Nutrimiromics: Role of microRNAs and Nutrition in Modulating Inflammation and Chronic Diseases
Abstract
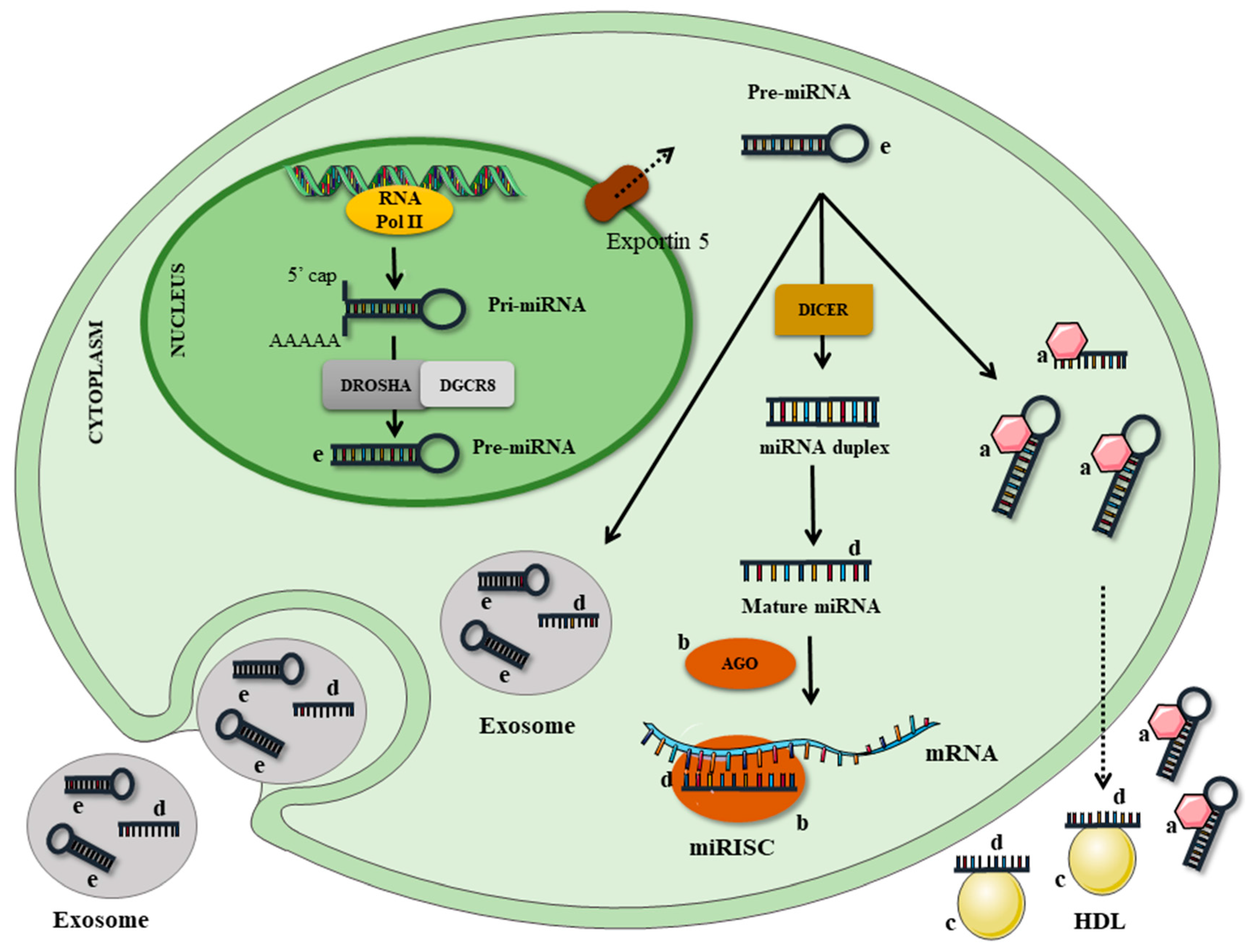
:1. Introduction
2. Inflammation and microRNA
3. microRNA, Nutrients, Inflammation and Chronic Diseases
3.1. Resveratrol
3.2. Saturated Fatty Acids (SFA)
3.3. Polyunsaturated Fatty Acids (PUFA)
3.4. Curcumin
3.5. Quercetin
3.6. Catechins
3.7. Minerals
3.8. Vitamins
4. Food-Derived microRNAs
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cominetti, C.; Horst, M.A.; Rogero, M.M. Brazilian Society for Food and Nutrition position statement: Nutrigenetic tests. Nutrire 2017, 42, 10. [Google Scholar] [CrossRef]
- Huntzinger, E.; Izaurralde, E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, S.; Tong, Y.; Steitz, J.A. Switching from repression to activation: MicroRNAs can up-regulate translation. Science 2007, 318, 1931–1934. [Google Scholar] [CrossRef] [PubMed]
- Osella, M.; Riba, A.; Testori, A.; Corà, D.; Caselle, M. Interplay of microRNA and epigenetic regulation in the human regulatory network. Front. Genet. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zheng, G.; Dong, D. Coordinated action of histone modification and microRNA regulations in human genome. Gene 2015, 570, 277–281. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Xia, J.; Zhao, Z. Functional complementation between transcriptional methylation regulation and post-transcriptional microRNA regulation in the human genome. BMC Genom. 2011, 12. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 2015, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Cloonan, N.; Wani, S.; Xu, Q.; Gu, J.; Lea, K.; Heater, S. MicroRNAs and their functions isomiRs function cooperatively to target commom biological pathways. Genome Biol. 2011. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef] [PubMed]
- Barile, L.; Moccetti, T.; Marbán, E.; Vassalli, G. Roles of exosomes in cardioprotection. Eur. Heart J. 2016. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Mao, X.; Ji, Q. miR-146a in PBMCs modulates Th1 function in patients with acute coronary syndrome. Immunol. Cell Biol. 2010, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhao, J.; Evan, G.; Xiao, C.; Cheng, Y.; Xiao, J. Circulating microRNAs: novel biomarkers for cardiovascular diseases. J. Mol. Med. 2012, 90, 865–875. [Google Scholar] [CrossRef] [PubMed]
- Vrijens, K.; Bollati, V.; Nawrot, T.S. MicroRNAs as potential signatures of environmental exposure or effect: A systematic review. Environ. Health Perspect. 2015, 123, 399. [Google Scholar] [CrossRef] [PubMed]
- Tili, E.; Michaille, J.-J.; Adair, B.; Alder, H.; Limagne, E.; Taccioli, C.; Ferracin, M.; Delmas, D.; Latruffe, N.; Croce, C.M. Resveratrol decreases the levels of miR-155 by upregulating miR-663, a microRNA targeting JunB and JunD. Carcinogenesis 2010, 31, 1561–1566. [Google Scholar] [CrossRef] [PubMed]
- Vinciguerra, M.; Sgroi, A.; Veyrat-Durebex, C.; Rubbia-Brandt, L.; Buhler, L.H.; Foti, M. Unsaturated fatty acids inhibit the expression of tumor suppressor phosphatase and tensin homolog (PTEN) via microRNA-21 up-regulation in hepatocytes. Hepatology 2009, 49, 1176–1184. [Google Scholar] [CrossRef] [PubMed]
