Sex-Specific Muscular Maturation Responses Following Prenatal Exposure to Methylation-Related Micronutrients in Pigs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals, Diets, Sample Collection
2.2. Phenotype Data Analyses
2.3. RNA Isolation and cDNA Synthesis
2.4. Primer Sequences and Primer Validation
2.5. Microfluidic High-Throughput qPCR
2.6. Transcript Data Preprocessing and Analysis
2.7. Validation of Microfluidic High-Throughput qPCRs
3. Results
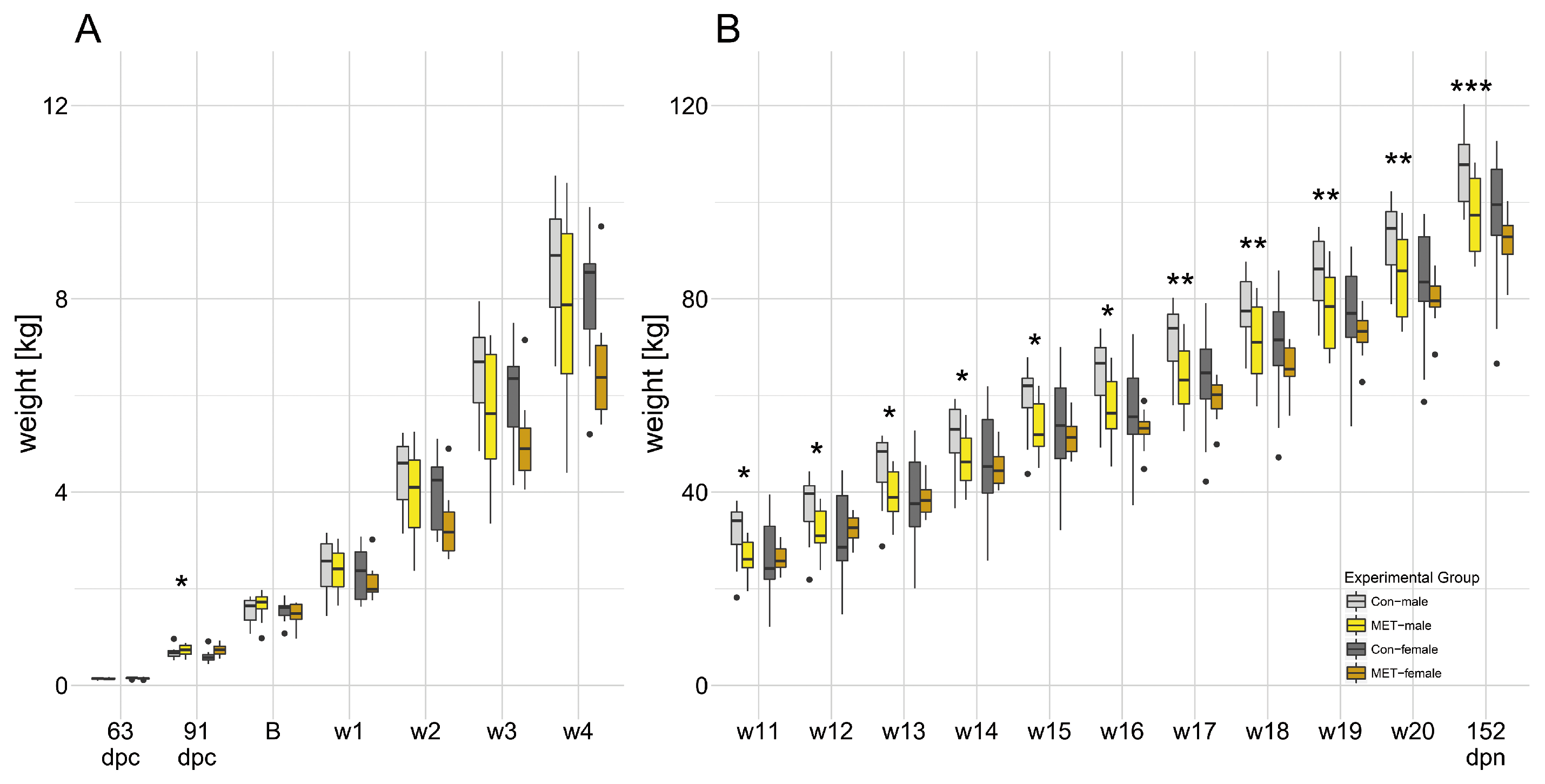
3.1. Maternal Supplementation with Methylation-Related Micronutrients Affected Fetal Weight and Live Weight in Males
3.2. Increased Percent Lean Mass in MET-Males
3.3. Gene Expression Pattern Specific for Maternal Diet and Sex
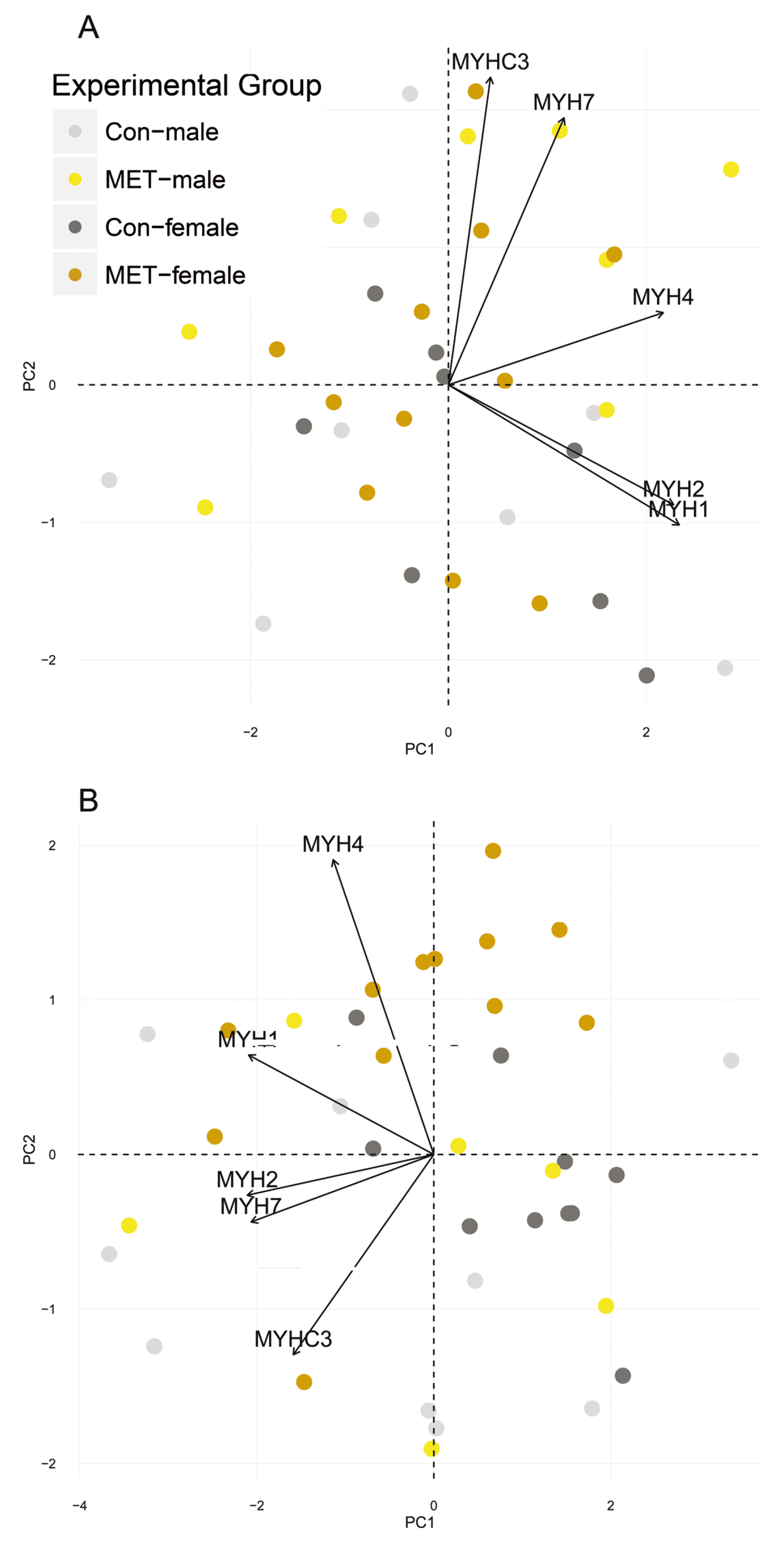
3.4. Myosin Heavy Chain Isoforms
3.5. Verification of qPCR Results
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| B | Birth |
| CON | Standard diet |
| dpc | days post conception |
| dpn | days post natum |
| FC | Fold change |
| IN | Insemination |
| MET | Standard diet supplemented with methylating micronutrients |
| nd | not detected |
| ns | not significant |
| PCA | Principle component analysis |
| qPCR | Quantitative polymerase chain reaction |
| w | week |
References
- Wolff, G.; Kodell, R.; Moore, S.; Cooney, C. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 1998, 12, 949–957. [Google Scholar] [PubMed]
- Waterland, R. Assessing the effects of high methionine intake on DNA methylation. J. Nutr. 2006, 136 (Suppl. S6), 1706S–1710S. [Google Scholar] [PubMed]
- Schaible, T.; Harris, R.; Dowd, S.; Smith, C.; Kellermayer, R. Maternal methyl-donor supplementation induces prolonged murine offspring colitis susceptibility in association with mucosal epigenetic and microbiomic changes. Hum. Mol. Genet. 2011, 20, 1687–1696. [Google Scholar] [CrossRef] [PubMed]
- Giudicelli, F.; Brabant, A.; Grit, I.; Parnet, P.; Amarger, V. Excess of methyl donor in the perinatal period reduces postnatal leptin secretion in rat and interacts with the effect of protein content in diet. PLoS ONE 2013, 8, e68268. [Google Scholar] [CrossRef] [PubMed]
