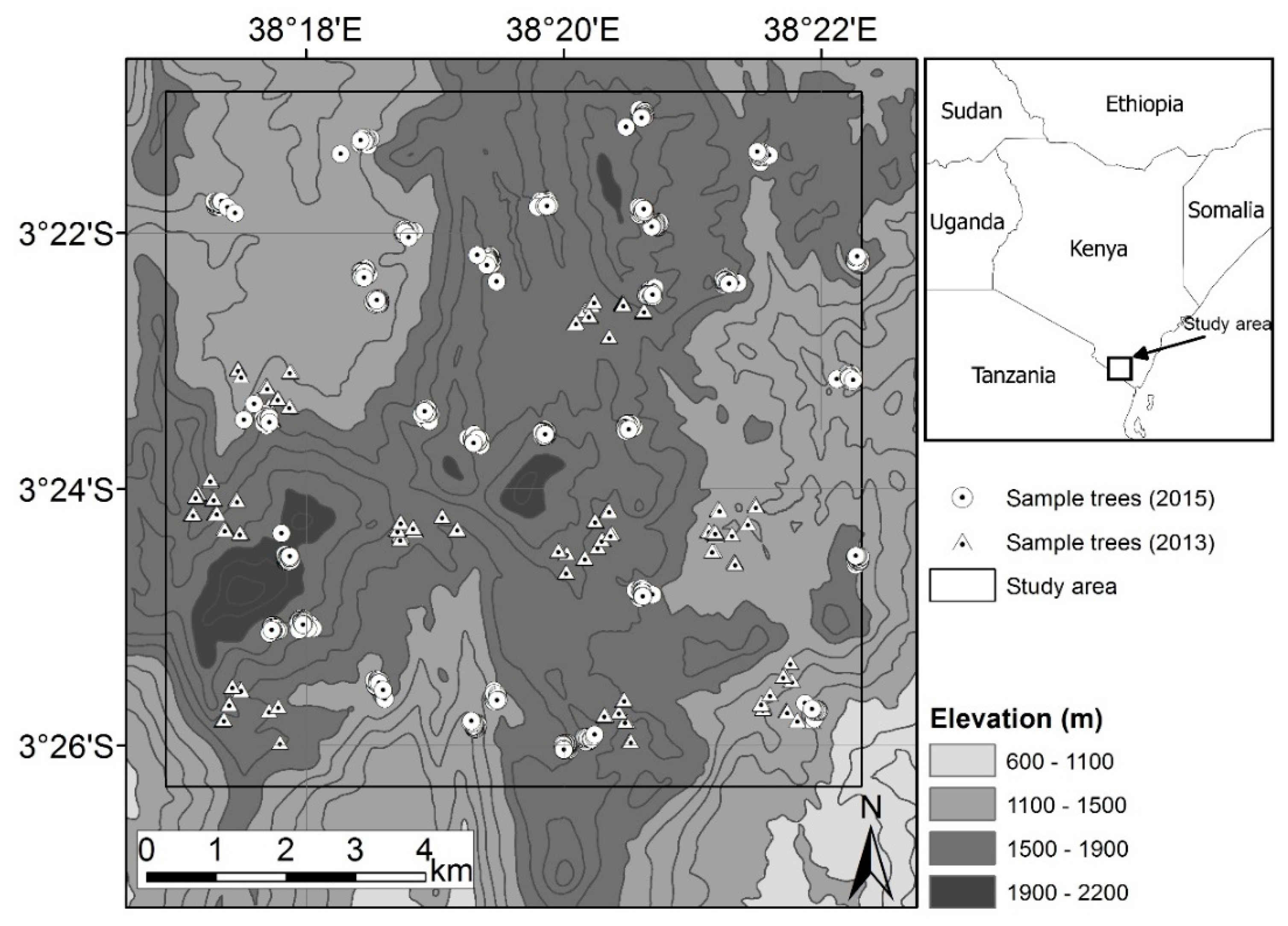
2.1. Study Site
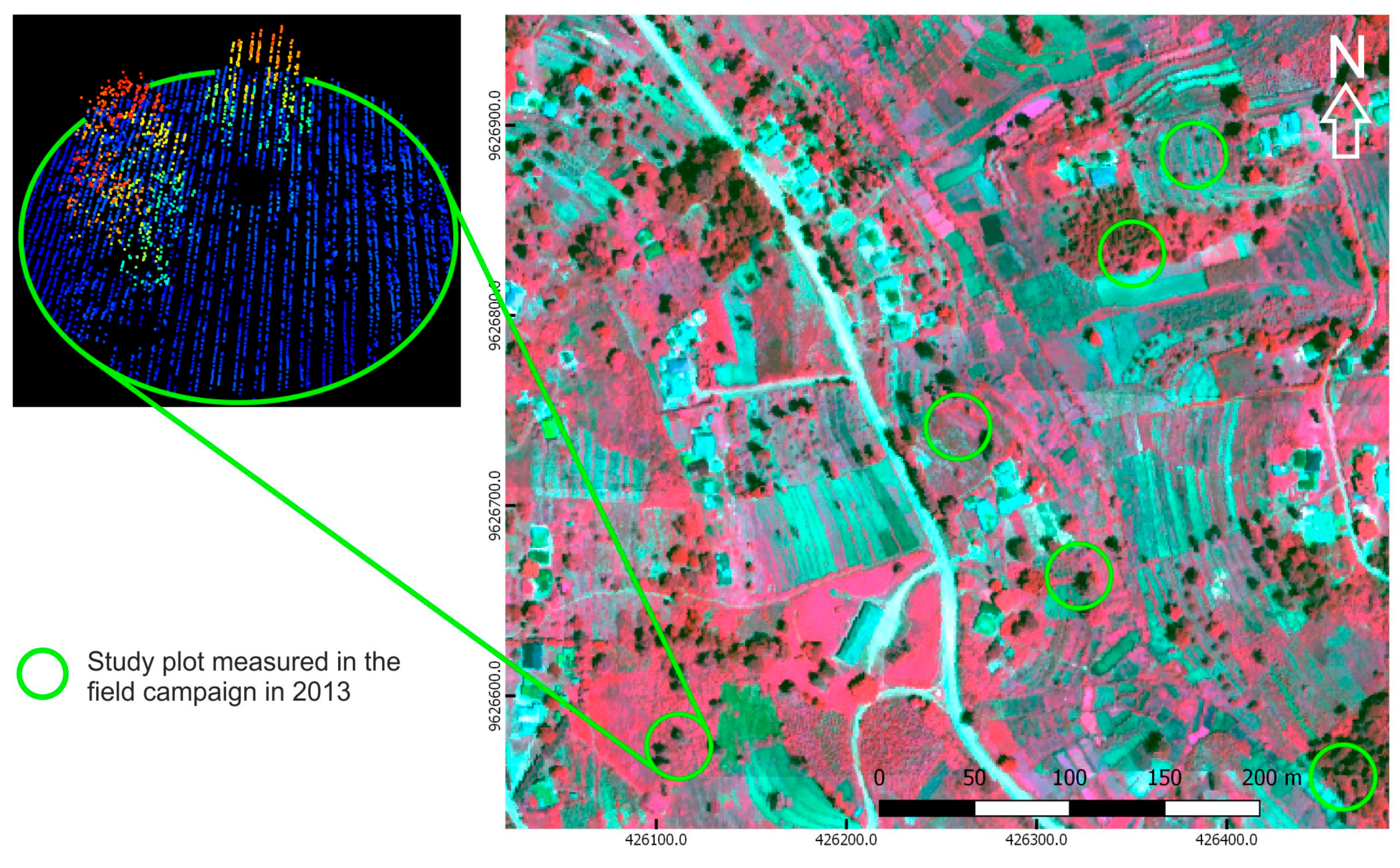
2.2. Field Data
2.3. Remote Sensing Data
2.4. Remote Sensing Data Preprocessing
2.5. Segmentation and Preparing Training Data
2.6. Minimum Noise Fraction Transformation
2.7. Narrowband Vegetation Indices
2.8. Point cloud Features
2.9. Feature Selection
2.10. Classification Methods
2.11. Measures of Performance
2.12. Jeffries–Matusita Distance
2.13. Statistical Significance Tests
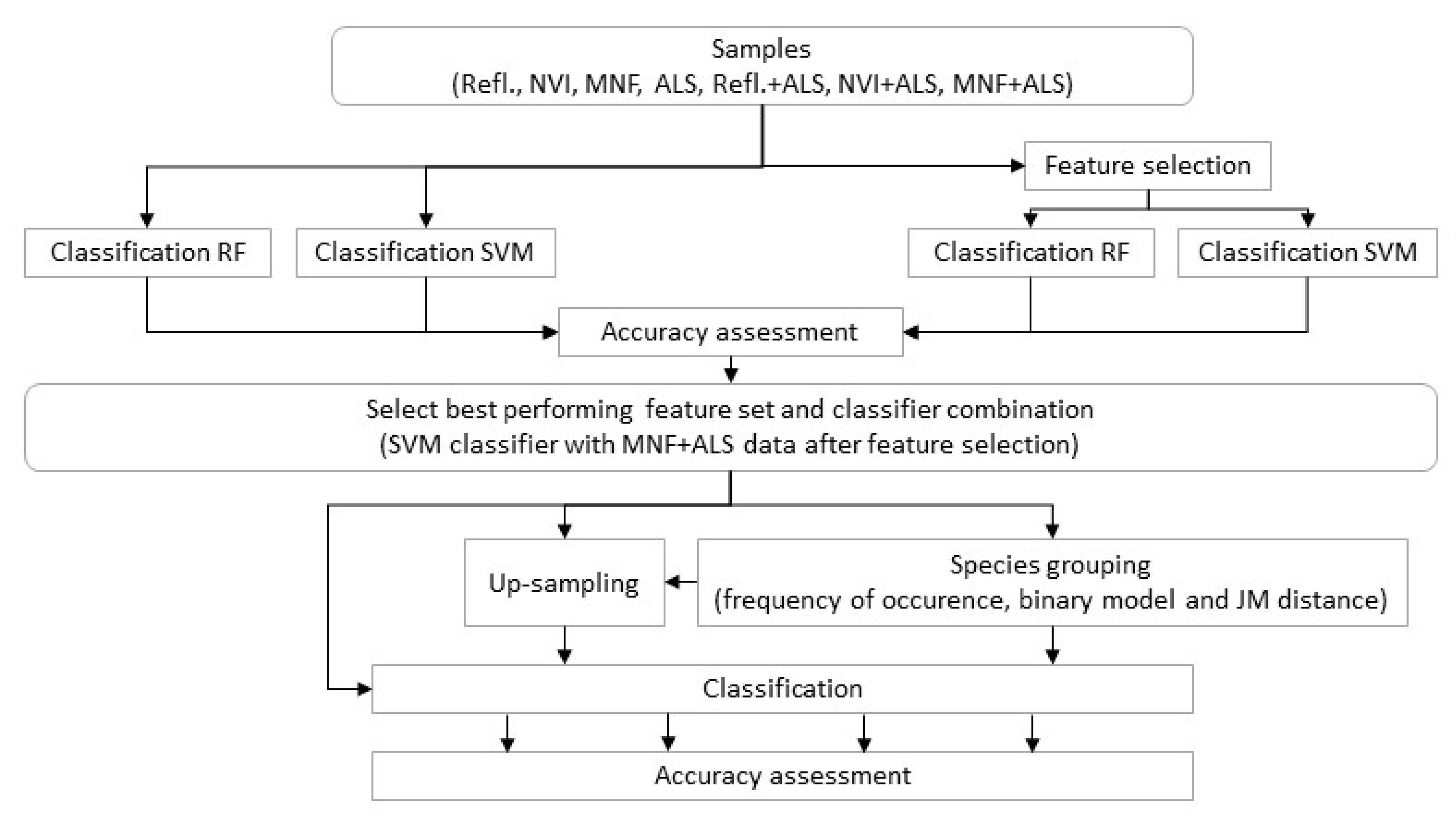
2.14. Classification Trials
3. Results
3.1. Comparison of Feature Sets and Classifiers
3.2. Feature Selection
3.3. Jeffries–Matusita Distance
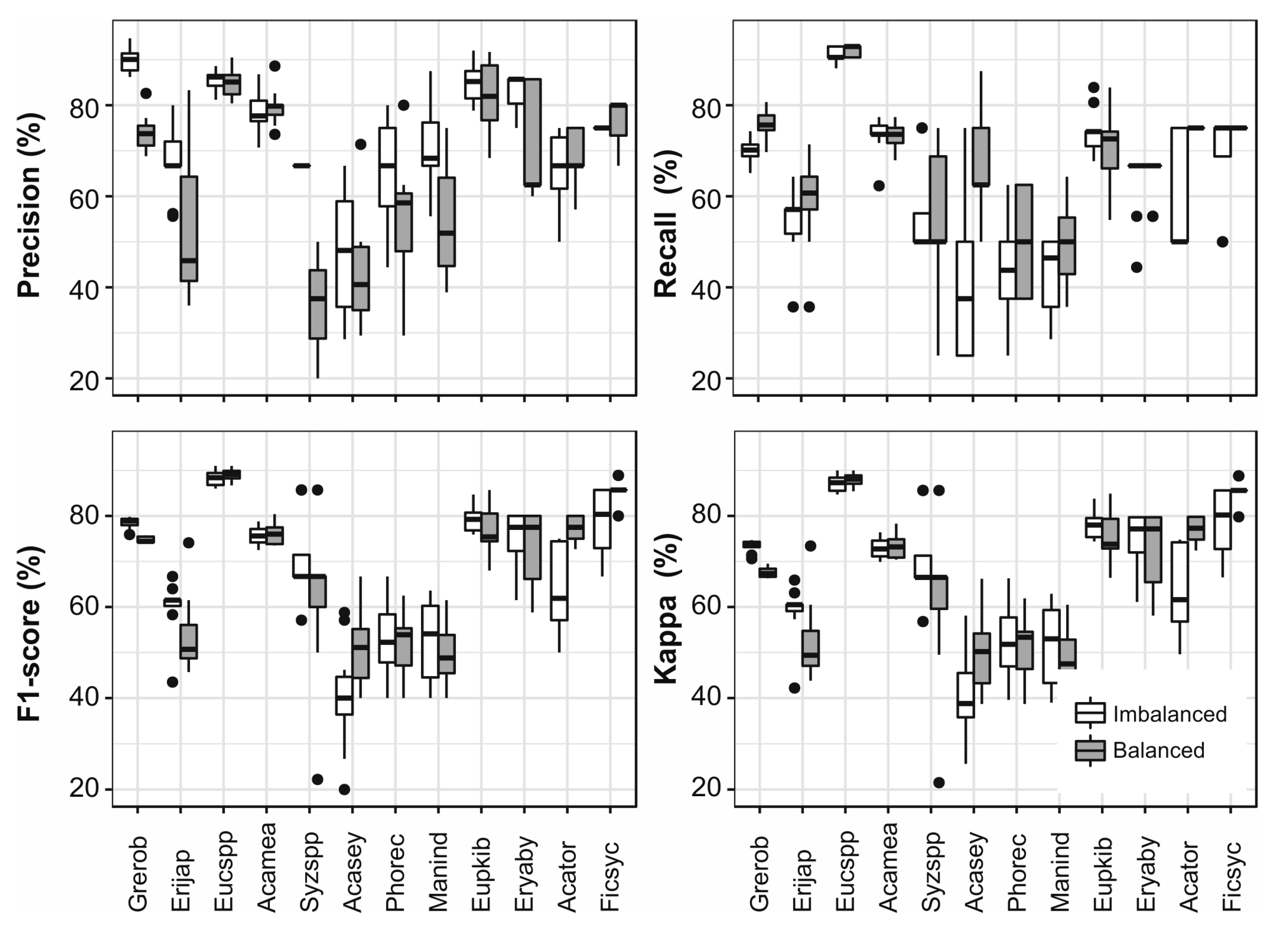
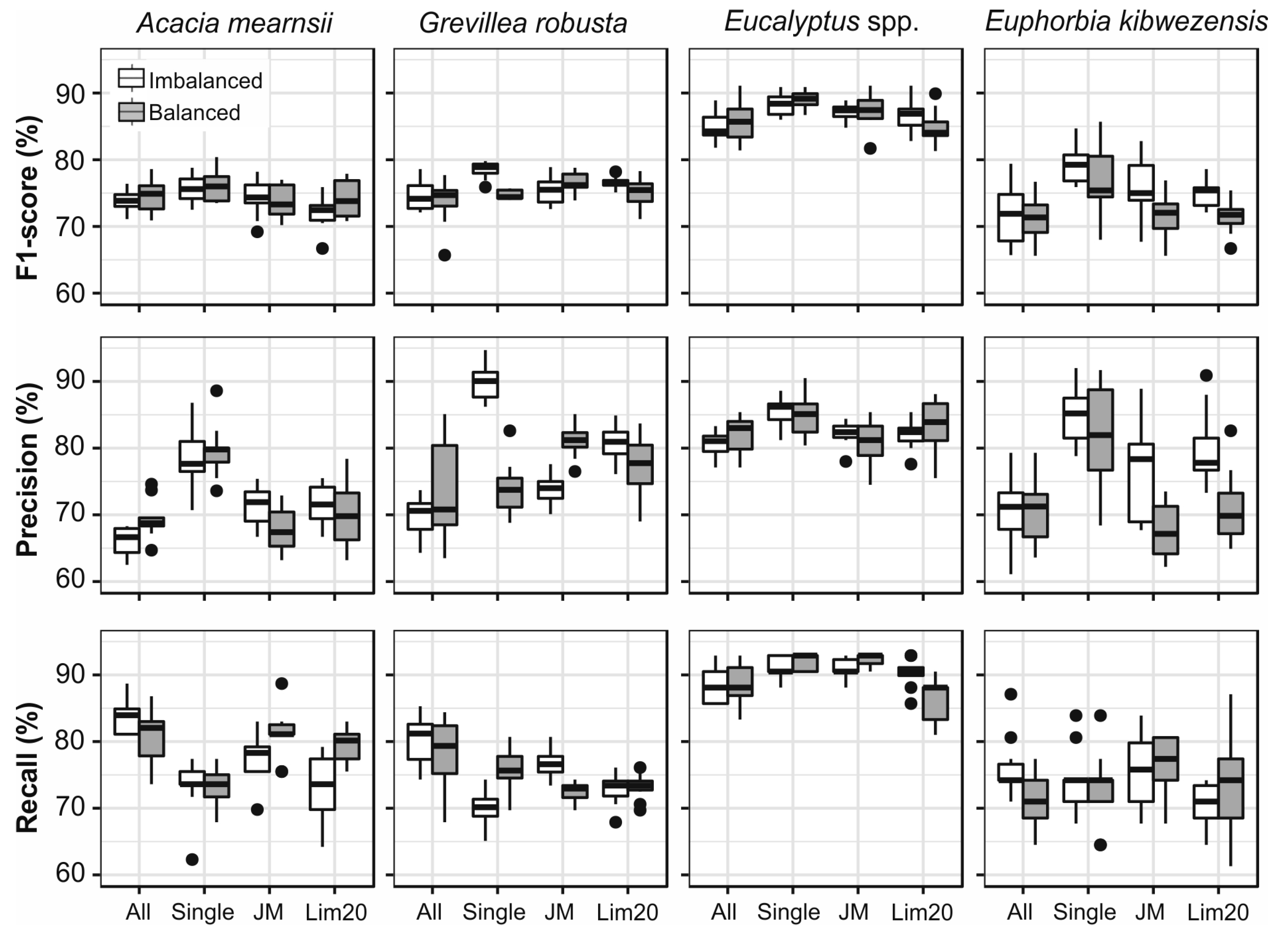
3.4. Data Balancing
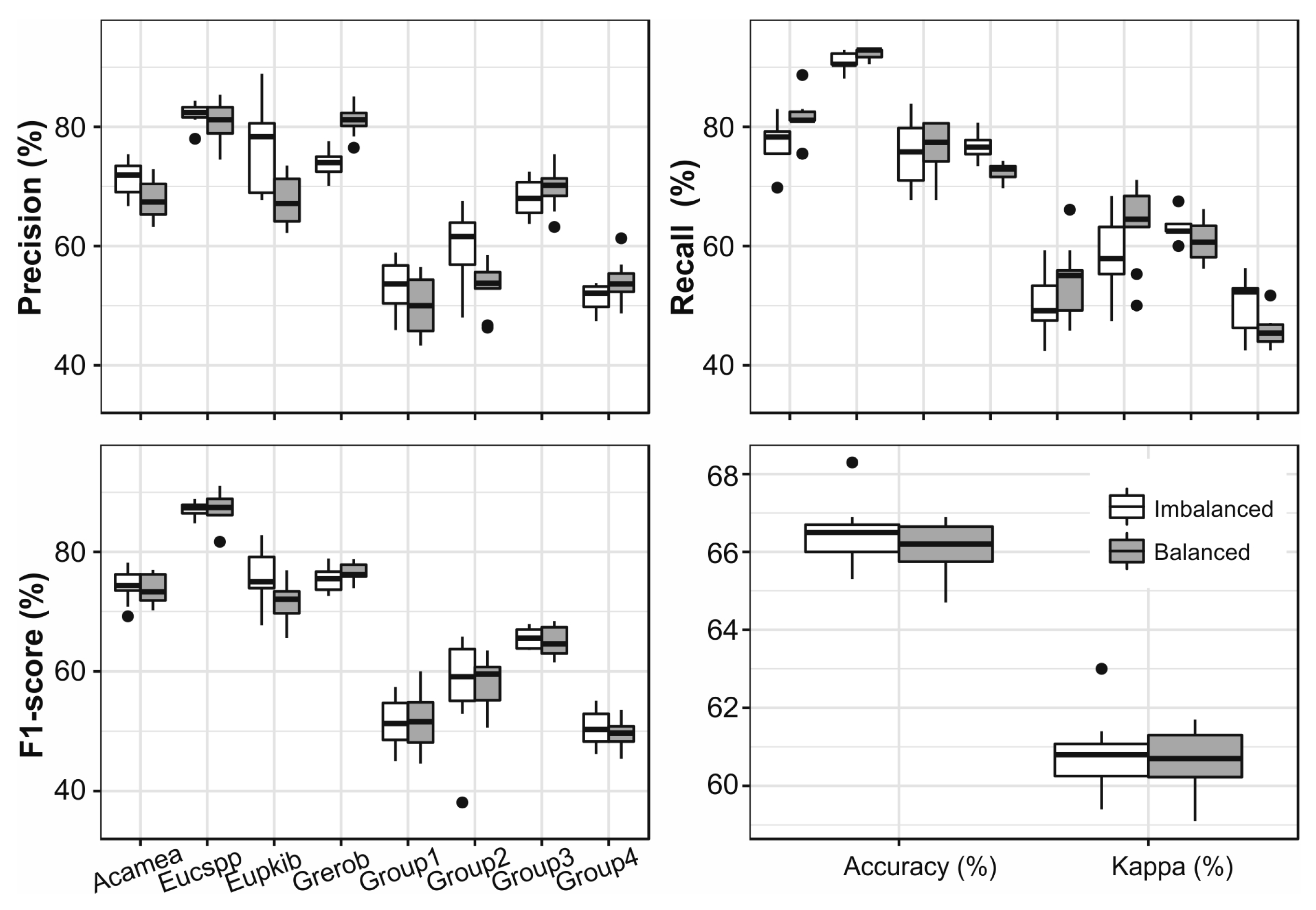
3.5. Grouping by Frequency
3.6. Single Species Classfication
3.7. Grouping Species Based on Jeffries–Matusita Distance
3.8. Comparison of Different Aproaches
4. Discussion
4.1. Impacts of Classifier, Feature Selection and Data Fusion
4.2. The Impact of Up-Sampling and Grouping of Species on the Classification Results
4.3. Evaluation of the Quality of Airborne Data, Field Measurements and Segmentation
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gibbs, H.K.; Ruesch, A.S.; Achard, F.; Clayton, M.K.; Holmgren, P.; Ramankutty, N.; Foley, J.A. Tropical forests were the primary sources of new agricultural land in the 1980s and 1990s. Proc. Natl. Acad. Sci. USA 2010, 107, 16732–16737. [Google Scholar] [CrossRef] [PubMed]
- Brink, A.B.; Bodart, C.; Brodsky, L.; Defourney, P.; Ernst, C.; Donney, F.; Lupi, A.; Tuckova, K. Anthropogenic pressure in East Africa—Monitoring 20 years of land cover changes by means of medium resolution satellite data. Int. J. Appl. Earth Obs. Geoinf. 2014, 28, 60–69. [Google Scholar] [CrossRef]
- Mbow, C.; van Noordwijk, M.; Prabhu, R.; Simons, T. Knowledge gaps and research needs concerning agroforestry’s contribution to Sustainable Development Goals in Africa. Curr. Opin. Environ. Sustain. 2014, 6, 162–170. [Google Scholar] [CrossRef]
- Luedeling, E.; Kindt, R.; Huth, N.I.; Koenig, K. Agroforestry systems in a changing climate—Challenges in projecting future performance. Curr. Opin. Environ. Sustain. 2014, 6, 1–7. [Google Scholar] [CrossRef]
