Modulation of miRNAs by Vitamin C in Human Bone Marrow Stromal Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Human BMSCs (hBMSCs)
2.2. Osteogenic and Adipogenic Differentiation Assays
2.3. Vitamin C Treatment, Gene Expression Analysis and Intracellular Vitamin C Estimation
2.4. Microrna Array and Bioinformatics Analysis
3. Results
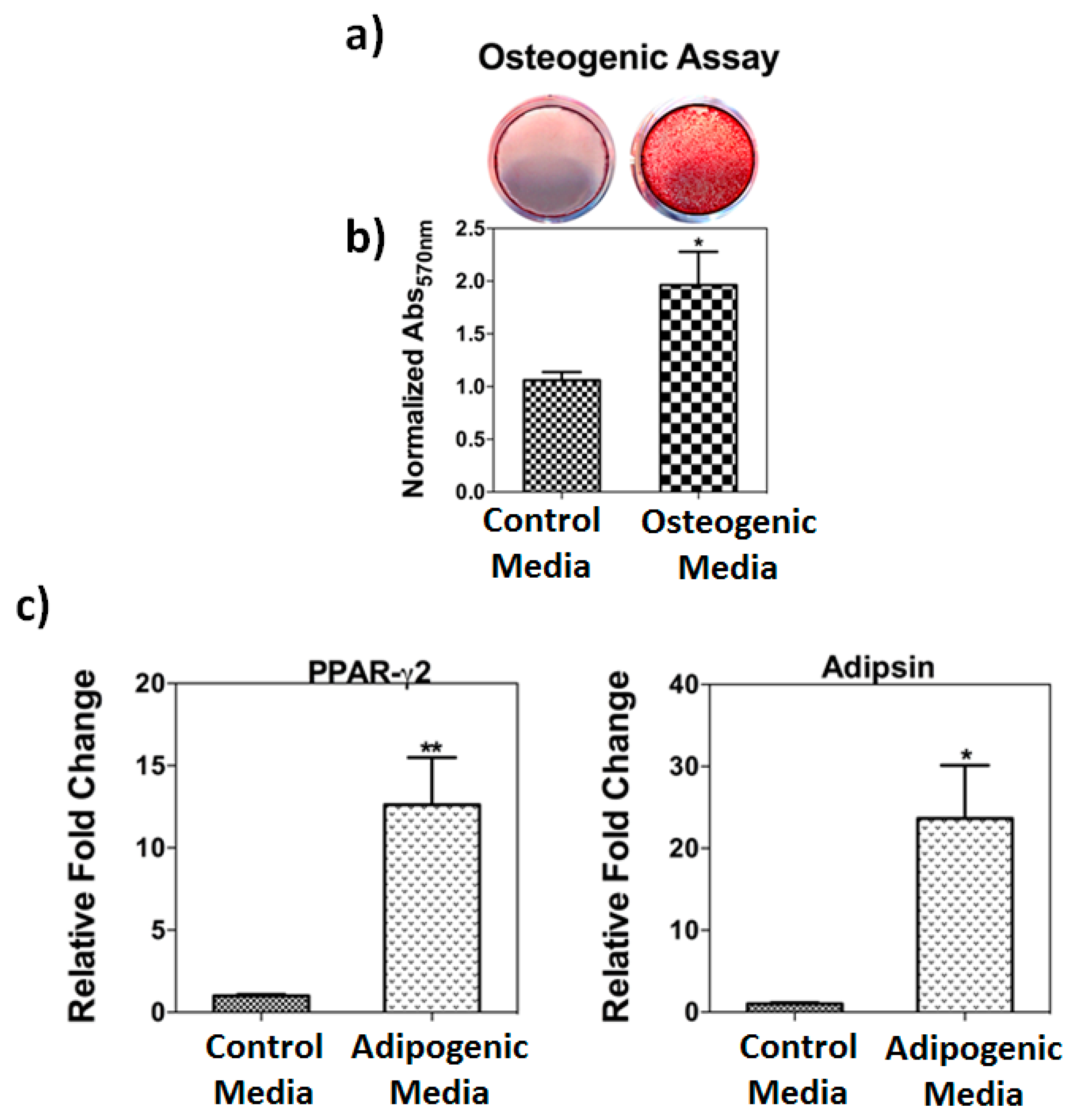
3.1. Osteogenic and Adipogenic Differentiation Assays
3.2. Vitamin C Regulates Musculoskeletal-Related Gene Expression in Hbmscs
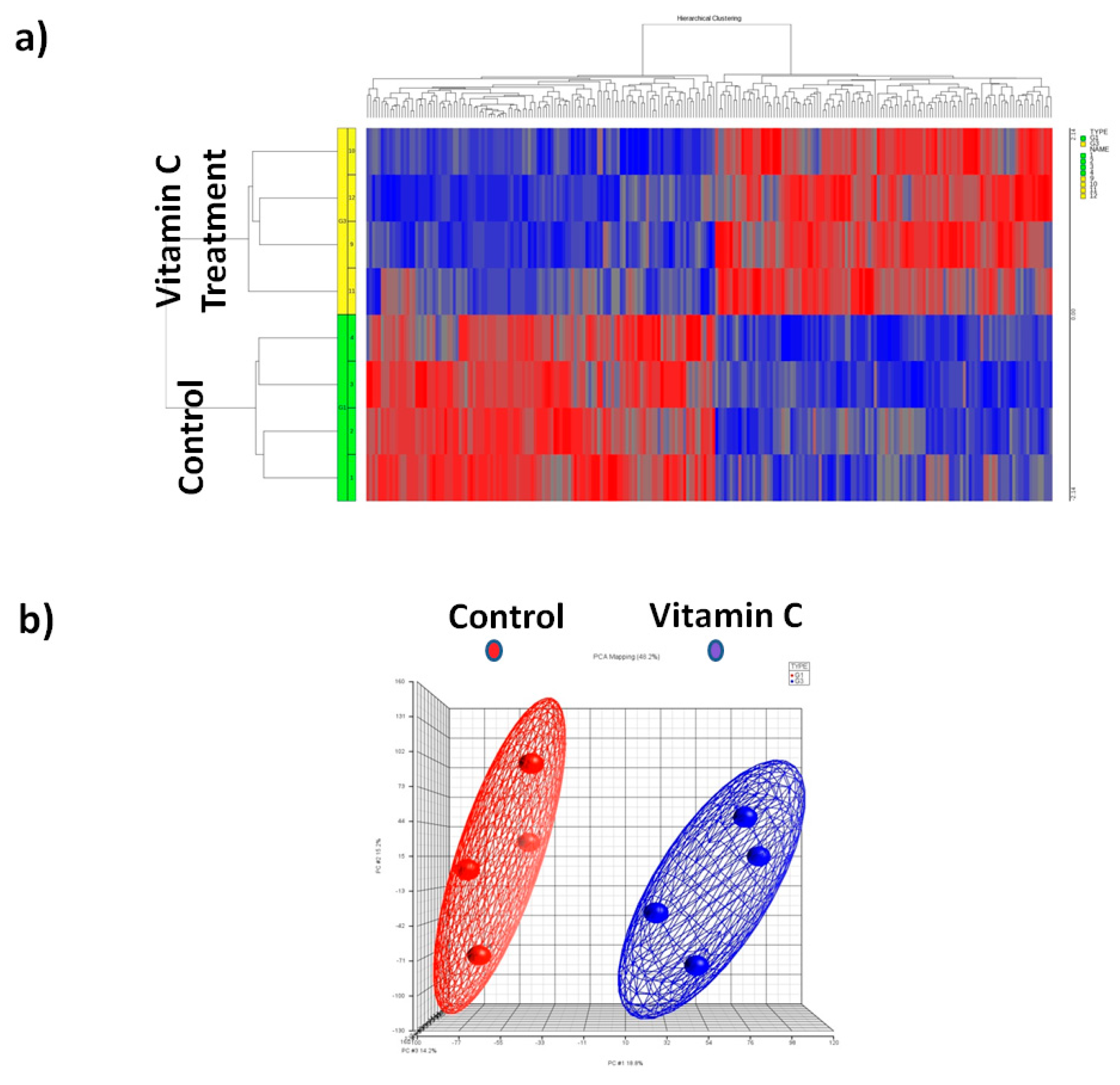
3.3. Global Mirna Expression Profile Following Vitamin C
3.4. Principal Component Analysis (PCA)
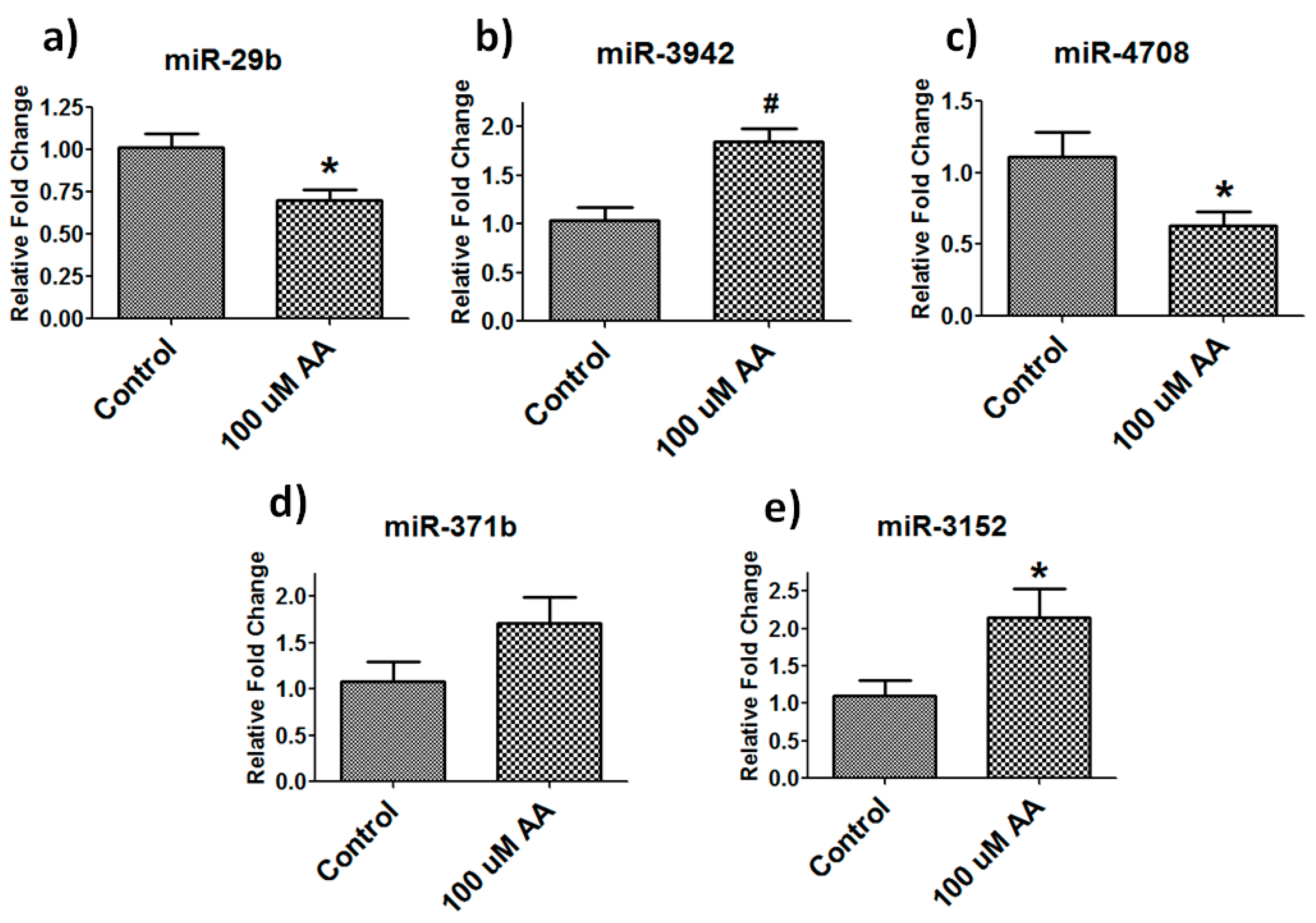
3.5. Validation of Differentially Expressed Mirnas
3.6. Signaling Pathway Predictions
3.7. Bioinformatics Mirna Target Prediction
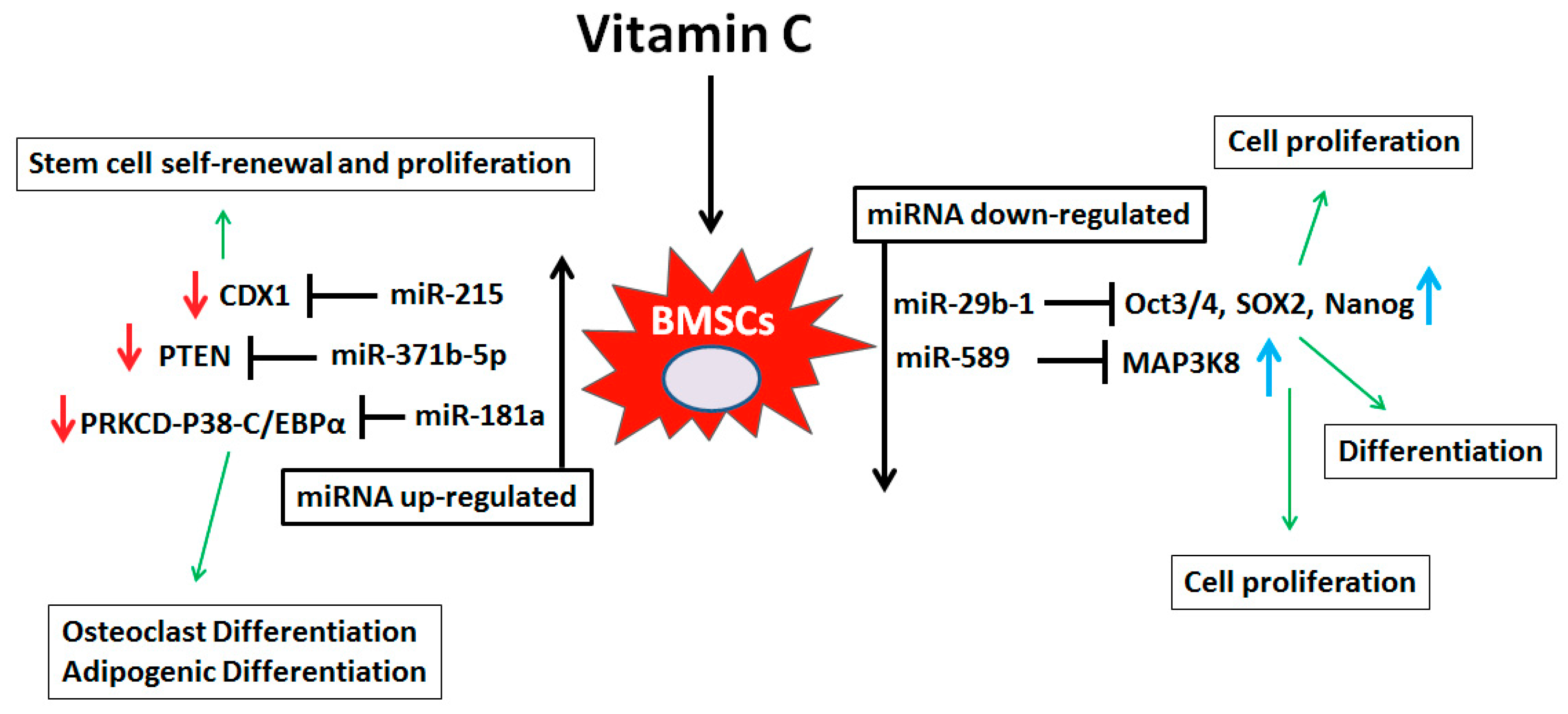
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgements
Author Contributions
Conflicts of Interest
References
- Jones, D.L.; Wagers, A.J. No place like home: Anatomy and function of the stem cell niche. Nat. Rev. Mol. Cell Biol. 2008, 9, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wei, S.; Liu, C.; Song, M.; Wu, H.; Yang, Y. Regulation of the osteogenic and adipogenic differentiation of bone marrow-derived stromal cells by extracellular uridine triphosphate: The role of P2Y2 receptor and ERK1/2 signaling. Int. J. Mol. Med. 2016, 37, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.X.; Gong, Y.Y.; Zhou, G.D.; Liu, W.; Cao, Y.; Zhang, W.J. Chondrogenic differentiation of bone marrow-derived mesenchymal stem cells induced by acellular cartilage sheets. Biomaterials 2012, 33, 5832–5840. [Google Scholar] [CrossRef] [PubMed]
- Rahman, F.; Bordignon, B.; Culerrier, R.; Peiretti, F.; Spicuglia, S.; Djabali, M.; Landrier, J.F.; Fontes, M. Ascorbic acid drives the differentiation of mesoderm-derived embryonic stem cells. Involvement of p38 MAPK/CREB and SVCT2 transporter. Mol. Nutr. Food Res. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Kilberg, M.S.; Terada, N.; Shan, J. Influence of Amino Acid Metabolism on Embryonic Stem Cell Function and Differentiation. Adv. Nutr. 2016, 7, 780S–789S. [Google Scholar] [CrossRef] [PubMed]
- Ochocki, J.D.; Simon, M.C. Nutrient-sensing pathways and metabolic regulation in stem cells. J. Cell Biol. 2013, 203, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Khillan, J.S. Vitamin A/retinol and maintenance of pluripotency of stem cells. Nutrients 2014, 6, 1209–1222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakayori, N.; Kimura, R.; Osumi, N. Impact of lipid nutrition on neural stem/progenitor cells. Stem Cells Int. 2013, 2013, 973508. [Google Scholar] [CrossRef] [PubMed]
- Sangani, R.; Periyasamy-Thandavan, S.; Pathania, R.; Ahmad, S.; Kutiyanawalla, A.; Kolhe, R.; Bhattacharyya, M.H.; Chutkan, N.; Hunter, M.; Hill, W.D.; et al. The crucial role of vitamin C and its transporter (SVCT2) in bone marrow stromal cell autophagy and apoptosis. Stem Cell Res. 2015, 15, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.H.; Ryu, H.; Rhee, K.J.; Oh, J.E.; Baik, S.K.; Shim, K.Y.; Kong, J.H.; Hyun, S.Y.; Pack, H.S.; Im, C.; et al. l-Ascorbic acid 2-phosphate and fibroblast growth factor-2 treatment maintains differentiation potential in bone marrow-derived mesenchymal stem cells through expression of hepatocyte growth factor. Growth Factors 2015, 33, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Aghajanian, P.; Hall, S.; Wongworawat, M.D.; Mohan, S. The Roles and Mechanisms of Actions of Vitamin C in Bone: New Developments. J. Bone Miner. Res. 2015, 30, 1945–1955. [Google Scholar] [CrossRef] [PubMed]
