Prebiotics Modulate the Effects of Antibiotics on Gut Microbial Diversity and Functioning in Vitro
Abstract
:1. Introduction
2. Experimental Section
2.1. Faecal Sample Preparation
2.2. Batch Culture Fermentation
2.3. DNA Extraction and 16S Sequencing
2.4. SCFA Analyses
2.5. PYY Release Assays
2.6. Statistical Analyses
3. Results
3.1. Starting Composition

3.2. Changes in Bacterial Abundance
| Response Variable | Explanatory Term | Sum Squares | Mean Squares | Degrees of Freedom | F | p |
|---|---|---|---|---|---|---|
| Total bacteria | Diet | 0.159 | 0.079 | 2.22 | 3.86 | 0.0365 |
| Antibiotic | 0.239 | 0.120 | 2.22 | 5.81 | 0.0094 | |
| Time | 0.087 | 0.044 | 2.75 | 2.12 | 0.1274 | |
| Antibiotic:Time | 0.377 | 0.094 | 4.75 | 4.58 | 0.0023 | |
| Bacteroides | Diet | 0.668 | 0.334 | 2.27 | 6.34 | 0.0055 |
| Antibiotic | 1.006 | 0.503 | 2.27 | 9.54 | 0.0007 | |
| Time | 1.949 | 0.975 | 2.81 | 18.48 | <0.0001 | |
| Diet:Antibiotic | 0.355 | 0.089 | 4.27 | 1.68 | 0.1826 | |
| Diet:Time | 1.399 | 0.350 | 4.81 | 6.63 | 0.0001 | |
| Antibiotic:Time | 0.730 | 0.183 | 4.81 | 3.46 | 0.0116 | |
| Diet:Antibiotic:Time | 1.954 | 0.244 | 8.81 | 4.63 | 0.0001 | |
| Bifidobacterium | Diet | 0.048 | 0.024 | 2.27 | 0.70 | 0.5043 |
| Antibiotic | 0.156 | 0.078 | 2.27 | 2.29 | 0.1202 | |
| Time | 0.291 | 0.097 | 3.81 | 2.85 | 0.0423 | |
| Diet:Antibiotic | 0.087 | 0.022 | 4.27 | 0.64 | 0.6381 | |
| Diet:Time | 0.438 | 0.073 | 6.81 | 2.15 | 0.0565 | |
| Antibiotic:Time | 0.957 | 0.160 | 6.81 | 4.70 | 0.0004 | |
| Diet:Antibiotic:Time | 1.100 | 0.092 | 12.81 | 2.70 | 0.0041 |

3.3. Taxonomic Composition
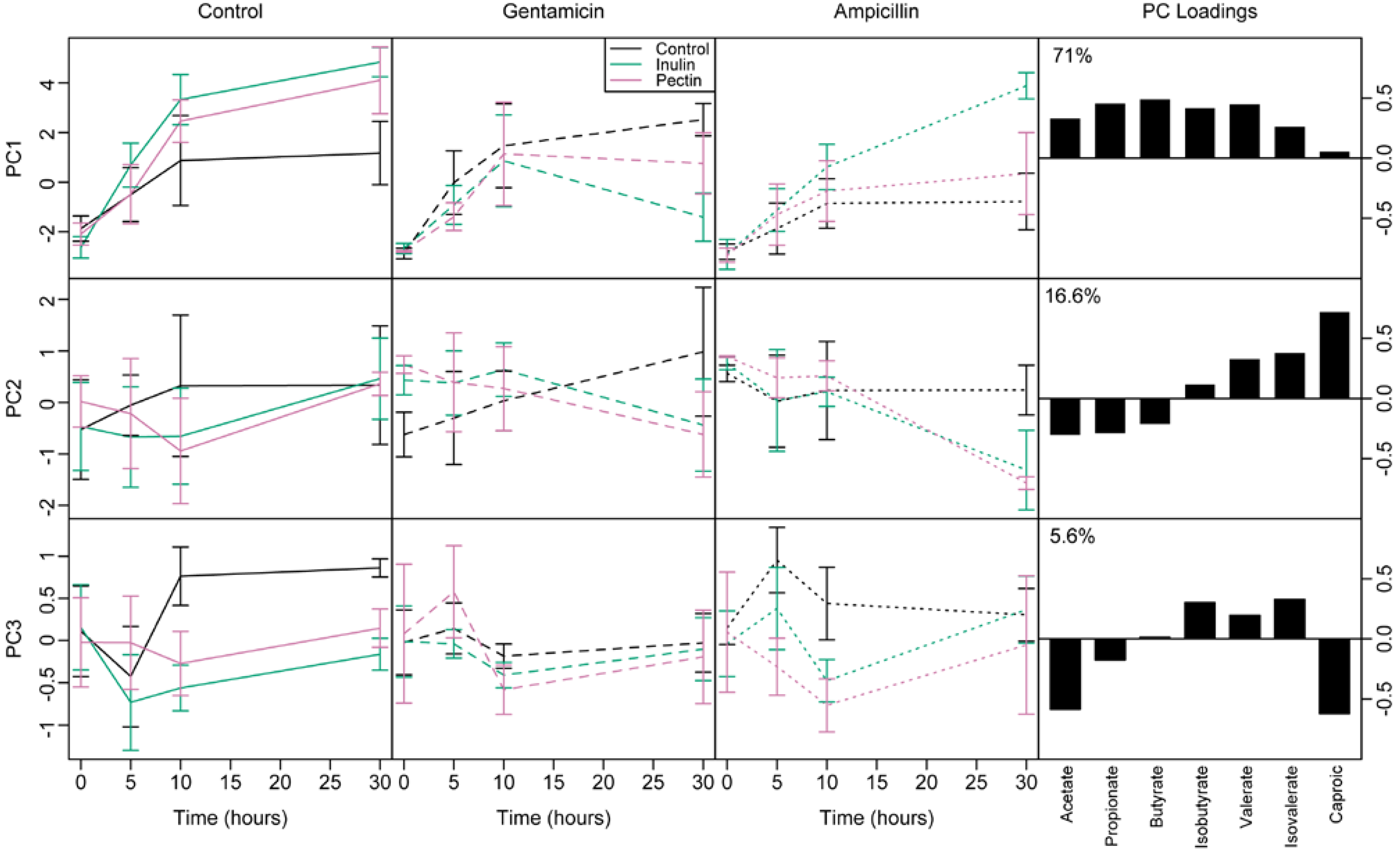
3.4. Short Chain Fatty Acids
3.5. Effect of Microbial Abundance and Composition on SCFA Production
3.6. PYY
4. Discussion
5. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Scott, K.P.; Gratz, S.W.; Sheridan, P.O.; Flint, H.J. The influence of diet on the gut microbiota. Pharmacol. Res. 2013, 69, 52–60. [Google Scholar] [CrossRef] [PubMed]
- LeBlanc, G.J.; Milani, C.; de Giori, G.S.; Sesma, F.; van Sinderen, D.; Ventura, M. Bacteria as vitamin suppliers to their host: A gut microbiota perspective. Curr. Opin. Biotechnol. 2013, 24, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Guarner, F.; Malagelada, J.R. Gut flora in health and disease. Lancet 2003, 361, 512–519. [Google Scholar] [CrossRef]
- Clarke, G.; Stilling, R.M.; Kennedy, P.J.; Stanton, C.; Cryan, J.F.; Dinan, T.G. Minireview: Gut microbiota: The neglected endocrine organ. Mol. Endocrinol. 2014, 28, 1221–1238. [Google Scholar] [CrossRef] [PubMed]
- Dethlefsen, L.; Huse, S.; Sogin, M.L.; Relman, D.A. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008, 6, 2383–2400. [Google Scholar] [CrossRef] [PubMed]
- Looft, T.; Allen, H.K. Collateral effects of antibiotics on mammalian gut microbiomes. Gut Microbes 2012, 3, 463–467. [Google Scholar] [CrossRef] [PubMed]