- Rodriguez-Suarez, J.A.; Soto, B.; Perez, R.; Diaz-Fierros, F. Influence of Eucalyptus globulus plantation growth on water table levels and low flows in a small catchment. J. Hydrol. 2011, 396, 321–326. [Google Scholar] [CrossRef]
- Lott, J.E.; Khan, A.A.H.; Black, C.R.; Ong, C.K. Water use in a Grevillea robusta-maize overstorey agroforestry system in semi-arid Kenya. For. Ecol. Manag. 2003, 180, 45–59. [Google Scholar] [CrossRef]
- Omoro, L.M.A.; Nair, P.K.R. Effects of mulching with multipurpose-tree prunings on soil and water run-off under semi-arid conditions in Kenya. Agrofor. Syst. 1993, 22, 225–239. [Google Scholar] [CrossRef]
- Thijs, K.W.; Aerts, R.; van de Moortele, P.; Aben, J.; Musila, W.; Pellikka, P.; Gulinck, H.; Muys, B. Trees in a human-modified tropical landscape: Species and trait composition and potential ecosystem services. Landsc. Urban Plan. 2015, 144, 49–58. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Latifi, H.; Sterenczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.T.; Straub, C.; Ghosh, A. Review of studies on tree species classification from remotely sensed data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- Graves, S.J.; Asner, G.P.; Martin, R.E.; Anderson, C.B.; Colgan, M.S.; Kalantari, L.; Bohlman, S.A. Tree species abundance predictions in a tropical agricultural landscape with a supervised classification model and imbalanced data. Remote Sens. 2016, 8, 161. [Google Scholar] [CrossRef]
- Feret, J.-B.; Asner, P.G. Tree species discrimination in tropical forests using airborne imaging spectroscopy. IEEE Trans. Geosci. Remote Sens. 2013, 51, 73–84. [Google Scholar] [CrossRef]
- Baldeck, C.A.; Asner, G.P. Improving remote species identification through efficient training data collection. Remote Sens. 2014, 6, 2682–2698. [Google Scholar] [CrossRef]
- Freeman, E.A.; Moisen, G.G.; Frescino, T.S. Evaluating effectiveness of down-sampling for stratified designs and unbalanced prevalence in Random Forest models of tree species distributions in Nevada. Ecol. Model. 2012, 233, 1–10. [Google Scholar] [CrossRef]
- Dalponte, M.; Ene, L.T.; Marconcini, M.; Gobakken, T.; Næsset, E. Semi-supervised SVM for individual tree crown species classification. ISPRS J. Photogramm. Remote Sens. 2015, 110, 77–87. [Google Scholar] [CrossRef]