- Fain, O. Musculoskeletal manifestations of scurvy. Joint Bone Spine 2005, 72, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Fulzele, S.; Chothe, P.; Sangani, R.; Chutkan, N.; Hamrick, M.; Bhattacharyya, M.; Prasad, P.D.; Zakhary, I.; Bowser, M.; Isales, C.; et al. Sodium-dependent vitamin C transporter SVCT2, expression and function in bone marrow stromal cells and in osteogenesis. Stem Cell Res. 2013, 10, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.M.; Seo, Y.K.; Yoon, H.H.; Song, K.Y.; Kwon, S.Y.; Lee, H.S.; Park, J.K. Effect of ascorbic acid on bone marrow-derived mesenchymal stem cell proliferation and differentiation. J. Biosci. Bioeng. 2008, 105, 586–594. [Google Scholar] [CrossRef] [PubMed]
- Potdar, P.D.; D’Souza, S.B. Ascorbic acid induces in vitro proliferation of human subcutaneous adipose tissue derived mesenchymal stem cells with upregulation of embryonic stem cell pluripotency markers Oct4 and SOX2. Hum. Cell 2010, 23, 152–155. [Google Scholar] [CrossRef] [PubMed]
- Naidu, K.A. Vitamin C in human health and disease is still a mystery? An overview. Nutr. J. 2003, 2, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drouin, G.; Godin, J.R.; Pagé, B. The genetics of vitamin C loss in vertebrates. Curr. Genomics 2011, 12, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Nishikimi, M.; Yagi, K. Molecular basis for the deficiency in humans of gulonolactone oxidase, a key enzyme for ascorbic acid biosynthesis. Am. J. Clin. Nutr. 1991, 54 (Suppl. 6), 1203S–1208S. [Google Scholar] [CrossRef] [PubMed]
- Chung, T.L.; Brena, R.M.; Kolle, G.; Grimmond, S.M.; Berman, B.P.; Laird, P.W.; Pera, M.F.; Wolvetang, E.J. Vitamin C promotes widespread yet specific DNA demethylation of the epigenome in human embryonic stem cells. Stem Cells 2010, 28, 1848–1855. [Google Scholar] [CrossRef] [PubMed]
- Blaschke, K.; Ebata, K.T.; Karimi, M.M.; Zepeda-Martínez, J.A.; Goyal, P.; Mahapatra, S.; Tam, A.; Laird, D.J.; Hirst, M.; Rao, A.; et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature 2013, 500, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Poal-Manresa, J.; Little, K.; Trueta, J. Some observations on the effects of vitamin C deficiency on bone. Br. J. Exp. Pathol. 1970, 51, 372–378. [Google Scholar] [PubMed]
- Kipp, D.E.; McElvain, M.; Kimmel, D.B.; Akhter, M.P.; Robinson, R.G.; Lukert, B.P. Scurvy results in decreased collagen synthesis and bone density in the guinea pig animal model. Bone 1996, 18, 281–288. [Google Scholar] [CrossRef]
- Chen, J.; Guo, L.; Zhang, L.; Wu, H.; Yang, J.; Liu, H.; Wang, X.; Hu, X.; Gu, T.; Zhou, Z.; et al. Vitamin C modulates TET1 function during somatic cell reprogramming. Nat. Genet. 2013, 45, 1504–1509. [Google Scholar] [CrossRef] [PubMed]
- Ivanyuk, D.; Budash, G.; Zheng, Y.; Gaspar, J.A.; Chaudhari, U.; Fatima, A.; Bahmanpour, S.; Grin, V.K.; Popandopulo, A.G.; Sachinidis, A.; et al. Ascorbic Acid-Induced Cardiac Differentiation of Murine Pluripotent Stem Cells: Transcriptional Profiling and Effect of a Small Molecule Synergist of Wnt/β-Catenin Signaling Pathway. Cell. Physiol. Biochem. 2015, 36, 810–830. [Google Scholar] [CrossRef] [PubMed]
- Weinhold, B. Epigenetics: The science of change. Environ. Health Perspect. 2006, 114, A160–A167. [Google Scholar] [CrossRef] [PubMed]