- Ortega, F.J.; Cardona-Alvarado, M.I.; Mercader, J.M.; Moreno-Navarrete, J.M.; Moreno, M.; Sabater, M.; Fuentes-Batllevell, N.; Ramírez-Chávez, E.; Ricart, W.; Molina-Torres, J.; et al. Circulating profiling reveals the effect of a polyunsaturated fatty acid-enriched diet on common microRNAs. J. Nutr. Biochem. 2015, 26, 1095–1101. [Google Scholar] [CrossRef] [PubMed]
- Maciel-Dominguez, A.; Swan, D.; Ford, D.; Hesketh, J. Selenium alters miRNA profile in an intestinal cell line: Evidence that miR-185 regulates expression of GPX2 and SEPSH2. Mol. Nutr. Food Res. 2013, 57, 2195–2205. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, Z.; Rasheed, N.; Al-Shaya, O. Epigallocatechin-3-O-gallate modulates global microRNA expression in interleukin-1β-stimulated human osteoarthritis chondrocytes: Potential role of EGCG on negative co-regulation of microRNA-140-3p and ADAMTS5. Eur. J. Nutr. 2017, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kronski, E.; Fiori, M.E.; Barbieri, O.; Astigiano, S.; Mirisola, V.; Killian, P.H.; Bruno, A.; Pagani, A.; Rovera, F.; Pfeffer, U.; et al. miR181b is induced by the chemopreventive polyphenol curcumin and inhibits breast cancer metastasis via down-regulation of the inflammatory cytokines CXCL1 and -2. Mol. Oncol. 2014, 8, 581–595. [Google Scholar] [CrossRef] [PubMed]
- Abbas, A.K.; Lichtman, A.H.H.; Pillai, S. Cellular and Molecular Immunology, 8th ed.; Elsevier Health Sciences: Philadelphia, PA, USA, 2014; ISBN 9780323286459. [Google Scholar]
- Murphy, K.; Weaver, C. Janeway’s Immunobiology, 9th ed.; Garland Science: New York, NY, USA, 2016. [Google Scholar]
- Libby, P. Inflammatory mechanisms: The molecular basis of inflammation and disease. Nutr. Rev. 2007, 65, S140–S146. [Google Scholar] [CrossRef] [PubMed]
- Medzhitov, R. Origin and physiological roles of inflammation. Nature 2008. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Ulevitch, R.J. Limiting inflammatory responses during activation of innate immunity. Nat. Immunol. 2005, 1198–1205. [Google Scholar] [CrossRef] [PubMed]
- O’Rourke, R.W. Inflammation, obesity, and the promise of immunotherapy for metabolic disease. Surg. Obes. Relat. Dis. 2013, 609–616. [Google Scholar] [CrossRef] [PubMed]
- Bondia-Pons, I.; Ryan, L.; Martinez, J.A. Oxidative stress and inflammation interactions in human obesity. J. Physiol. Biochem. 2012, 701–711. [Google Scholar] [CrossRef] [PubMed]
- Maskrey, B.H.; Megson, I.L.; Whitfield, P.D.; Rossi, A.G. Mechanisms of resolution of inflammation. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 1001–1006. [Google Scholar] [CrossRef] [PubMed]
- Medzhitov, R. Inflammation 2010: New adventures of an old flame. Cell 2010, 771–776. [Google Scholar] [CrossRef] [PubMed]
- Hotamisligil, G.S. Inflammation and metabolic disorders. Nature 2006, 444, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Mraz, M.; Haluzik, M. The role of adipose tissue immune cells in obesity and low-grade inflammation. J. Endocrinol. 2014, 222, R113–R127. [Google Scholar] [CrossRef] [PubMed]