- Vapnik, V. Statistical Learning Theory; John Wiley & Sons: New York, NY, USA, 1998. [Google Scholar]
- Breiman, L.E.O. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Belgiu, M.; Drăguţ, L. Random forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Ghosh, A.; Fassnacht, F.E.; Joshi, P.K.; Kochb, B. A framework for mapping tree species combining hyperspectral and LiDAR data: Role of selected classifiers and sensor across three spatial scales. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 49–63. [Google Scholar] [CrossRef]
- Ghosh, A.; Joshi, P.K. A comparison of selected classification algorithms for mappingbamboo patches in lower Gangetic plains using very high resolution WorldView 2 imagery. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 298–311. [Google Scholar] [CrossRef]
- Green, A.; Berman, M.; Switzer, P.; Craig, M.D. A transformation for ordering multispectral data in terms of image quality with implications for noise removal. IEEE Trans. Geosci. Remote Sens. 1988, 26, 65–74. [Google Scholar] [CrossRef]
- Piiroinen, R.; Heiskanen, J.; Mõttus, M.; Pellikka, P. Classification of crops across heterogeneous agricultural landscape in Kenya using AisaEAGLE imaging spectroscopy data. Int. J. Appl. Earth Obs. Geoinf. 2015, 39. [Google Scholar] [CrossRef]
- Jaetzold, R.; Schmidt, H.; Hornetz, B.; Shisanya, C. Farm Management Handbook of Kenya: Natural Conditions and Farm Management Information; Ministry of Agriculture, Kenya & German Agricultural Team; German Agency for Technical Cooperation: Rossdorf, Germany, 1983; Volume 2. [Google Scholar]
- Aerts, R.; Thijs, K.W.; Lehouck, V.; Beentje, H.; Bytebier, B.; Matthysen, E.; Gulinck, H.; Lens, L.; Muys, B. Woody plant communities of isolated Afromontane cloud forests in Taita Hills, Kenya. Plant Ecol. 2011, 212, 639–649. [Google Scholar] [CrossRef] [Green Version]
- Clark, B.; Pellikka, P. Landscape analysis using multi-scale segmentation and objectoriented classification. Recent Adv. Remote Sens. 2009, 323–341. [Google Scholar]
- Pellikka, P.K.E.; Clark, B.J.F.; Gosa, A.G.; Himberg, N.; Hurskainen, P.; Maeda, E.; Mwang’ombe, J.; Omoro, L.M.A.; Siljander, M. Agricultural expansion and its consequences in the Taita Hills, Kenya. Dev. Earth Surf. Process. 2013, 16, 165–179. [Google Scholar] [CrossRef]