- Gibson, G.R.; Roberfroid, M.B. Dietary modulation of the human colonic microbiota: Introducing the concept of prebiotics. J. Nutr. 1995, 125, 1401–1412. [Google Scholar] [PubMed]
- Walker, A.W.; Ince, J.; Duncan, S.H.; Webster, L.M.; Holtrop, G.; Ze, X.; Brown, D.; Stares, M.D.; Scott, P.; Bergerat, A.; et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011, 5, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Flint, H.J.; Scott, K.P.; Duncan, S.H.; Louis, P.; Forano, E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 2012, 3, 289–306. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.M.W.; de Souza, R.; Kendall, C.W.C.; Emam, A.; Jenkins, D.J.A. Colonic health: Fermentation and short chain fatty acids. J. Clin. Gastroenterol. 2006, 40, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Cummings, J.H.; Hill, M.J.; Bone, E.S.; Branch, W.J.; Jenkins, D.J.A. The effect of meat protein and dietary fiber on colonic function and metabolism; II. Bacterial metabolites in feces and urine. Am. J. Clin. Nutr. 1979, 32, 2094–2101. [Google Scholar] [PubMed]
- Sleeth, M.L.; Thompson, E.L.; Ford, H.E.; Zac-Varghese, S.E.K.; Frost, G. Free fatty acid receptor 2 and nutrient sensing: A proposed role for fibre, fermentable carbohydrates and short-chain fatty acids in appetite regulation. Nutr. Res. Rev. 2010, 23, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Karaki, S.; Mitsui, R.; Hayashi, H.; Kato, I.; Sugiya, H.; Iwanaga, T.; Furness, J.B.; Kuwahara, A. Short-chain fatty acid receptor, GPR43, is expressed by enteroendocrine cells and mucosal mast cells in rat intestine. Cell Tissue Res. 2006, 324, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Delzenne, N.M.; Neyrinck, A.M.; Baeckhed, F.; Cani, P.D. Targeting gut microbiota in obesity: Effects of prebiotics and probiotics. Nat. Rev. Endocrinol. 2011, 7, 639–646. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, D.S.; Pereira, M.A.; Kroenke, C.H.; Hilner, J.E.; van Horn, L.; Slattery, M.L.; Jacobs, D.R. Dietary fiber, weight gain, and cardiovascular disease risk factors in young adults. J. Am. Med. Assoc. 1999, 282, 1539–1546. [Google Scholar] [CrossRef]
- Maskarinec, G.; Takata, Y.; Pagano, I.; Carlin, L.; Goodman, M.T.; Le Marchand, L.; Nomura, A.M.Y.; Wilkens, L.R.; Kolonel, L.N. Trends and dietary determinants of overweight and obesity in a multiethnic population. Obesity 2006, 14, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Sarbini, S.R.; Kolida, S.; Naeye, T.; Einerhand, A.; Brison, Y.; Remaud-Simeon, M.; Monsan, P.; Gibson, G.R.; Rastall, R.A. In vitro fermentation of linear and alpha-1,2-branched dextrans by the human fecal microbiota. Appl. Environ. Microbiol. 2011, 77, 5307–5315. [Google Scholar] [CrossRef] [PubMed]