- Mikael, L.; Deng, L.; Paul, L.; Selhub, J.; Rozen, R. Moderately high intake of folic acid has a negative impact on mouse embryonic development. Birth Defects Res. A Clin. Mol. Teratol. 2013, 97, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Hoile, S.; Lillycrop, K.; Grenfell, L.; Hanson, M.; Burdge, G. Increasing the folic acid content of maternal or post-weaning diets induces differential changes in phosphoenolpyruvate carboxykinase mRNA expression and promoter methylation in rats. Br. J. Nutr. 2012, 108, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Pickell, L.; Brown, K.; Li, D.; Wang, X.; Deng, L.; Wu, Q.; Selhub, J.; Luo, L.; Jerome-Majewska, L.; Rozen, R. High intake of folic acid disrupts embryonic development in mice. Birth Defects Res. A Clin. Mol. Teratol. 2011, 91, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Oster, M.; Nuchchanart, W.; Trakooljul, N.; Murani, E.; Zeyner, A.; Wirthgen, E.; Hoeflich, A.; Ponsuksili, S.; Wimmers, K. Methylating micronutrient supplementation during pregnancy influences foetal hepatic gene expression and IGF signalling and increases foetal weight. Eur. J. Nutr. 2016, 55, 1717–1727. [Google Scholar] [CrossRef] [PubMed]
- Niculescu, M.; Zeisel, S. Diet, methyl donors and DNA methylation: interactions between dietary folate, methionine and choline. J. Nutr. 2002, 132 (Suppl. S8), 2333S–2335S. [Google Scholar] [PubMed]
- Braunschweig, M.; Jagannathan, V.; Gutzwiller, A.; Bee, G. Investigations on transgenerational epigenetic response down the male line in F2 pigs. PLoS ONE 2012, 7, e30583. [Google Scholar] [CrossRef] [PubMed]
- Bruggmann, R.; Jagannathan, V.; Braunschweig, M. In search of epigenetic marks in testes and sperm cells of differentially fed boars. PLoS ONE 2013, 8, e78691. [Google Scholar] [CrossRef] [PubMed]
- Wigmore, P.; Stickland, N. Muscle development in large and small pig fetuses. J. Anat. 1983, 137 Pt 2, 235–245. [Google Scholar] [PubMed]
- Hwang, S.; Kang, Y.; Sung, B.; Kim, M.; Kim, D.; Lee, Y.; Yoo, M.; Kim, C.; Chung, H.; Kim, N. Folic acid promotes the myogenic differentiation of C2C12 murine myoblasts through the Akt signaling pathway. Int. J. Mol. Med. 2015, 36, 1073–1080. [Google Scholar] [CrossRef] [PubMed]
- Theys, N.; Bouckenooghe, T.; Ahn, M.T.; Remacle, C.; Reusens, B. Maternal low-protein diet alters pancreatic islet mitochondrial function in a sex-specific manner in the adult rat. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009, 297, R1516–R1525. [Google Scholar] [CrossRef] [PubMed]
- Gallou-Kabani, C.; Gabory, A.; Tost, J.; Karimi, M.; Mayeur, S.; Lesage, J.; Boudadi, E.; Gross, M.; Taurelle, J.; Vige, A.; et al. Sex- and diet-specific changes of imprinted gene expression and DNA methylation in mouse placenta under a high-fat diet. PLoS ONE 2010, 5, e14398. [Google Scholar] [CrossRef] [PubMed]