- Burgess, N.D.; Butynski, T.M.; Cordeiro, N.J.; Doggart, N.H.; Fjeldså, J.; Howell, K.M.; Kilahama, F.B.; Loader, S.P.; Lovett, J.C.; Mbilinyi, B.; et al. The biological importance of the Eastern Arc Mountains of Tanzania and Kenya. Biol. Conserv. 2007, 134, 209–231. [Google Scholar] [CrossRef]
- Pellikka, P.K.E.; Lötjönen, M.; Siljander, M.; Lens, L. Airborne remote sensing of spatiotemporal change (1955–2004) in indigenous and exotic forest cover in the Taita Hills, Kenya. Int. J. Appl. Earth Obs. Geoinf. 2009, 11, 221–232. [Google Scholar] [CrossRef]
- Hohenthal, J.; Räsänen, M.; Owidi, E.; Andersson, B.; Minoia, P.; Pellikka, P.K.E. Community and Institutional Perspectives on Water Management and Environmental Changes in the Taita Hills, Kenya; University of Helsinki: Helsinki, Finland, 2015; ISBN 9789515113412. [Google Scholar]
- Mugedo, J.Z.A.; Waterman, P.G. Sources of tannin: Alternatives to wattle (Acacia mearnsii) among indigenous Kenyan species. Econ. Bot. 1992, 46, 55–63. [Google Scholar] [CrossRef]
- Boudiaf, I.; Le Roux, C.; Baudoin, E.; Galiana, A.; Beddiar, A.; Prin, Y.; Duponnois, R. Soil Bradyrhizobium population response to invasion of a natural Quercus suber forest by the introduced nitrogen-fixing tree Acacia mearnsii in El Kala National Park, Algeria. Soil Biol. Biochem. 2014, 70, 162–165. [Google Scholar] [CrossRef]
- Engeman, R.M.; Sugihara, R.T.; Pank, L.F.; Dusenberry, W.E. A comparison of plotless density estimators using Monte Carlo simulation. Ecology 1994, 75, 1769–1779. [Google Scholar] [CrossRef]
- Khosravipour, A.; Skidmore, A.K.; Isenburg, M.; Wang, T.; Hussin, Y. Generating pit-free canopy height models from airborne lidar. Photogramm. Eng. Remote Sens. 2014, 80, 863–872. [Google Scholar] [CrossRef]
- Richter, R.; Schläpfer, D. Geo-atmospheric processing of airborne imaging spectrometry data. Part 2: Atmospheric/topographic correction. Int. J. Remote Sens. 2002, 23, 2631–2649. [Google Scholar] [CrossRef]
- Valbuena, R. Integrating airborne laser scanning with data from global navigation satellite systems and optical sensors. In Forestry Applications of Airborne Laser Scanning: Concepts and Case Studies; Maltamo, M., Naesset, E., Vauhkonen, J., Eds.; Springer: Berlin, Germany, 2014; Volume 27, pp. 63–88. [Google Scholar]