- Olsson, B.; Nord, C.E.; Wadstrom, T. Formation of beta-lactamase in Bacteroides fragilis—Cell bound and extracellular activity. Antimicrob. Agents Chemother. 1976, 9, 727–735. [Google Scholar] [CrossRef] [PubMed]
- Bryan, L.E.; Kowand, S.K.; van den Elzen, H.M. Mechanism of amino glycoside antibiotic resistance in anaerobic bacteria Clostridium perfringens and Bacteroides fragilis. Antimicrob. Agents Chemother. 1979, 15, 7–13. [Google Scholar] [CrossRef] [PubMed]
- D’Aimmo, M.R.; Modesto, M.; Biavati, B. Antibiotic resistance of lactic acid bacteria and Bifidobacterium spp. isolated from dairy and pharmaceutical products. Int. J. Food Microbiol. 2007, 115, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Kheadr, E.; Dabour, N.; Le Lay, C.; Lacroix, C.; Fliss, I. Antibiotic susceptibility profile of bifidobacteria as affected by oxgall, acid, and hydrogen peroxide stress. Antimicrob. Agents Chemother. 2007, 51, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Onumpai, C.; Kolida, S.; Bonnin, E.; Rastall, R.A. Microbial utilization and selectivity of pectin fractions with various structures. Appl. Environ. Microbiol. 2011, 77, 5747–5754. [Google Scholar] [CrossRef] [PubMed]
- Meyer, D.; Stasse-Wolthuis, M. The bifidogenic effect of inulin and oligofructose and its consequences for gut health. Eur. J. Clin. Nutr. 2009, 63, 1277–1289. [Google Scholar] [CrossRef] [PubMed]
- Kolida, S.; Tuohy, K.M.; Gibson, G.R. Prebiotic effects of inulin and oligofructose. Br. J. Nutr. 2002, 87, S193–S197. [Google Scholar] [CrossRef] [PubMed]
- Costabile, A.; Kolia, S.; Klinder, A.; Gietl, E.; Baeuerlein, M.; Frohberg, C.; Landschuetze, V.; Gibson, G.R. A double-blind, placebo-controlled, cross-over study to establish the bifidogenic effect of a very-long-chain inulin extracted from globe artichoke (Cynara scolymus) in healthy human subjects. Br. J. Nutr. 2010, 104, 1007–1017. [Google Scholar] [CrossRef] [PubMed]
- Daims, H.; Bruhl, A.; Amann, R.; Schleifer, K.H.; Wagner, M. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: Development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 1999, 22, 434–444. [Google Scholar] [CrossRef]
- Manz, W.; Amann, R.; Ludwig, W.; Vancanneyt, M.; Schleifer, K.H. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiol. UK 1996, 142, 1097–1106. [Google Scholar] [CrossRef]
- Langendijk, P.S.; Schut, F.; Jansen, G.J.; Raangs, G.C.; Kamphuis, G.R.; Wilkinson, M.H.F.; Welling, G.W. Quantitative fluoresecence in situ hydridisation of Bifidobacterium spp. with genus specific16S ribosomal-RNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 1995, 61, 3069–3075. [Google Scholar] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2014; Available online: http://www.R-project.org/ (accessed on 30 October 2012).