- Fu, G.; Ren, A.; Qiu, Y.; Zhang, Y. Epigenetic Regulation of Osteogenic Differentiation of Mesenchymal Stem Cells. Curr. Stem Cell Res. Ther. 2016, 11, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Yannarelli, G.; Pacienza, N.; Cuniberti, L.; Medin, J.; Davies, J.; Keating, A. Brief report: The potential role of epigenetics on multipotent cell differentiation capacity of mesenchymal stromal cells. Stem Cells 2013, 31, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Challen, G.A.; Sun, D.; Jeong, M.; Luo, M.; Jelinek, J.; Berg, J.S.; Bock, C.; Vasanthakumar, A.; Gu, H.; Xi, Y.; et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat. Genet. 2011, 44, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Arnsdorf, E.J.; Tummala, P.; Castillo, A.B.; Zhang, F.; Jacobs, C.R. The epigenetic mechanism of mechanically induced osteogenic differentiation. J. Biomech. 2010, 43, 2881–2886. [Google Scholar] [CrossRef] [PubMed]
- Alexanian, A.R. Epigenetic modifiers promote efficient generation of neural-like cells from bone marrow-derived mesenchymal cells grown in neural environment. J. Cell. Biochem. 2007, 100, 362–371. [Google Scholar] [CrossRef] [PubMed]
- Sangani, R.; Periyasamy-Thandavan, S.; Kolhe, R.; Bhattacharyya, M.H.; Chutkan, N.; Hunter, M.; Isales, C.; Hamrick, M.; Hill, W.D.; Fulzele, S. MicroRNAs-141 and 200a regulate the SVCT2 transporter in bone marrow stromal cells. Mol. Cell. Endocrinol. 2015, 410, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Ebata, K.T.; Mesh, K.; Liu, S.; Bilenky, M.; Fekete, A.; Acker, M.G.; Hirst, M.; Garcia, B.A.; Ramalho-Santos, M. Vitamin C induces specific demethylation of H3K9me2 in mouse embryonic stem cells via Kdm3a/b. Epigenet. Chromatin 2017, 10, 36. [Google Scholar] [CrossRef] [PubMed]
- Abbey, D.; Seshagiri, P.B. Ascorbic acid-mediated enhanced cardiomyocytes differentiation of mouse ES-cells involves interplay of DNA methylation and multiple-signals. Differentiation 2017, 96, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Eid, W.; Abdel-Rehim, W. Vitamin C promotes pluripotency of human induced pluripotent stem cells via the histone demethylase JARID1A. Biol. Chem. 2016, 397, 1205–1213. [Google Scholar] [CrossRef] [PubMed]
- Camarena, V.; Wang, G. The epigenetic role of vitamin C in health and disease. Cell. Mol. Life Sci. 2016, 73, 1645–1658. [Google Scholar] [CrossRef] [PubMed]
- Young, J.I.; Züchner, S.; Wang, G. Regulation of the Epigenome by Vitamin C. Annu. Rev. Nutr. 2015, 35, 545–564. [Google Scholar] [CrossRef] [PubMed]
- Yin, R.; Mao, S.Q.; Zhao, B.; Chong, Z.; Yang, Y.; Zhao, C.; Zhang, D.; Huang, H.; Gao, J.; Li, Z.; et al. Ascorbic acid enhances Tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. J. Am. Chem. Soc. 2013, 135, 10396–10403. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, F.E.; Lopez-Martinez, A.F. MicroRNAs in human diseases. Recent Pat. DNA Gene Seq. 2010, 4, 142–154. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Santos, M.C.; Aragón-Raygoza, A.; Espinal-Centeno, A.; Arteaga-Vázquez, M.; Cruz-Hernández, A.; Bako, L.; Cruz-Ramírez, A. The Role of microRNAs in Animal Cell Reprogramming. Stem Cells Dev. 2016, 25, 1035–1049. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, J.; Ruohola-Baker, H. Regulation of stem cell populations by microRNAs. Adv. Exp. Med. Biol. 2013, 786, 329–351. [Google Scholar] [PubMed]
- Luo, W.; Nie, Q.; Zhang, X. MicroRNAs involved in skeletal muscle differentiation. J. Genet. Genomics 2013, 40, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Yi, R.; Fuchs, E. MicroRNAs and their roles in mammalian stem cells. J. Cell Sci. 2011, 124, 1775–1783. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, G.; Giuffrida, R.; Lo Furno, D.; Parrinello, N.L.; Forte, S.; Gulino, R.; Colarossi, C.; Schinocca, L.R.; Giuffrida, R.; Cardile, V.; et al. Potential Effect of CD271 on Human Mesenchymal Stromal Cell Proliferation and Differentiation. Int. J. Mol. Sci. 2015, 16, 15609–15624. [Google Scholar] [CrossRef] [PubMed]