- Galic, S.; Oakhill, J.S.; Steinberg, G.R. Adipose tissue as an endocrine organ. Mol. Cell. Endocrinol. 2010, 316, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Bastos, D.H.M.; Rogero, M.M.; Arêas, J.A.G. Mecanismos de ação de compostos bioativos dos alimentos no contexto de processos inflamatórios relacionados à obesidade. Arq. Bras. Endocrinol. Metab. 2009, 646–656. [Google Scholar] [CrossRef]
- Shah, A.; Mehta, N.; Reilly, M.P. Adipose inflammation, insulin resistance, and cardiovascular disease. JPEN J. Parenter Enter. Nutr. 2008, 32, 638–644. [Google Scholar] [CrossRef] [PubMed]
- Lumeng, C.N.; Saltiel, A.R. Inflammatory links between obesity and metabolic disease. J. Clin. Investig. 2011, 2111–2117. [Google Scholar] [CrossRef] [PubMed]
- Geloneze, B.; Umeda, L.M.; Vasquez, A.C.J. Fisiologia e morfologia do tecido adiposo humano. In Tratado de Obesidade; Mancini, M.C., Ed.; GEN: Rio de Janeiro, Brazil, 2015; pp. 117–136. [Google Scholar]
- Greenberg, A.S.; Obin, M.S. Obesity and the role of adipose tissue in inflammation and metabolism. Am. J. Clin. Nutr. 2006, 83, 461S–465S. [Google Scholar] [PubMed]
- Tilg, H.; Moschen, A.R. Adipocytokines: Mediators linking adipose tissue, inflammation and immunity. Nat. Rev. Immunol. 2006, 6, 772–783. [Google Scholar] [CrossRef] [PubMed]
- Goossens, G.H. The role of adipose tissue dysfunction in the pathogenesis of obesity-related insulin resistance. Physiol. Behav. 2008, 94, 206–218. [Google Scholar] [CrossRef] [PubMed]
- Maury, E.; Brichard, S.M. Adipokine dysregulation, adipose tissue inflammation and metabolic syndrome. Mol. Cell. Endocrinol. 2010, 314, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Elks, C.M.; Francis, J. Central adiposity, systemic inflammation, and the metabolic syndrome. Curr. Hypertens. Rep. 2010, 12, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Kahn, S.E.; Hull, R.L.; Utzschneider, K.M. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 2006, 840–846. [Google Scholar] [CrossRef] [PubMed]
- Savini, I.; Catani, M.V.; Evangelista, D.; Gasperi, V.; Avigliano, L. Obesity-associated oxidative stress: Strategies finalized to improve redox state. Int. J. Mol. Sci. 2013, 10497–10538. [Google Scholar] [CrossRef] [PubMed]
- Vincent, H.K.; Taylor, A.G. Biomarkers and potential mechanisms of obesity-induced oxidant stress in humans. Int. J. Obes. 2006, 400–418. [Google Scholar] [CrossRef] [PubMed]
- Pal, M.; Febbraio, M.A.; Lancaster, G.I. The roles of c-Jun NH2-terminal kinases (JNKs) in obesity and insulin resistance. J. Physiol. 2016, 267–279. [Google Scholar] [CrossRef] [PubMed]
- Baud, V.; Karin, M. Is NF-κB a good target for cancer therapy? Hopes and pitfalls. Nat. Rev. Drug Discov. 2009, 33–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vallabhapurapu, S.; Karin, M. Regulation and function of NF-κB transcription factors in the immune system. Annu. Rev. Immunol. 2009, 27, 693–733. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.M.; Rao, D.S.; Baltimore, D. microRNA regulation of inflammatory responses. Annu. Rev. Immunol. 2012, 30, 295–312. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Schober, A. MicroRNA regulation of macrophages in human pathologies. Cell. Mol. Life Sci. 2016, 3473–3495. [Google Scholar] [CrossRef] [PubMed]