- Baldeck, C.A.; Asner, G.P.; Martin, R.E.; Anderson, C.B.; Knapp, E.; Kellner, J.R.; Wright, S.J. Operational tree species mapping in a diverse tropical forest with airborne imaging spectroscopy. PLoS ONE 2015. [Google Scholar] [CrossRef] [PubMed]
- Dalponte, M.; Coomes, D.A. Tree-centric mapping of forest carbon density from airborne laser scanning and hyperspectral data. Methods Ecol. Evol. 2016, 7, 1236–1245. [Google Scholar] [CrossRef] [PubMed]
- Roussel, J.-R.; Auty, D. lidR: Airborne LiDAR Data Manipulation and Visualization for Forestry Applications, Version 1.2.0. Available online: https://rdrr.io/cran/lidR/ (accessed on 13 June 2017).
- R Core Team. R: A Language and Environment for Statistical Computing; Version 3.4.0; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- RSI. ENVI User’s Guide; Research Systems, Inc.: Boulder, CO, USA, 2004; pp. 1–1150. [Google Scholar]
- Zhang, C.; Xie, Z. Combining object-based texture measures with a neural network for vegetation mapping in the Everglades from hyperspectral imagery. Remote Sens. Environ. 2012, 124, 310–320. [Google Scholar] [CrossRef]
- Roberts, D.A.; Roth, K.L.; Perroy, R.L. Hyperspectral vegetation indices. In Hyperspectral Remote Sensing of Vegetation; Thenkabail, P.S., Lyon, P.S., Huete, J.G., Eds.; CRC press: Boca Raton, FL, USA, 2011; pp. 309–328. [Google Scholar]
- Genuer, R.; Poggi, J.-M.; Tuleau-Malot, C. Variable selection using random forests. Pattern Recognit. Lett. 2010, 31, 2225–2236. [Google Scholar] [CrossRef]
- Genuer, R.; Poggi, J.-M.; Tuleau-Malot, C. VSURF: An R package for variable selection using random forests. R J. 2015, 7, 19–33. [Google Scholar]
- Kuhn, M. Building Predictive Models in R Using the caret Package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- Karatzoglou, A.; Smola, A.; Hornik, K.; Zeileis, A. kernlab—An S4 Package for Kernel Methods in R. J. Stat. Softw. 2004, 11, 1–20. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R News 2002, 2, 18–22. [Google Scholar] [CrossRef]
- Congalton, R.G. A review of assessing the accuracy of classifications of remotely sensed data. Remote Sens. Environ. 1991, 37, 35–46. [Google Scholar] [CrossRef]
- Dalponte, M.; Ørka, H.O. varSel: Sequential Forward Floating Selection Using Jeffries-Matusita Distance. Available online: https://rdrr.io/cran/varSel/ (accessed on 5 November 2016).