- Frost, G.S.; Walton, G.E.; Swann, J.R.; Psichas, A.; Costabile, A.; Johnson, L.P.; Sponheimer, M.; Gibson, G.R.; Barraclough, T.G. Impacts of plant-based foods in Ancestral Hominin diets on the metabolism and function of gut microbiota in vitro. Mbio 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Psichas, A.; Sleeth, M.L.; Murphy, K.G.; Brooks, L.; Bewick, G.A.; Hanyaloglu, A.C.; Ghatei, M.A.; Bloom, S.R.; Frost, G. The short chain fatty acid propionate stimulates GLP-1 and PYY secretion via free fatty acid receptor 2 in rodents. Int. J. Obes. 2014. [Google Scholar] [CrossRef] [PubMed]
- Koren, O.; Knights, D.; Gonzalez, A.; Waldron, L.; Segata, N.; Knight, R.; Huttenhower, C.; Ley, R.E. A guide to enterotypes across the human body: Meta-analysis of microbial community structures in human microbiome datasets. PLoS Comput. Biol. 2013, 9, e1002863. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, M.; Jeroen, R.; Pelletier, E.; Le Paslier, D.; Yamuda, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Stearns, J.C.; Lynch, M.D.J.; Senadheera, D.B.; Tenenbaum, H.C.; Goldberg, M.B.; Cvitkovitch, D.G.; Croitoru, K.; Moreno-Hagelsieb, G.; Neufeld, J.D. Bacterial biogeography of the human digestive tract. Sci. Rep. 2011, 1, 170. [Google Scholar] [CrossRef] [PubMed]
- Sim, K.; Cox, M.J.; Wopereis, H.; Martin, R.; Knol, J.; Li, M.; Cookson, W.O.C.M.; Moffatt, M.F.; Kroll, J.S. Improved detection of bifidobacteria with optimised 16S rRNA-gene based pyrosequencing. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Milani, C.; Hevia, A.; Foroni, E.; Duranti, S.; Turroni, F.; Lugli, G.A.; Sanchez, B.; Martin, R.; Gueimonde, M.; van Sinderen, D.; et al. Assessing the fecal microbiota: An optimized ion torrent 16S rRNA gene-based analysis protocol. PLoS ONE 2013, 8, e68739. [Google Scholar] [CrossRef] [PubMed]
- Sonnenburg, E.D.; Zheng, H.; Joglekar, P.; Higginbottom, S.K.; Firbank, S.J.; Bolam, D.N.; Sonnenburg, J.L. Specificity of polysaccharide use in intestinal Bacteroides species determines diet-induced microbiota alterations. Cell 2010, 141, 1241–1256. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Paganelli, F.L.; Bierschenk, D.; Kuipers, A.; Bonten, M.J.M.; Willems, R.J.L.; van Schaik, W. Genome-wide identification of ampicillin resistance determinants in Enterococcus faecium. PLoS Genet. 2012, 8. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Peterson, D.A.; Gordon, J.I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 2006, 124, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Fouhy, F.; Guinane, C.M.; Hussey, S.; Wall, R.; Ryan, C.A.; Dempsey, E.M.; Murphy, B.; Ross, R.P.; Fitzgerald, G.F.; Stanton, C.; et al. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob. Agents Chemother. 2012, 56, 5811–5820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maurice, C.F.; Haiser, H.J.; Turnbaugh, P.J. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 2013, 152, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Farias, C.; Slezak, K.; Fuller, Z.; Duncan, A.; Holtrop, G.; Louis, P. Effect of inulin on the human gut microbiota: Stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br. J. Nutr. 2009, 101, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Moens, F.; Riviere, A.; Selak, M.; de Vuyst, L. Inulin-type fructan degradation capacity of interesting butyrate-producing colon bacteria and cross-feeding interactions of Faecalibacterium prausnitzi DSM 17677T with bifidobacteria. Arch. Public Health 2014, 72, 6. [Google Scholar] [CrossRef]
- Macfarlane, G.T.; Gibson, G.R.; Beatty, E.; Cummings, J.H. Estimation of short chain fatty acid production from protein by human intestinal bacteria based on branched-chain fatty acid measurements. FEMS Microbiol. Ecol. 1992, 101, 81–88. [Google Scholar] [CrossRef]
- Falony, G.; Lazidou, K.; Verschaeren, A.; Weckx, S.; Maes, D.; de Vuyst, L. In vitro kinetic analysis of fermentation of prebiotic inulin-yype fructans by Bifidobacterium species reveals four different phenotypes. Appl. Environ. Microbiol. 2009, 75, 454–461. [Google Scholar] [CrossRef] [PubMed]
- Reichardt, N.; Duncan, S.H.; Young, P.; Belenguer, A.; Leitch, C.M.; Scott, K.P.; Flint, H.J.; Louis, P. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 2014, 8, 1323–1335. [Google Scholar] [CrossRef] [PubMed]
- Lesniewska, V.; Rowland, I.; Cani, P.D.; Neyrinck, A.M.; Delzenne, N.M.; Naughton, P.J. Effect on components of the intestinal microflora and plasma neuropeptide levels of feeding Lactobacillus delbrueckii, Bifidobacterium lactis, and inulin to adult and elderly rats. Appl. Environ. Microbiol. 2006, 72, 6533–6538. [Google Scholar] [CrossRef] [PubMed]
- Windey, K.; de Preter, V.; Verbeke, K. Relevance of protein fermentation to gut health. Mol. Nutr. Food Res. 2012, 56, 184–196. [Google Scholar] [CrossRef] [PubMed]
- Goodman, A.L.; Kallstrom, G.; Faith, J.J.; Reyes, A.; Moore, A.; Dantas, G.; Gordon, J.I. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc. Natl. Acad. Sci. USA 2011, 108, 6252–6257. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Johnson, L.P.; Walton, G.E.; Psichas, A.; Frost, G.S.; Gibson, G.R.; Barraclough, T.G. Prebiotics Modulate the Effects of Antibiotics on Gut Microbial Diversity and Functioning in Vitro. Nutrients 2015, 7, 4480-4497. https://doi.org/10.3390/nu7064480
Johnson LP, Walton GE, Psichas A, Frost GS, Gibson GR, Barraclough TG. Prebiotics Modulate the Effects of Antibiotics on Gut Microbial Diversity and Functioning in Vitro. Nutrients. 2015; 7(6):4480-4497. https://doi.org/10.3390/nu7064480
Chicago/Turabian StyleJohnson, Laura P., Gemma E. Walton, Arianna Psichas, Gary S. Frost, Glenn R. Gibson, and Timothy G. Barraclough. 2015. "Prebiotics Modulate the Effects of Antibiotics on Gut Microbial Diversity and Functioning in Vitro" Nutrients 7, no. 6: 4480-4497. https://doi.org/10.3390/nu7064480