- Contreras, J.; Rao, D.S. MicroRNAs in inflammation and immune responses. Leukemia 2012, 404–413. [Google Scholar] [CrossRef] [PubMed]
- Hulsmans, M.; De Keyzer, D.; Holvoet, P. MicroRNAs regulating oxidative stress and inflammation in relation to obesity and atherosclerosis. FASEB 2011, 2515–2527. [Google Scholar] [CrossRef] [PubMed]
- Chang, R.C.-A.; Ying, W.; Bazer, F.W.; Zhou, B. MicroRNAs control macrophage formation and activation: The inflammatory link between obesity and cardiovascular diseases. Cells 2014, 702–712. [Google Scholar] [CrossRef] [PubMed]
- Klöting, N.; Berthold, S.; Kovacs, P.; Schön, M.R.; Fasshauer, M.; Ruschke, K.; Stumvoll, M.; Blüher, M. MicroRNA expression in human omental and subcutaneous adipose tissue. PLoS ONE 2009, e4699. [Google Scholar] [CrossRef] [PubMed]
- Iliopoulos, D.; Jaeger, S.A.; Hirsch, H.A.; Bulyk, M.L.; Struhl, K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol. Cell 2010, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Sheedy, F.J.; Palsson-McDermott, E.; Hennessy, E.J.; Martin, C.; O’Leary, J.J.; Ruan, Q.; Johnson, D.S.; Chen, Y.; O’Neill, L.A. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat. Immunol. 2010, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Ganesh, K.; Khanna, S.; Sen, C.K.; Roy, S. Engulfment of apoptotic cells by macrophages: A role of microRNA-21 in the resolution of wound inflammation. J. Immunol. 2014, 1120–1129. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.-M.; Lin, K.-Y.; Chen, Y.-Q. Diverse functions of miR-125 family in different cell contexts. J. Hematol. Oncol. 2013. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Tang, Y.; Qu, B.; Cui, H.; Wang, S.; Wang, L.; Luo, X.; Huang, X.; Li, J.; Chen, S.; et al. MicroRNA-125a contributes to elevated inflammatory chemokine RANTES levels via targeting KLF13 in systemic lupus erythematosus. Arthritis Rheumatol. 2010, 3425–3435. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.; Zhu, S.; Dai, D.; Liu, Z.; Li, D.; Li, B.; Gagliani, N.; Zheng, Y.; Tang, Y.; Weirauch, M.T. Weirauch MT: MiR-125a targets effector programs to stabilize Treg-mediated immune homeostasis. Nat. Commun. 2015. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-M.; Kim, T.S.; Jo, E.-K. MiR-146 and miR-125 in the regulation of innate immunity and inflammation. BMB Rep. 2016, 49, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.L.; Shi, C.M.; Xu, G.F.; Chen, L.; Zhu, L.L.; Zhu, L.; Guo, X.R.; Xu, M.Y.; Ji, C.B. TNF-α, IL-6, and leptin increase the expression of miR-378, an adipogenesis-related microRNA in human adipocytes. Cell Biochem. Biophys. 2014, 771–776. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.; Wang, J.; Xie, W.; Lyu, Q.; Wu, J.; He, J.; Qiu, W.; Xu, N.; Zhang, Y. MiR-378a-3p enhances adipogenesis by targeting mitogen-activated protein kinase 1. Biochem. Biophys. Res. Commun. 2015, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Ishida, M.; Shimabukuro, M.; Yagi, S.; Nishimoto, S.; Kozuka, C.; Fukuda, D.; Soeki, T.; Masuzaki, H.; Tsutsui, M.; Sata, M. MicroRNA-378 regulates adiponectin expression in adipose tissue: A new plausible mechanism. PLoS ONE 2014, e111537. [Google Scholar] [CrossRef] [PubMed]