- Cuthbert, R.J.; Giannoudis, P.V.; Wang, X.N.; Nicholson, L.; Pawson, D.; Lubenko, A.; Tan, H.B.; Dickinson, A.; McGonagle, D.; Jones, E. Examining the feasibility of clinical grade CD271 + enrichment of mesenchymal stromal cells for bone regeneration. PLoS ONE 2015, 10, e0117855. [Google Scholar] [CrossRef] [PubMed]
- Kolhe, R.; Hunter, M.; Liu, S.; Jadeja, R.N.; Pundkar, C.; Mondal, A.K.; Mendhe, B.; Drewry, M.; Rojiani, M.V.; Liu, Y.; et al. Gender-specific differential expression of exosomal miRNA in synovial fluid of patients with osteoarthritis. Sci. Rep. 2017, 7, 2029. [Google Scholar] [CrossRef] [PubMed]
- Tuan, R.S.; Boland, G.; Tuli, R. Adult mesenchymal stem cells and cell-based tissue engineering. Arthritis Res. Ther. 2003, 5, 32–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, X.L.; Xu, M.J.; Li, Z.H.; Pan, Q.; Fu, J.H. Effects of vitamin E on expressions of eight microRNAs in the liver of Nile tilapia (Oreochromis niloticus). Fish Shellfish Immunol. 2013, 34, 1470–1475. [Google Scholar] [CrossRef] [PubMed]
- Gaedicke, S.; Zhang, X.; Schmelzer, C.; Lou, Y.; Doering, F.; Frank, J.; Rimbach, G. Vitamin E dependent microRNA regulation in rat liver. FEBS Lett. 2008, 582, 3542–3546. [Google Scholar] [CrossRef] [PubMed]
- Merrigan, S.L.; Kennedy, B.N. Vitamin D receptor agonists regulate ocular developmental angiogenesis and modulate expression of dre-miR-21 and VEGF. Br. J. Pharmacol. 2017, 174, 2636–2651. [Google Scholar] [CrossRef] [PubMed]
- Arboleda, J.F.; Urcuqui-Inchima, S. Vitamin D-Regulated MicroRNAs: Are They Protective Factors against Dengue Virus Infection? Adv. Virol. 2016, 2016, 1016840. [Google Scholar] [CrossRef] [PubMed]
- Drummond, M.J.; Glynn, E.L.; Fry, C.S.; Dhanani, S.; Volpi, E.; Rasmussen, B.B. Essential amino acids increase microRNA-499, -208b, and -23a and downregulate myostatin and myocyte enhancer factor 2C mRNA expression in human skeletal muscle. J. Nutr. 2009, 139, 2279–2284. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.M.; Lim, S.M.; Yoo, J.A.; Woo, M.J.; Cho, K.H. Consumption of high-dose vitamin C (1250 mg per day) enhances functional and structural properties of serum lipoprotein to improve anti-oxidant, anti-atherosclerotic, and anti-aging effects via regulation of anti-inflammatory microRNA. Food Funct. 2015, 6, 3604–3612. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Han, Z.; Li, Q.; Wu, Y.; Shi, X.; Ai, Z.; Du, J.; Li, W.; Guo, Z.; Zhang, Y. Vitamin C induces a pluripotent state in mouse embryonic stem cells by modulating microRNA expression. FEBS J. 2015, 282, 685–699. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Ku, S.Y.; Rosenwaks, Z.; Liu, H.C.; Chi, S.W.; Kang, J.S.; Lee, W.J.; Jung, K.C.; Kim, S.H.; Choi, Y.M.; et al. MicroRNA expression profiles are altered by gonadotropins and vitamin C status during in vitro follicular growth. Reprod. Sci. 2010, 17, 1081–1089. [Google Scholar] [PubMed]
- Di Fiore, R.; Drago-Ferrante, R.; Pentimalli, F.; Di Marzo, D.; Forte, I.M.; D’Anneo, A.; Carlisi, D.; De Blasio, A.; Giuliano, M.; Tesoriere, G.; et al. MicroRNA-29b-1 impairs in vitro cell proliferation, self-renewal and chemoresistance of human osteosarcoma 3AB-OS cancer stem cells. Int. J. Oncol. 2014, 45, 2013–2023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Jiang, P.; Shuai, L.; Chen, K.; Li, Z.; Zhang, Y.; Jiang, Y.; Li, X. miR-589-5p inhibits MAP3K8 and suppresses CD90(+) cancer stem cells in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2016, 35, 176. [Google Scholar] [CrossRef] [PubMed]