- Penailillo, R.; Guajardo, A.; Llanos, M.; Hirsch, S.; Ronco, A. Folic acid supplementation during pregnancy induces sex-specific changes in methylation and expression of placental 11beta-hydroxysteroid dehydrogenase 2 in rats. PLoS ONE 2015, 10, e0121098. [Google Scholar] [CrossRef] [PubMed]
- Goodspeed, D.; Seferovic, M.; Holland, W.; Mcknight, R.; Summers, S.; Branch, D.W.; Lane, R.; Aagaard, K. Essential nutrient supplementation prevents heritable metabolic disease in multigenerational intrauterine growth-restricted rats. FASEB J. 2015, 29, 807–819. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Ford, S.; Means, W.; Hess, B.; Nathanielsz, P.; Du, M. Maternal nutrient restriction affects properties of skeletal muscle in offspring. J. Physiol. 2006, 575 Pt 1, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Wimmers, K.; Ngu, N.; Jennen, D.; Tesfaye, D.; Murani, E.; Schellander, K.; Ponsuksili, S. Relationship between myosin heavy chain isoform expression and muscling in several diverse pig breeds. J. Anim. Sci. 2008, 86, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Institute of Medicine (US) Standing Committee on the Scientific Evaluation of Dietary Reference Intakes. Dietary Reference Intakes for Thiamin, Riboflavin, Niacin, Vitamin B6, Folate, Vitamin B12, Pantothenic Acid, Biotin, and Choline; National Academies Press: Washington, DC, USA, 1998. [Google Scholar]
- Kira, J.; Tobimatsu, S.; Goto, I. Vitamin B12 metabolism and massive-dose methyl vitamin B12 therapy in Japanese patients with multiple sclerosis. Intern. Med. 1994, 33, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Izumi, Y.; Kaji, R. Clinical trials of ultra-high-dose methylcobalamin in ALS. Brain Nerve 2007, 59, 1141–1147. [Google Scholar] [PubMed]
- Evans, A.; O’Doherty, J. Endocrine changes and management factors affecting puberty in gilts. Livestock Prod. Sci. 2001, 68, 1–12. [Google Scholar] [CrossRef]
- Szostak, B.; Stasiak, A.; Przykaza, L. The effect of naked oats (Avena nuda L.) used in feeding gilts on their sexual activity. Archiv fuer Tierzucht 2015, 58, 7–11. [Google Scholar] [CrossRef]
- Livak, K.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2 Delta Delta C(T) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3. [Google Scholar] [CrossRef] [Green Version]
- Storey, J.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [PubMed]
- Rees, W.; Hay, S.; Cruickshank, M. An imbalance in the methionine content of the maternal diet reduces postnatal growth in the rat. Metabolism 2006, 55, 763–770. [Google Scholar] [CrossRef] [PubMed]