- Chou, W.-W.; Wang, Y.-T.; Liao, Y.-C.; Chuang, S.-C.; Wang, S.-N.; Juo, S.-H.H. Decreased microRNA-221 is associated with high levels of TNF-α in human adipose tissue-derived mesenchymal stem cells from obese woman. Cell. Physiol. Biochem. 2013, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Creemers, E.E.; Tijsen, A.J.; Pinto, Y.M. Circulating microRNAs: Novel biomarkers and extracellular communicators in cardiovascular disease? Circ. Res. 2012, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Hoekstra, M.; van der Lans, C.A.C.; Halvorsen, B.; Gullestad, L.; Kuiper, J.; Aukrust, P.; van Berkel, T.J.C.; Biessen, E.A.L. The peripheral blood mononuclear cell microRNA signature of coronary artery disease. Biochem. Biophys. Res. Commun. 2010, 792–797. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Nazari-Jahantigh, M.; Neth, P.; Weber, C.; Schober, A. MicroRNA-126, -145, and-155: A therapeutic triad in atherosclerosis? Arterioscler. Thromb. Vasc. Biol. 2013, 449–454. [Google Scholar] [CrossRef] [PubMed]
- Weber, C.; Schober, A.; Zernecke, A. MicroRNAs in arterial remodelling, inflammation and atherosclerosis. Curr. Drug Targets 2010, 11, 950–956. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Zhang, D.; Chen, S.; Liu, X.; Lin, L.; Huang, X.; Guo, Z.; Liu, J.; Wang, Y.; Yuan, W. Endothelial enriched microRNAs regulate angiotensin II-induced endothelial inflammation and migration. Atherosclerosis 2011, 215, 286–293. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Shi, C.; Manduchi, E.; Civelek, M.; Davies, P.F. MicroRNA-10a regulation of proinflammatory phenotype in athero-susceptible endothelium in vivo and in vitro. Proc. Natl. Acad. Sci. USA 2010, 107, 13450–13455. [Google Scholar] [CrossRef] [PubMed]
- Villeneuve, L.M.; Kato, M.; Reddy, M.A.; Wang, M.; Lanting, L.; Natarajan, R. Enhanced levels of microRNA-125b in vascular smooth muscle cells of diabetic db/db mice lead to increased inflammatory gene expression by targeting the histone methyltransferase Suv39h1. Diabetes 2010, 59, 2904–2915. [Google Scholar] [CrossRef] [PubMed]
- Nakamachi, Y.; Kawano, S.; Takenokuchi, M.; Nishimura, K.; Sakai, Y.; Chin, T.; Saura, R.; Kurosaka, M.; Kumagai, S. MicroRNA-124a is a key regulator of proliferation and monocyte chemoattractant protein 1 secretion in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Arthritis Rheumatol. 2009, 60, 1294–1304. [Google Scholar] [CrossRef] [PubMed]
- Aranha, M.M.; Santos, D.M.; Solá, S.; Steer, C.J.; Rodrigues, C.M.P. miR-34a regulates mouse neural stem cell differentiation. PLoS ONE 2011, 6, e21396. [Google Scholar] [CrossRef] [PubMed]
- Strum, J.C.; Johnson, J.H.; Ward, J.; Xie, H.; Field, J.; Hester, A.; Alford, A.; Waters, K.M. MicroRNA 132 regulates nutritional stress-induced chemokine production through repression of SirT1. Mol. Endocrinol. 2009, 23, 1876–1884. [Google Scholar] [CrossRef] [PubMed]
- Mechtler, P.; Singhal, R.; Kichina, J.V.; Bard, J.E.; Buck, M.J.; Kandel, E.S. MicroRNA analysis suggests an additional level of feedback regulation in the NF-κB signaling cascade. Oncotarget 2015, 6, 17097. [Google Scholar] [CrossRef] [PubMed]