- Quan, Y.; Wang, Z.; Gong, L.; Peng, X.; Richard, M.A.; Zhang, J.; Fornage, M.; Alcorn, J.L.; Wang, D. Exosome miR-371b-5p promotes proliferation of lung alveolar progenitor type II cells by using PTEN to orchestrate the PI3K/Akt signaling. Stem Cell Res. Ther. 2017, 8, 138. [Google Scholar] [CrossRef] [PubMed]
- Su, R.; Lin, H.S.; Zhang, X.H.; Yin, X.L.; Ning, H.M.; Liu, B.; Zhai, P.F.; Gong, J.N.; Shen, C.; Song, L.; et al. MiR-181 family: Regulators of myeloid differentiation and acute myeloid leukemia as well as potential therapeutic targets. Oncogene 2015, 34, 3226–3239. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.F.; Hara, T.; Francis, P.; Li, X.L.; Bilke, S.; Zhu, Y.; Pineda, M.; Subramanian, M.; Bodmer, W.F.; Lal, A. The CDX1-microRNA-215 axis regulates colorectal cancer stem cell differentiation. Proc. Natl. Acad. Sci. USA 2015, 112, E1550–E1558. [Google Scholar] [CrossRef] [PubMed]
- Pustylnik, S.; Fiorino, C.; Nabavi, N.; Zappitelli, T.; da Silva, R.; Aubin, J.E.; Harrison, R.E. EB1 levels are elevated in ascorbic Acid (AA)-stimulated osteoblasts and mediate cell-cell adhesion-induced osteoblast differentiation. J. Biol. Chem. 2013, 288, 22096–22110. [Google Scholar] [CrossRef] [PubMed]
- Jubiz, W.; Ramirez, M. Effect of vitamin C on the absorption of levothyroxine in patients with hypothyroidism and gastritis. J. Clin. Endocrinol. Metab. 2014, 99, E1031–E1034. [Google Scholar] [CrossRef] [PubMed]
- Williams, G.R.; Bassett, J.H.D. Thyroid diseases and bone health. J. Endocrinol. Investig. 2017, 2, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Williams, G.R. Thyroid hormone actions in cartilage and bone. Eur. Thyroid. J. 2013, 2, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Bayerle-Eder, M.; Pleiner, J.; Mittermayer, F.; Schaller, G.; Roden, M.; Waldhäusl, W.; Bieglmayer, C.; Wolzt, M. Effect of systemic vitamin C on free fatty acid-induced lipid peroxidation. Diabetes Metab. 2004, 30, 433–439. [Google Scholar] [CrossRef]
- Ginter, E.; Ondreicka, R.; Bobek, P.; Simko, V. The influence of chronic vitamin C deficiency on fatty acid composition of blood serum, liver triglycerides and cholesterol esters in guinea pigs. J. Nutr. 1969, 99, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.A.; Haque, M.; Akbar, M. Role of Polyunsaturated Fatty Acids and Their Metabolites on Stem Cell Proliferation and Differentiation. Adv. Neurobiol. 2016, 12, 367–380. [Google Scholar] [PubMed]
- Das, U.N. Influence of polyunsaturated fatty acids and their metabolites on stem cell biology. Nutrition 2011, 27, 21–25. [Google Scholar] [CrossRef] [PubMed]

| MicroRNA ID/Probeset ID | Fold-Change | p-Value |
|---|---|---|
| hsa-miR-3651_st | −3.76469 | 0.000718 |
| hsa-miR-4485_st | −3.70276 | 0.002619 |
| hsa-miR-1275_st | −3.00098 | 0.022393 |
| hsa-miR-4708-5p_st | −2.94613 | 0.002456 |
| hsa-miR-3197_st | −2.86102 | 0.048879 |
| hsa-miR-720_st | −2.81658 | 0.019125 |
| hsa-miR-210_st | −2.75739 | 0.001763 |
| hsa-miR-29b-1-star_st | −2.71192 | 0.032586 |
| hsa-miR-4284_st | −2.49253 | 0.003259 |
| hsa-miR-4479_st | −2.39325 | 0.026759 |
| hsa-miR-3175_st | −2.37271 | 0.013122 |
| hsa-miR-4730_st | −2.36644 | 0.009171 |
| hsa-miR-23a-star_st | −2.26148 | 0.001843 |
| hsa-miR-4321_st | −2.25569 | 0.024075 |
| hp_hsa-mir-3676_st | −2.13436 | 0.008900 |
| hsa-miR-4787-3p_st | −2.05594 | 0.003156 |
| hsa-miR-574-5p_st | −2.02613 | 0.019283 |
| hsa-miR-4492_st | −1.99221 | 0.011873 |
| hsa-miR-345_st | −1.96585 | 0.019316 |
| hsa-miR-1270_st | −1.90283 | 0.047187 |