- Maloney, C.; Hay, S.; Rees, W. The effects of feeding rats diets deficient in folic acid and related methyl donors on the blood pressure and glucose tolerance of the offspring. Br. J. Nutr. 2009, 101, 1333–1340. [Google Scholar] [CrossRef] [PubMed]
- Roberfroid, D.; Huybregts, L.; Lanou, H.; Habicht, J.; Henry, M.; Meda, N.; Kolsteren, P. Prenatal micronutrient supplements cumulatively increase fetal growth. J. Nutr. 2012, 142, 548–554. [Google Scholar] [CrossRef] [PubMed]
- Owens, J. Endocrine and substrate control of fetal growth: Placental and maternal influences and insulin-like growth factors. Reprod. Fertil. Dev. 1991, 3, 501–517. [Google Scholar] [CrossRef] [PubMed]
- Fowden, A. Endocrine regulation of fetal growth. Reprod. Fertil. Dev. 1995, 7, 351–363. [Google Scholar] [CrossRef] [PubMed]
- White, R.; Biérinx, A.; Gnocchi, V.; Zammit, P. Dynamics of muscle fibre growth during postnatal mouse development. BMC Dev. Biol. 2010, 10, 21. [Google Scholar] [CrossRef] [PubMed]
- Le Grand, F.; Rudnicki, M. Skeletal muscle satellite cells and adult myogenesis. Curr. Opin. Cell Biol. 2007, 19, 628–633. [Google Scholar] [CrossRef] [PubMed]
- Berkes, C.; Tapscott, S. MyoD and the transcriptional control of myogenesis. Semin. Cell Dev. Biol. 2005, 16, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Knapp, J.; Davie, J.; Myer, A.; Meadows, E.; Olson, E.; Klein, W. Loss of myogenin in postnatal life leads to normal skeletal muscle but reduced body size. Development 2006, 133, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Braun, T.; Gautel, M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat. Rev. Mol. Cell Biol. 2011, 12, 349–361. [Google Scholar] [CrossRef] [PubMed]
- Huot, P.; Dodington, D.; Mollard, R.; Reza-López, S.; Sánchez-Hernández, D.; Cho, C.; Kuk, J.; Ward, W.; Anderson, G. High Folic Acid Intake during Pregnancy Lowers Body Weight and Reduces Femoral Area and Strength in Female Rat Offspring. J. Osteoporos. 2013, 2013, 154109. [Google Scholar] [CrossRef] [PubMed]
- Hotamisligil, G.; Johnson, R.; Distel, R.; Ellis, R.; Papaioannou, V.; Spiegelman, B. Uncoupling of obesity from insulin resistance through a targeted mutation in aP2, the adipocyte fatty acid binding protein. Science 1996, 274, 1377–1379. [Google Scholar] [CrossRef] [PubMed]
- Furuhashi, M.; Saitoh, S.; Shimamoto, K.; Miura, T. Fatty Acid-Binding Protein 4 (FABP4): Pathophysiological Insights and Potent Clinical Biomarker of Metabolic and Cardiovascular Diseases. Clin. Med. Insights Cardiol. 2015, 8 (Suppl. S3), 23–33. [Google Scholar] [CrossRef] [PubMed]