- Ballanti, L.; Blesius, L.; Hines, E.; Kruse, B. Tree species classification using hyperspectral imagery: A comparison of two classifiers. Remote Sens. 2016, 8, 445. [Google Scholar] [CrossRef]
- Foody, G.M. Thematic map comparison: evaluating the statistical significance of differences in classification accuracy. Photogramm. Eng. Remote Sens. 2004, 70, 627–633. [Google Scholar] [CrossRef]
- Dietterich, T.G. Approximate statistical tests for comparing supervised classi cation learning algorithms. Neural Comput. 1998, 10, 1895–1923. [Google Scholar] [CrossRef] [PubMed]
- Jones, T.G.; Coops, N.C.; Sharma, T. Assessing the utility of airborne hyperspectral and LiDAR data for species distribution mapping in the coastal Pacific Northwest, Canada. Remote Sens. Environ. 2010, 114, 2841–2852. [Google Scholar] [CrossRef]
- Hovi, A.; Korhonen, L.; Vauhkonen, J.; Korpela, I. LiDAR waveform features for tree species classification and their sensitivity to tree- and acquisition related parameters. Remote Sens. Environ. 2016, 173, 224–237. [Google Scholar] [CrossRef]
- Van Der Linde, J.A.; Roux, J.; Wingfield, M.J.; Six, D.L. Die-off of giant Euphorbia trees in South Africa: Symptoms and relationships to climate. S. Afr. J. Bot. 2012, 83, 172–185. [Google Scholar] [CrossRef]
- Landmann, T.; Piiroinen, R.; Makori, D.M.; Abdel-Rahman, E.M.; Makau, S.; Pellikka, P.; Raina, S.K. Application of hyperspectral remote sensing for flower mapping in African savannas. Remote Sens. Environ. 2015, 166, 50–60. [Google Scholar] [CrossRef]
- Colgan, M.S.; Baldeck, C.A.; Féret, J.-B.; Asner, G.P. Mapping savanna tree species at ecosystem scales using support vector machine classification and BRDF correction on airborne hyperspectral and LiDAR data. Remote Sens. 2012, 4, 3462–3480. [Google Scholar] [CrossRef]
- Clark, M.L.; Roberts, D.A. Species-Level Differences in Hyperspectral Metrics among Tropical Rainforest Trees as Determined by a Tree-Based. Remote Sens. 2012, 4, 1820–1855. [Google Scholar] [CrossRef]


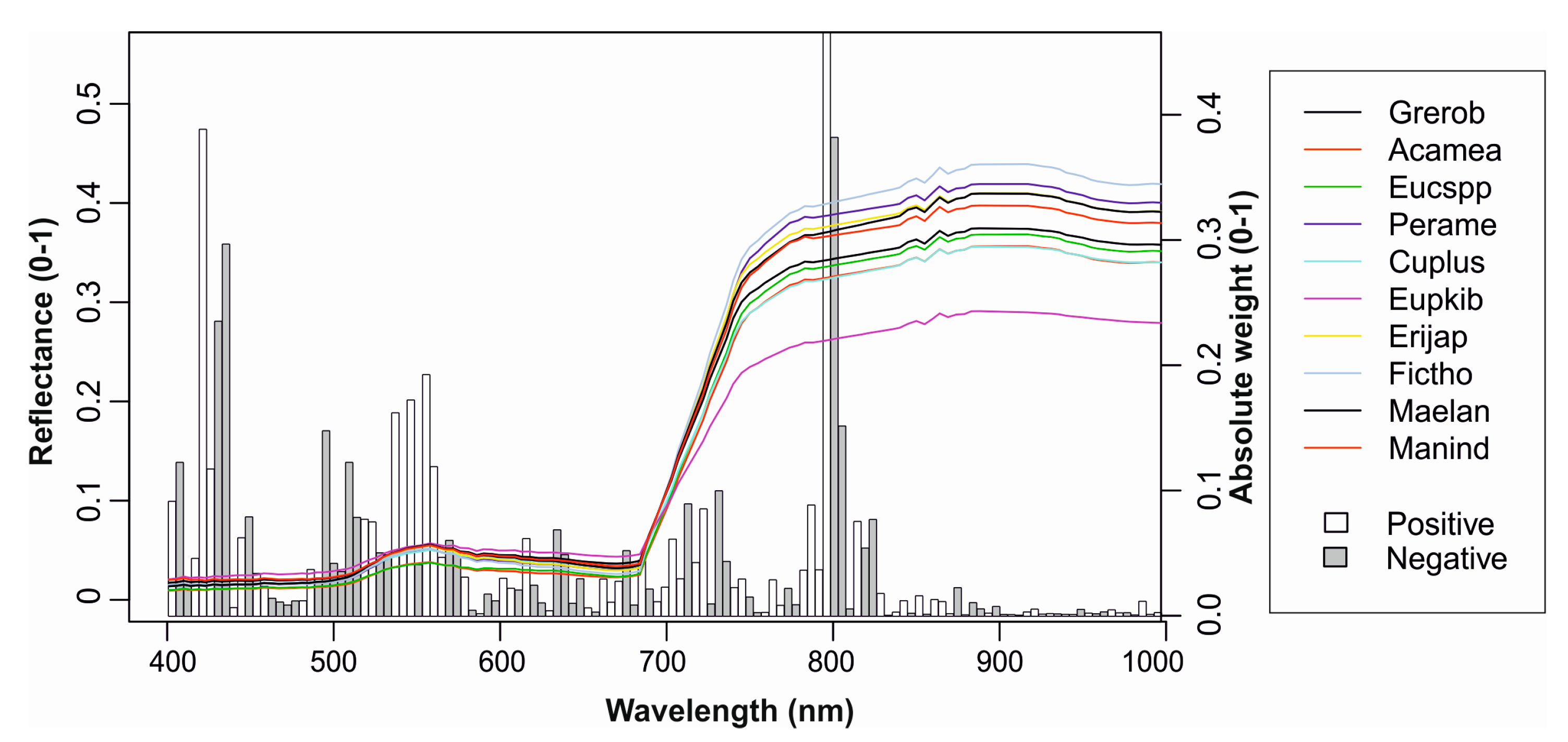