| hsa-miR-4697-5p_st | −1.83134 | 0.039445 |
| hsa-miR-4433_st | −1.80613 | 0.005357 |
| hp_hsa-mir-4477a_st | 1.83543 | 0.0351021 |
| hp_hsa-mir-548ag-2_st | 1.83747 | 0.0217069 |
| hsa-miR-4727-3p_st | 1.86033 | 0.0446828 |
| hsa-miR-335_st | 1.86288 | 0.0415722 |
| hsa-miR-202_st | 1.87834 | 0.00218913 |
| hsa-miR-4436a_st | 1.96484 | 0.00880198 |
| hp_hsa-mir-532_st | 1.98369 | 0.035662 |
| hsa-miR-3942-5p_st | 1.98754 | 0.002823 |
| hsa-miR-3163_st | 2.0011 | 0.019565 |
| hp_hsa-mir-548f-1_st | 2.01894 | 0.026558 |
| hsa-miR-92a-2-star_st | 2.02393 | 0.003107 |
| hsa-miR-548a-3p_st | 2.02776 | 0.003973 |
| hsa-miR-3121-3p_st | 2.03509 | 0.007681 |
| hsa-miR-3201_st | 2.07949 | 0.006952 |
| hsa-miR-4657_st | 2.07988 | 0.027179 |
| hsa-miR-4704-5p_st | 2.10426 | 0.002415 |
| hsa-miR-1825_st | 2.1387 | 0.028925 |
| hsa-miR-550a-star_st | 2.16711 | 0.017221 |
| hsa-miR-1323_st | 2.21591 | 0.006721 |
| hsa-miR-3927_st | 2.23141 | 0.002801 |
| hsa-miR-509-3-5p_st | 2.2723 | 0.003025 |
| hsa-miR-4423-3p_st | 2.27939 | 0.030500 |
| hsa-miR-890_st | 2.34751 | 0.035107 |
| hsa-miR-4773_st | 2.41459 | 0.002370 |
| hsa-miR-371b-5p_st | 2.42868 | 0.038520 |
| hsa-miR-3128_st | 2.49419 | 0.014536 |
| hsa-miR-1272_st | 2.59151 | 0.033417 |
| hsa-miR-4659a-3p_st | 2.60253 | 0.005995 |
| hsa-miR-377_st | 2.64791 | 0.003494 |
| hsa-miR-550b_st | 2.68252 | 0.013600 |
| hsa-miR-20b-star_st | 2.76807 | 0.004101 |
| hsa-miR-3152-3p_st | 3.2214 | 0.002098 |
| hsa-miR-1208_st | 5.02485 | 0.010838 |
| hsa-miR-4529-3p_st | 12.0104 | 0.009329 |
| (a) | |||
| KEGG Pathway | p-Value | Number of Genes Involved | Number of miRNAs Involved |
| Prion diseases | 4.07 × 10−10 | 7 | 7 |
| Morphine addiction | 4.80 × 10−5 | 32 | 17 |
| Amino sugar and nucleotide sugar metabolism | 0.002142 | 16 | 11 |
| Thyroid hormone signaling pathway | 0.002958 | 37 | 17 |
| Cell adhesion molecules (CAMs) | 0.006179 | 43 | 16 |
| Endocytosis | 0.009192 | 63 | 18 |
| Oxytocin signaling pathway | 0.011941 | 53 | 21 |
| Melanogenesis | 0.013774 | 35 | 17 |
| MAPK signaling pathway | 0.013774 | 77 | 22 |
| GABAergic synapse | 0.02274 | 32 | 18 |
| Vasopressin-regulated water reabsorption | 0.035605 | 16 | 11 |
| Adrenergic signaling in cardiomyocytes | 0.035605 | 48 | 22 |
| Glutamatergic synapse | 0.03746 | 34 | 13 |
| Biosynthesis of unsaturated fatty acids | 0.040121 | 7 | 5 |
| Circadian entrainment | 0.046058 | 37 | 16 |
| (b) | |||
| KEGG Pathway | p-Value | Number of Genes Involved | Number of miRNAs Involved |
| Mucin type O-Glycan biosynthesis | 1.69 × 10−11 | 11 | 7 |
| Fatty acid biosynthesis | 1.86 × 10−8 | 1 | 1 |
| Glycosphingolipid biosynthesis-lacto and neolacto series | 6.27 × 10−5 | 9 | 8 |
| Fatty acid metabolism | 0.000184 | 7 | 6 |
| Adherens junction | 0.000623 | 25 | 18 |
| Biotin metabolism | 0.010804 | 1 | 1 |
| Arrhythmogenic right ventricular cardiomyopathy | 0.014001 | 21 | 18 |
| Caffeine metabolism | 0.027365 | 2 | 2 |
| Thyroid hormone signaling pathway | 0.027365 | 32 | 21 |
| (a) | |||
| GO Category | p-Value | Genes | miRNAs |
| organelle | 1.15 × 10−87 | 2073 | 24 |
| ion binding | 7.43 × 10−46 | 1270 | 25 |
| cellular nitrogen compound metabolic process | 7.82 × 10−45 | 1006 | 24 |
| biosynthetic process | 1.56 × 10−32 | 856 | 24 |
| small molecule metabolic process | 1.56 × 10−17 | 489 | 24 |
| cellular protein modification process | 3.92 × 10−15 | 475 | 24 |
| neurotrophin TRK receptor signaling pathway | 2.05 × 10−14 | 70 | 18 |
| synaptic transmission | 2.05 × 10−14 | 121 | 21 |
| nucleic acid binding transcription factor activity | 1.32 × 10−13 | 231 | 24 |