- Bodine, S.; Stitt, T.; Gonzalez, M.; Kline, W.; Stover, G.; Bauerlein, R.; Zlotchenko, E.; Scrimgeour, A.; Lawrence, J.; Glass, D.; Yancopoulos, G. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat. Cell Biol. 2001, 3, 1014–1019. [Google Scholar] [CrossRef] [PubMed]
- Ceci, M.; Ross, J.; Condorelli, G. Molecular determinants of the physiological adaptation to stress in the cardiomyocyte: A focus on AKT. J. Mol. Cell. Cardiol. 2004, 37, 905–912. [Google Scholar] [CrossRef] [PubMed]
- Kalbe, C.; Lösel, D.; Block, J.; Lefaucheur, L.; Brüssow, K.; Bellmann, O.; Pfuhl, R.; Puppe, B.; Otten, W.; Metges, C.; et al. Moderate high or low maternal protein diets change gene expression but not the phenotype of skeletal muscle from porcine fetuses. Domest. Anim. Endocrinol. 2016, 58, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Berard, J.; Bee, G. Effects of dietary l-arginine supplementation to gilts during early gestation on foetal survival, growth and myofiber formation. Animal 2010, 4, 1680–1687. [Google Scholar] [CrossRef] [PubMed]
- Dwyer, C.; Stickland, N.; Fletcher, J. The influence of maternal nutrition on muscle fiber number development in the porcine fetus and on subsequent postnatal growth. J. Anim. Sci. 1994, 72, 911–917. [Google Scholar] [PubMed]
- Murani, E.; Muraniova, M.; Ponsuksili, S.; Schellander, K.; Wimmers, K. Identification of genes differentially expressed during prenatal development of skeletal muscle in two pig breeds differing in muscularity. BMC Dev. Biol. 2007, 7, 109. [Google Scholar] [CrossRef] [PubMed]
- Uaesoontrachoon, K.; Yoo, H.; Tudor, E.; Pike, R.; Mackie, E.; Pagel, C. Osteopontin and skeletal muscle myoblasts: Association with muscle regeneration and regulation of myoblast function in vitro. Int. J. Biochem. Cell Biol. 2008, 40, 2303–2314. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, J.; Perry, R.; Asakura, A.; Rudnicki, M. MyoD induces myogenic differentiation through cooperation of its NH2- and COOH-terminal regions. J. Cell Biol. 2005, 171, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Kahles, F.; Findeisen, H.; Bruemmer, D. Osteopontin: A novel regulator at the cross roads of inflammation, obesity and diabetes. Mol. Metab. 2014, 3, 384–393. [Google Scholar] [CrossRef] [PubMed]