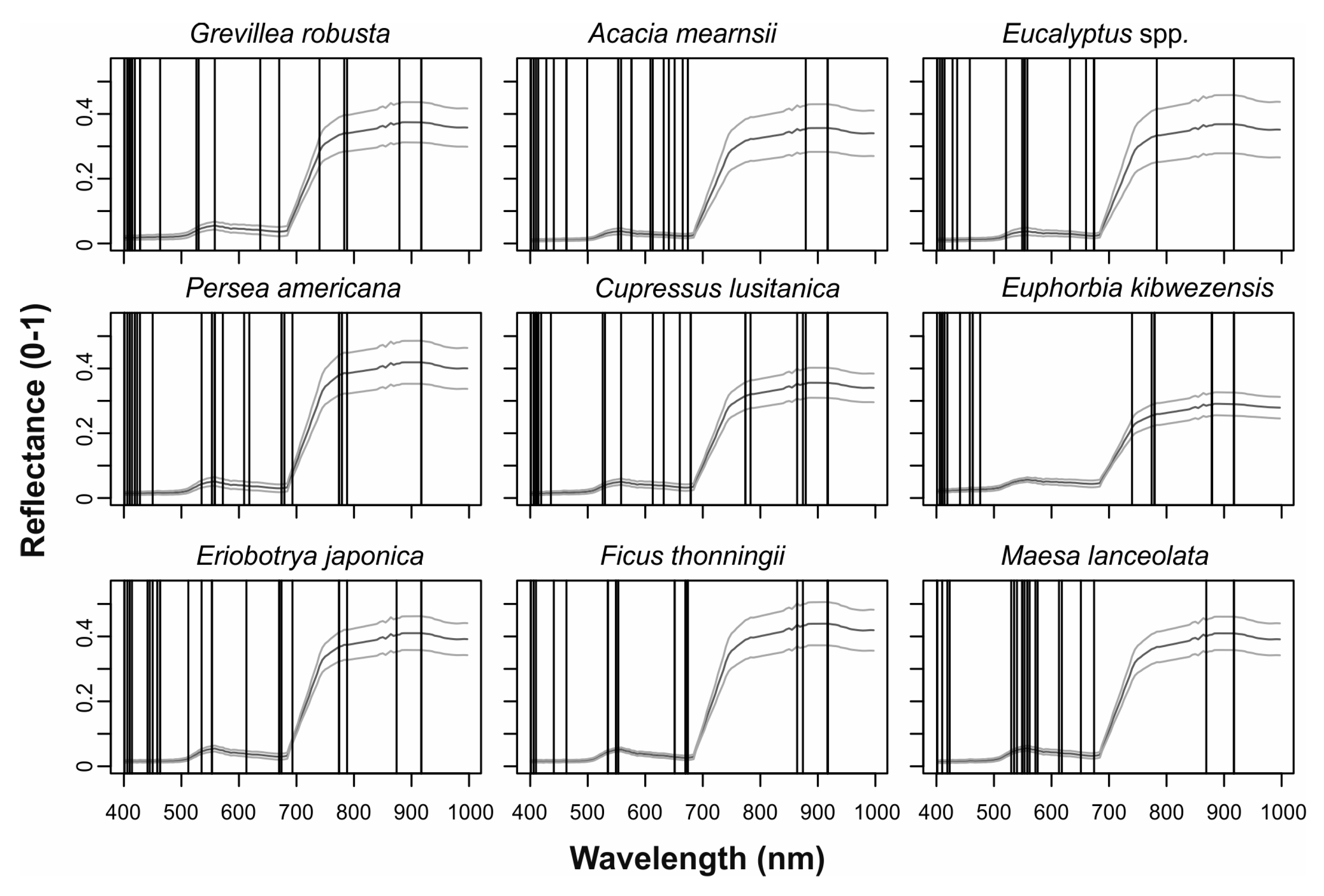
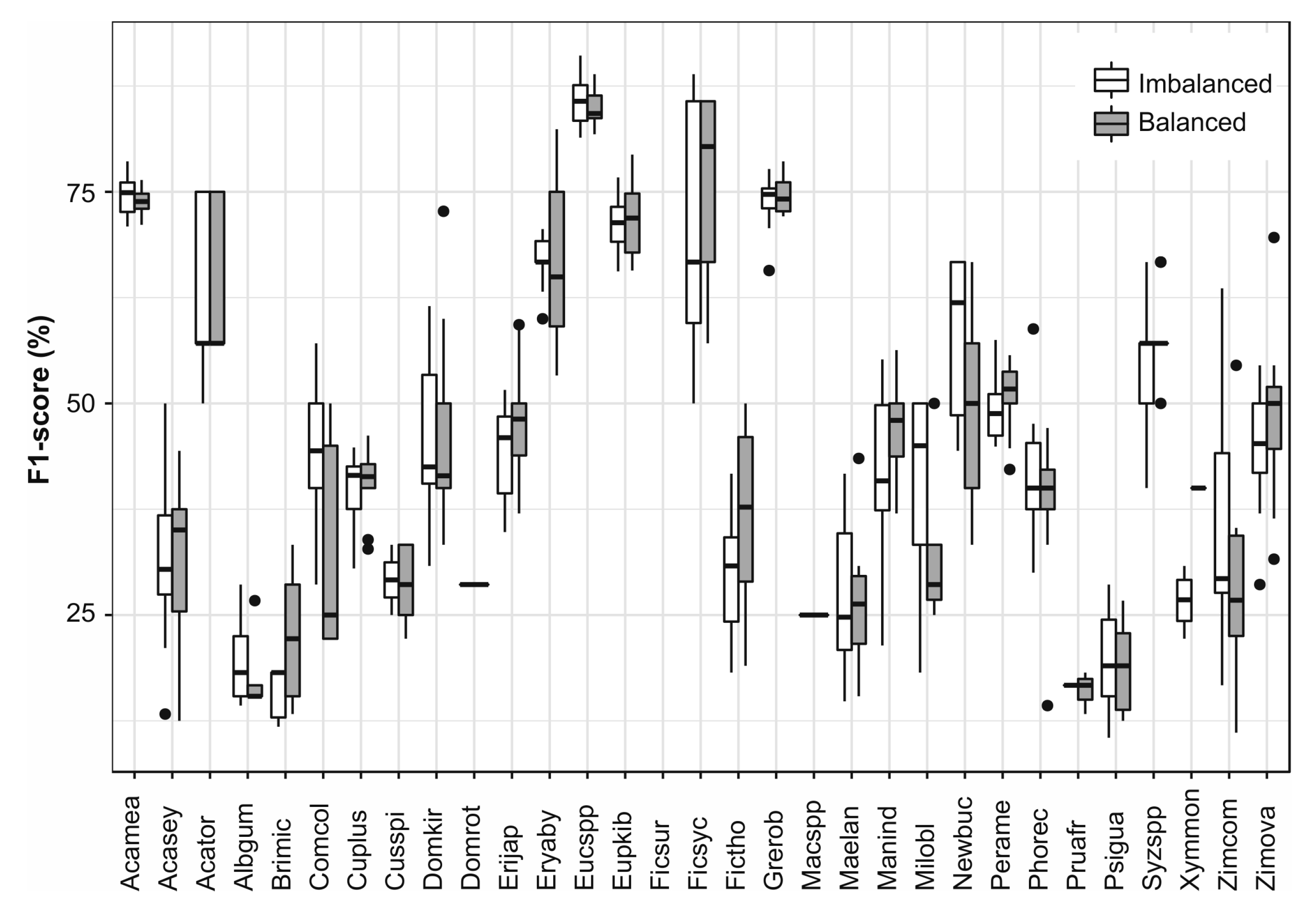
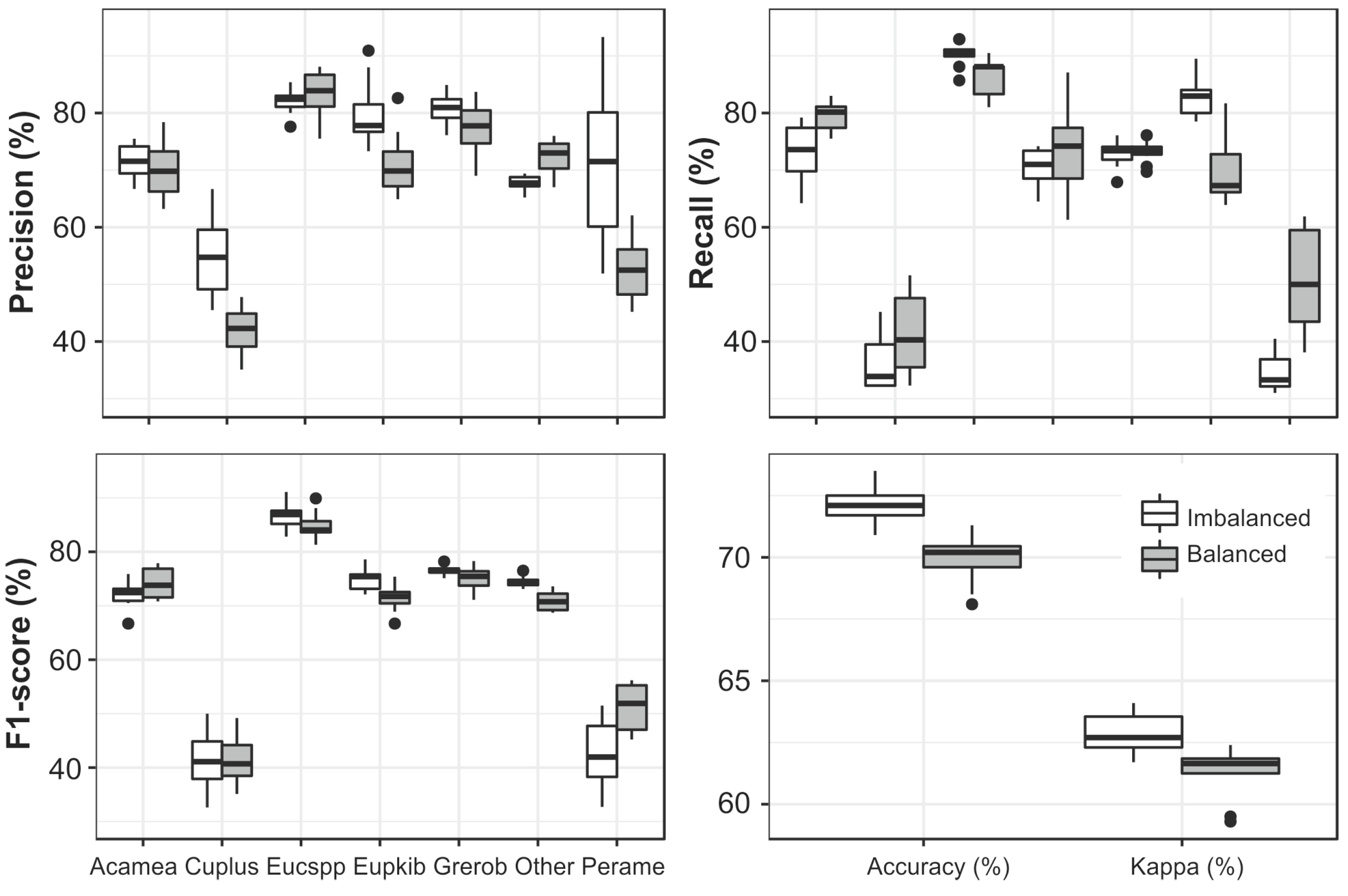
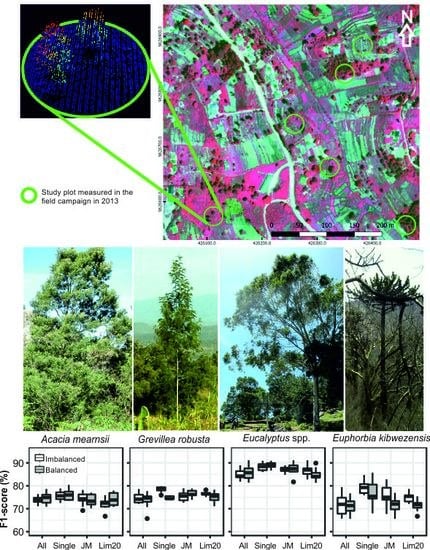
| Species | Abbreviation | Type | Crowns | Pixels |
|---|---|---|---|---|
| Grevillea robusta | Grerob | exotic | 109 | 5485 |
| Acacia mearnsii | Acamea | exotic | 53 | 2437 |
| Eucalyptus spp. | Eucspp | exotic | 42 | 2577 |
| Persea americana | Perame | exotic | 42 | 1989 |
| Cupressus lusitanica | Cuplus | exotic | 31 | 1641 |
| Euphorbia kibwezensis | Eupkib | native | 31 | 1181 |
| Eriobotrya japonica | Erijap | exotic | 14 | 516 |
| Ficus thonningii | Fictho | native | 14 | 948 |
| Maesa lanceolata | Maelan | native | 14 | 612 |
| Mangifera indica | Manind | exotic | 14 | 668 |
| Zimmermania ovata | Zimova | native | 13 | 674 |
| Zimmermania commiphora | Zimcom | native | 11 | 426 |
| Psidium guajava | Psigua | exotic | 10 | 459 |
| Erythrina abyssinica | Eryaby | native | 9 | 415 |
| Acacia seyal | Acasey | native | 8 | 417 |
| Phoenix reclinata | Phorec | native | 8 | 442 |
| Albizia gummifera | Albgum | native | 7 | 387 |
| Prunus africana | Pruafr | native | 7 | 410 |
| Bridelia micrantha | Brimic | native | 6 | 335 |
| Dombeya kirkii | Domkir | native | 6 | 138 |
| Ficus sur | Ficsur | native | 6 | 311 |
| Combretum collinum | Comcol | native | 5 | 234 |
| Cussonia spicata | Cusspi | native | 5 | 215 |
| Macademia spp. | Macspp | exotic | 5 | 122 |
| Millettia oblata | Milobl | native | 5 | 401 |
| Acacia tortilis | Acator | native | 4 | 90 |
| Dombeya rotundifolia | Domrot | native | 4 | 169 |
| Ficus sycomorus | Ficsyc | native | 4 | 228 |
| Newtonia buchananii | Newbuc | native | 4 | 385 |
| Syzygium spp. | Syzspp | native | 4 | 174 |
| Xymalos monospora | Xymmon | native | 4 | 123 |
| Support Vector Machine | Random Forest | ||||
|---|---|---|---|---|---|
| Feature Set | Accuracy | Kappa | Accuracy | Kappa | |
| Refl. | 37.9 | 30.9 | 31.7 | 21.6 | |
| NVI | 45.5 | 37.5 | 35.9 | 25.8 | |
| MNF | 53.3 | 46.8 | 51.3 | 44.8 | |
| ALS | 31.7 | 21.6 | 30.7 | 21.8 | |