| cellular protein metabolic process | 2.55 × 10−11 | 106 | 20 |
| cell-cell signaling | 1.27 × 10−10 | 162 | 24 |
| catabolic process | 1.50 × 10−10 | 390 | 24 |
| cellular component assembly | 2.41 × 10−9 | 272 | 23 |
| molecular_function | 2.43 × 10−9 | 3342 | 25 |
| gene expression | 2.95 × 10−9 | 116 | 22 |
| symbiosis, encompassing mutualism through parasitism | 4.66 × 10−9 | 111 | 21 |
| post-translational protein modification | 4.93 × 10−9 | 46 | 17 |
| cellular_component | 5.00 × 10−9 | 3383 | 25 |
| blood coagulation | 1.28 × 10−8 | 101 | 20 |
| viral process | 2.94 × 10−8 | 97 | 21 |
| protein binding transcription factor activity | 7.72 × 10−8 | 110 | 22 |
| macromolecular complex assembly | 1.45 × 10−7 | 185 | 23 |
| enzyme binding | 1.74 × 10−7 | 261 | 23 |
| Fc-epsilon receptor signaling pathway | 5.08 × 10−7 | 38 | 15 |
| response to stress | 1.46 × 10−6 | 437 | 25 |
| toll-like receptor TLR1:TLR2 signaling pathway | 2.79 × 10−6 | 21 | 10 |
| toll-like receptor TLR6:TLR2 signaling pathway | 2.79 × 10−6 | 21 | 10 |
| toll-like receptor 10 signaling pathway | 3.62 × 10−6 | 20 | 10 |
| membrane organization | 5.32 × 10−6 | 121 | 23 |
| energy reserve metabolic process | 8.12 × 10−6 | 30 | 13 |
| TRIF-dependent toll-like receptor signaling pathway | 1.46 × 10−5 | 21 | 10 |
| protein complex assembly | 1.95 × 10−5 | 158 | 23 |
| MyD88-independent toll-like receptor signaling pathway | 2.89 × 10−5 | 22 | 11 |
| protein complex | 3.71 × 10−5 | 708 | 24 |
| toll-like receptor 5 signaling pathway | 3.75 × 10−5 | 20 | 10 |
| toll-like receptor 9 signaling pathway | 4.84 × 10−5 | 21 | 10 |
| Golgi lumen | 5.79 × 10−5 | 26 | 11 |
| O-glycan processing | 7.15 × 10−5 | 18 | 12 |
| immune system process | 0.000127 | 309 | 25 |
| mitotic cell cycle | 0.00013 | 74 | 19 |
| cytoskeletal protein binding | 0.000137 | 155 | 22 |
| generation of precursor metabolites and energy | 0.000171 | 74 | 18 |
| regulation of rhodopsin mediated signaling pathway | 0.000201 | 12 | 10 |
| toll-like receptor 4 signaling pathway | 0.000201 | 26 | 11 |
| inositol phosphate metabolic process | 0.000219 | 17 | 9 |
| nucleobase-containing compound catabolic process | 0.000385 | 168 | 24 |
| toll-like receptor 2 signaling pathway | 0.000449 | 21 | 10 |
| platelet degranulation | 0.000563 | 20 | 15 |
| glycosaminoglycan metabolic process | 0.000573 | 26 | 13 |
| platelet activation | 0.000643 | 44 | 18 |
| (b) | |||
| GO Category | p-Value | Genes | miRNAs |
| organelle | 1.67 × 10−95 | 2091 | 47 |
| ion binding | 3.27 × 10−55 | 1303 | 47 |
| cellular nitrogen compound metabolic process | 7.65 × 10−47 | 1010 | 46 |
| biosynthetic process | 1.19 × 10−35 | 866 | 46 |
| gene expression | 7.04 × 10−23 | 151 | 38 |
| cellular protein modification process | 1.18 × 10−22 | 507 | 46 |
| small molecule metabolic process | 7.20 × 10−18 | 488 | 45 |
| nucleic acid binding transcription factor activity | 1.08 × 10−12 | 227 | 43 |
| molecular_function | 2.35 × 10−11 | 3343 | 48 |
| cellular_component | 8.85 × 10−10 | 3375 | 48 |
| cellular protein metabolic process | 1.27 × 10−9 | 101 | 35 |
| enzyme binding | 1.33 × 10−9 | 272 | 43 |
| protein binding transcription factor activity | 5.19 × 10−9 | 114 | 37 |
| cellular component assembly | 9.08 × 10−9 | 268 | 42 |
| viral process | 1.39 × 10−8 | 98 | 31 |
| blood coagulation | 2.54 × 10−8 | 100 | 35 |
| symbiosis, encompassing mutualism through parasitism | 6.42 × 10−8 | 107 | 32 |
| neurotrophin TRK receptor signaling pathway | 9.66 × 10−8 | 56 | 28 |
| catabolic process | 9.95 × 10−8 | 371 | 44 |