- Nomiyama, T.; Perez-Tilve, D.; Ogawa, D.; Gizard, F.; Zhao, Y.; Heywood, E.; Jones, K.; Kawamori, R.; Cassis, L.; Tschöp, M.; et al. Osteopontin mediates obesity-induced adipose tissue macrophage infiltration and insulin resistance in mice. J. Clin. Investig. 2007, 117, 2877–2888. [Google Scholar] [CrossRef] [PubMed]
- Hovdenak, N.; Haram, K. Influence of mineral and vitamin supplements on pregnancy outcome. Eur. J. Obstet. Gynecol. Reprod. Biol. 2012, 164, 127–132. [Google Scholar] [CrossRef] [PubMed]


| Methylating Micronutrient | Sow CON Diet | Sow MET Diet |
|---|---|---|
| Methionine, mg | 2050 | 4700 |
| Choline, mg | 500 | 2230 |
| Folic acid, mg | 3 | 92.2 |
| Vitamin B6, mg | 3 | 1180 |
| Vitamin B12, μg | 31 | 5930 |
| Zinc, mg | 21.8 | 149 |
| Gene | Males 63 dpc | Females 63 dpc | Males 91 dpc | Females 91 dpc | Males 150 dpn | Females 150 dpn | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p | FC 1 | p | FC 1 | p | FC 1 | p | FC 1 | p | FC 1 | p | FC 1 | |
| Akt1 | 0.026 * | +1.40 | 0.002 | +1.51 | ns | ns | 0.046 | +1.29 | ns | ns | ns | ns |
| FABP3 | ns | ns | ns | ns | ns | ns | 0.001 | +1.38 | ns | ns | ns | ns |
| FABP4 | ns | ns | ns | ns | ns | ns | 0.031 | +1.39 | ns | ns | ns | ns |
| FBXO32 | ns | ns | 0.038 | +1.33 | ns | ns | ns | ns | ns | ns | ns | ns |
| FLT1 | ns | ns | ns | ns | ns | ns | 0.017 | +1.41 | ns | ns | ns | ns |
| FLT4 | 0.029 * | +1.36 | ns | ns | ns | ns | 0.018 | +1.55 | ns | ns | ns | ns |
| FST | ns | ns | ns | ns | ns | ns | 0.026 | −2.65 | ns | ns | ns | ns |
| GALK1 | ns | ns | ns | ns | ns | ns | 0.035 | +1.25 | ns | ns | ns | ns |
| GHR | ns | ns | 0.023 | +1.54 | ns | ns | ns | ns | ns | ns | ns | ns |
| GLUT1 | ns | ns | ns | ns | ns | ns | 0.024 | +1.37 | ns | ns | ns | ns |
| GLUT4 | ns | ns | ns | ns | ns | ns | 0.025 | +1.30 | ns | ns | ns | ns |
| GSK3b | ns | ns | ns | ns | ns | ns | 0.023 | +1.38 | 0.029 * | +1.41 | ns | ns |
| HGF | ns | ns | 0.006 | +1.61 | ns | ns | ns | ns | ns | ns | ns | ns |
| HSD11B1 | ns | ns | ns | ns | ns | ns | 0.013 | +1.40 | ns | ns | ns | ns |
| IGFBP5 | ns | ns | ns | ns | ns | ns | 0.027 | +1.26 | ns | ns | ns | ns |
| KDR | ns | ns | ns | ns | ns | ns | 0.018 | +1.32 | ns | ns | ns | ns |
| MAT2A | ns | ns | 0.023 | +1.22 | ns | ns | ns | ns | ns | ns | ns | ns |
| MAT2B | ns | ns | ns | ns | ns | ns | 0.025 | +1.25 | ns | ns | ns | ns |
| MET | ns | ns | ns | ns | ns | ns | ns | ns | 0.023 * | +1.54 | 0.026 * | −1.50 |
| MSTN | ns | ns | 0.029 | +1.56 | ns | ns | ns | ns | ns | ns | ns | ns |
| MYF5 | ns | ns | 0.035 | +1.29 | ns | ns | ns | ns | ns | ns | ns | ns |