| Refl.+ALS | 42.9 | 35.7 | 42.9 | 35.6 | |
| NVI+ALS | 43.5 | 37.2 | 44.3 | 37.5 | |
| MNF + ALS | 57.1 | 52.1 | 54.1 | 48.2 | |
| Feature Set | No Var. | Feature Names |
|---|---|---|
| Refl. | 19 | R406, R401, R553, R549, R414, R562, R419, R572, R769, R717, R576, R530, R526, R521, R581, R458, R688, R632, R674 |
| NVI | 8 | ACI, ARI, CIred edge, PRI, PSSR, mCARI, CRI1, EVI |
| MNF | 10 | MNF9, MNF1, MNF5, MNF7, MNF6, MNF4, MNF8, MNF10, MNF14, MNF2 |
| ALS | 5 | HD, MADmedian, P95, AADmedian, min |
| Refl. + ALS | 15 | HD, max, MADmedian, MADmean, AADmedian, R406, R562, R558, min, R414, R423, R726, R731, R540, R774 |
| NVI + ALS | 17 | ACI, HD, ARI, P95, max, MADmedian, CARI, AADmedian, AADmean, CRI2, CIred edge, HC, min, PRI, PSSR, SR, VIgreen |
| MNF + ALS | 13 | MNF9, HD, MNF5, MNF1, MNF4, MNF8, MNF7, MNF6, MNF11, MNF12, P95, MNF10, MNF14 |
| R = reflectance, MNF = minimum noise fraction | ||
| Group | Species in the Group | Samples |
|---|---|---|
| 1 | Macspp, Maelan, Erijap, Domrot, Xymmon, ComCol, Zimova | 59 |
| 2 | Zimcom, Eryaby, Acasey, Acator, Domkir | 38 |
| 3 | Perame, Ficsur, Phorec, Manind, Brimic, Syzspp | 80 |
| 4 | Psigua, Cusspi, Cuplus, Pruafr, Ficsyc, Newbuc, Fictho, Milobl, Albgum | 87 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piiroinen, R.; Heiskanen, J.; Maeda, E.; Viinikka, A.; Pellikka, P. Classification of Tree Species in a Diverse African Agroforestry Landscape Using Imaging Spectroscopy and Laser Scanning. Remote Sens. 2017, 9, 875. https://doi.org/10.3390/rs9090875
Piiroinen R, Heiskanen J, Maeda E, Viinikka A, Pellikka P. Classification of Tree Species in a Diverse African Agroforestry Landscape Using Imaging Spectroscopy and Laser Scanning. Remote Sensing. 2017; 9(9):875. https://doi.org/10.3390/rs9090875
Chicago/Turabian StylePiiroinen, Rami, Janne Heiskanen, Eduardo Maeda, Arto Viinikka, and Petri Pellikka. 2017. "Classification of Tree Species in a Diverse African Agroforestry Landscape Using Imaging Spectroscopy and Laser Scanning" Remote Sensing 9, no. 9: 875. https://doi.org/10.3390/rs9090875