| cell death | 3.44 × 10−7 | 195 | 40 |
| protein complex | 5.91 × 10−7 | 724 | 46 |
| enzyme regulator activity | 7.70 × 10−7 | 179 | 36 |
| platelet activation | 9.11 × 10−7 | 52 | 25 |
| membrane organization | 2.77 × 10−6 | 122 | 36 |
| post-translational protein modification | 3.62 × 10−6 | 40 | 23 |
| Fc-gamma receptor signaling pathway involved in phagocytosis | 4.22 × 10−6 | 22 | 13 |
| cytoskeletal protein binding | 1.60 × 10−5 | 160 | 37 |
| nucleoplasm | 1.79 × 10−5 | 233 | 43 |
| cytosol | 2.00 × 10−5 | 523 | 43 |
| cell junction organization | 2.35 × 10−5 | 42 | 24 |
| nucleobase-containing compound catabolic process | 3.39 × 10−5 | 174 | 41 |
| transmembrane transporter activity | 3.63 × 10−5 | 219 | 38 |
| macromolecular complex assembly | 3.63 × 10−5 | 172 | 39 |
| toll-like receptor TLR1:TLR2 signaling pathway | 5.04 × 10−5 | 19 | 15 |
| toll-like receptor TLR6:TLR2 signaling pathway | 5.04 × 10−5 | 19 | 15 |
| TRIF-dependent toll-like receptor signaling pathway | 5.32 × 10−5 | 20 | 16 |
| toll-like receptor 10 signaling pathway | 6.86 × 10−5 | 18 | 15 |
| Fc-epsilon receptor signaling pathway | 8.73 × 10−5 | 33 | 19 |
| cellular component disassembly involved in execution phase of apoptosis | 0.000104 | 15 | 11 |
| vitamin metabolic process | 0.000237 | 21 | 19 |
| MyD88-independent toll-like receptor signaling pathway | 0.000351 | 20 | 16 |
| homeostatic process | 0.000351 | 168 | 41 |
| mitotic cell cycle | 0.000368 | 72 | 34 |
| protein complex assembly | 0.000392 | 150 | 37 |
| water-soluble vitamin metabolic process | 0.000403 | 19 | 17 |
| toll-like receptor 5 signaling pathway | 0.000491 | 18 | 15 |
| energy reserve metabolic process | 0.000538 | 26 | 18 |
| glycerophospholipid biosynthetic process | 0.000692 | 23 | 14 |
| activation of signaling protein activity involved in unfolded protein response | 0.000692 | 18 | 16 |
| transcription initiation from RNA polymerase II promoter | 0.000793 | 52 | 25 |
| toll-like receptor 9 signaling pathway | 0.001896 | 18 | 15 |
| miRNA | No. of Targets | Stem Cell Related Genes | |
|---|---|---|---|
| MiRDB Scan | Target Scan | Common Targets | |
| hsa-miR-3619-5p | 788 | 5948 | PPARGC1B, RUNX3, DLX3, TGFBRAP1, TNFAIP1, TRAF1, TRAF3, TRAF5, TNFAIP8L1, MMP24, SOX8, QSOX2, WNT3 |
| hsa-miR-548a-3p | 947 | 6717 | BMPR2, TGFBR3, TGFBR1, TAB2, TWISTNB, CDC42BPB, IL6R, TRAF6, SMAD4, SMAD1, SMAD5, MMP2, WNT3 |
| hsa-miR-3942-5p | 437 | 3758 | DLX1, BMPR2, BMP2K, TGFB2, IL6R, TNFRSF11A, TRAF3, SMAD1 |
| hsa-miR-4741 | 253 | 4549 | TAB2, SMURF1, SNIP1 |
| hsa-miR-1825 | 253 | 4160 | PPARD, RUNX2, TGFBRAP1, TGFBR1, CD40, SOX6 |
| hsa-miR-1208 | 304 | 4651 | TGFB2, TWISTNB, SMURF1, MMP16, LEPROT |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kolhe, R.; Mondal, A.K.; Pundkar, C.; Periyasamy-Thandavan, S.; Mendhe, B.; Hunter, M.; Isales, C.M.; Hill, W.D.; Hamrick, M.W.; Fulzele, S. Modulation of miRNAs by Vitamin C in Human Bone Marrow Stromal Cells. Nutrients 2018, 10, 186. https://doi.org/10.3390/nu10020186
Kolhe R, Mondal AK, Pundkar C, Periyasamy-Thandavan S, Mendhe B, Hunter M, Isales CM, Hill WD, Hamrick MW, Fulzele S. Modulation of miRNAs by Vitamin C in Human Bone Marrow Stromal Cells. Nutrients. 2018; 10(2):186. https://doi.org/10.3390/nu10020186
Chicago/Turabian StyleKolhe, Ravindra, Ashis K. Mondal, Chetan Pundkar, Sudharsan Periyasamy-Thandavan, Bharati Mendhe, Monte Hunter, Carlos M. Isales, William D. Hill, Mark W. Hamrick, and Sadanand Fulzele. 2018. "Modulation of miRNAs by Vitamin C in Human Bone Marrow Stromal Cells" Nutrients 10, no. 2: 186. https://doi.org/10.3390/nu10020186