| MYF6 | ns | ns | ns | ns | 0.043 | −1.38 | ns | ns | ns | ns | ns | ns |
| MYH2 | ns | ns | ns | ns | ns | ns | 0.044 | +1.69 | ns | ns | ns | ns |
| MyoD1 | nd | nd | nd | nd | ns | ns | ns | ns | 0.009 | +2.03 | ns | ns |
| Myogenin | ns | ns | ns | ns | ns | ns | ns | ns | 0.003 | +1.86 | ns | ns |
| NCAPD2 | ns | ns | 0.032 | +1.29 | ns | ns | ns | ns | 0.030 * | +1.43 | ns | ns |
| Pax7 | ns | ns | ns | ns | ns | ns | ns | ns | 0.003 | +1.84 | ns | ns |
| PC | ns | ns | ns | ns | ns | ns | 0.011 | +1.59 | ns | ns | ns | ns |
| PDGFA | ns | ns | ns | ns | 0.047 | −1.29 | ns | ns | 0.028 * | +1.48 | ns | ns |
| PDPK1 | ns | ns | 0.013 | +1.28 | ns | ns | ns | ns | ns | ns | ns | ns |
| PFKM | ns | ns | 0.008 | +1.31 | ns | ns | ns | ns | ns | ns | ns | ns |
| PIK3CA | ns | ns | ns | ns | ns | ns | ns | ns | 0.018 * | +1.54 | ns | ns |
| PIK3CD | ns | ns | ns | ns | ns | ns | 0.041 | +1.47 | ns | ns | ns | ns |
| PIK3CG | ns | ns | ns | ns | ns | ns | 0.022 | +1.42 | 0.039 * | +1.60 | ns | ns |
| PPARA | ns | ns | 0.032 | +1.28 | ns | ns | 0.023 | +1.39 | ns | ns | ns | ns |
| PPARD | ns | ns | ns | ns | 0.013 | −1.50 | ns | ns | ns | ns | ns | ns |
| PPARGC1A | ns | ns | 0.040 | +1.46 | ns | ns | ns | ns | ns | ns | ns | ns |
| PRKAA2 | ns | ns | 0.023 | +1.37 | ns | ns | ns | ns | ns | ns | ns | ns |
| SPP1 | ns | ns | ns | ns | ns | ns | ns | ns | ns | ns | 0.043 * | +4.34 |
| VEGFB | 0.025 * | +1.14 | 0.012 | +1.12 | ns | ns | ns | ns | ns | ns | ns | ns |
| VEGFC | ns | ns | ns | ns | 0.032 | −1.51 | ns | ns | ns | ns | ns | ns |
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oster, M.; Trakooljul, N.; Reyer, H.; Zeyner, A.; Muráni, E.; Ponsuksili, S.; Wimmers, K. Sex-Specific Muscular Maturation Responses Following Prenatal Exposure to Methylation-Related Micronutrients in Pigs. Nutrients 2017, 9, 74. https://doi.org/10.3390/nu9010074
Oster M, Trakooljul N, Reyer H, Zeyner A, Muráni E, Ponsuksili S, Wimmers K. Sex-Specific Muscular Maturation Responses Following Prenatal Exposure to Methylation-Related Micronutrients in Pigs. Nutrients. 2017; 9(1):74. https://doi.org/10.3390/nu9010074
Chicago/Turabian StyleOster, Michael, Nares Trakooljul, Henry Reyer, Annette Zeyner, Eduard Muráni, Siriluck Ponsuksili, and Klaus Wimmers. 2017. "Sex-Specific Muscular Maturation Responses Following Prenatal Exposure to Methylation-Related Micronutrients in Pigs" Nutrients 9, no. 1: 74. https://doi.org/10.3390/nu9010074