- Cruz, K.J.C.; Oliveira, A.R.S.; Morais, J.B.S.; Severo, J.S.; Marreiro, D.N. The role of MicroRNAs on adipogenesis, chronic low grade inflammation and insulin resistance in obesity. Nutrition 2016. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Becker Buscaglia, L.E.; Barker, J.R.; Li, Y. MicroRNAs in NF-κB signaling. J. Mol. Cell Biol. 2011, 3, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Imaizumi, T.; Tanaka, H.; Tajima, A.; Yokono, Y.; Matsumiya, T.; Yoshida, H.; Tsuruga, K.; Aizawa-Yashiro, T.; Hayakari, R.; Inoue, I. IFN-γ and TNF-α synergistically induce microRNA-155 which regulates TAB2/IP-10 expression in human mesangial cells. Am. J. Nephrol. 2010, 32, 462–468. [Google Scholar] [CrossRef] [PubMed]
- Androulidaki, A.; Iliopoulos, D.; Arranz, A.; Doxaki, C.; Schworer, S.; Zacharioudaki, V.; Margioris, A.N.; Tsichlis, P.N.; Tsatsanis, C. The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity 2009, 31, 220–231. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.M.; Kahn, D.; Gibson, W.S.J.; Round, J.L.; Scholz, R.L.; Chaudhuri, A.A.; Kahn, M.E.; Rao, D.S.; Baltimore, D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity 2010, 33, 607–619. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.M.; Taganov, K.D.; Boldin, M.P.; Cheng, G.; Baltimore, D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. USA 2007, 104, 1604–1609. [Google Scholar] [CrossRef] [PubMed]
- Boldin, M.P.; Taganov, K.D.; Rao, D.S.; Yang, L.; Zhao, J.L.; Kalwani, M.; Garcia-Flores, Y.; Luong, M.; Devrekanli, A.; Xu, J. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J. Exp. Med. 2011. [Google Scholar] [CrossRef] [PubMed]
- Latruffe, N.; Lançon, A.; Frazzi, R.; Aires, V.; Delmas, D.; Michaille, J.J.; Djouadi, F.; Bastin, J.; Cherkaoui-Malki, M. Exploring new ways of regulation by resveratrol involving miRNAs, with emphasis on inflammation. Ann. N. Y. Acad. Sci. 2015, 1348, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Lancon, A.; Kaminski, J.; Tili, E.; Michaille, J.-J.; Latruffe, N. Control of MicroRNA expression as a new way for resveratrol to deliver its beneficial effects. J. Agric. Food Chem. 2012, 60, 8783–8789. [Google Scholar] [CrossRef] [PubMed]
- Takashina, M.; Inoue, S.; Tomihara, K.; Tomita, K.; Hattori, K.; Zhao, Q.-L.; Suzuki, T.; Noguchi, M.; Ohashi, W.; Hattori, Y. Different effect of resveratrol to induction of apoptosis depending on the type of human cancer cells. Int. J. Oncol. 2017, 50, 787–797. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xie, C.; Zhuang, J.; Li, H.; Yao, Y.; Shao, C.; Wang, H. Resveratrol attenuates inflammation in the rat heart subjected to ischemia-reperfusion: Role of the TLR4/NF-κB signaling pathway. Mol. Med. Rep. 2015, 11, 1120–1126. [Google Scholar] [CrossRef] [PubMed]
- Gambini, J.; Inglés, M.; Olaso, G.; Lopez-Grueso, R.; Bonet-Costa, V.; Gimeno-Mallench, L.; Mas-Bargues, C.; Abdelaziz, K.M.; Gomez-Cabrera, M.C.; Vina, J. Properties of resveratrol: In vitro and in vivo studies about metabolism, bioavailability, and biological effects in animal models and humans. Oxid. Med. Cell. Longev. 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Jun, M.; Ahn, M.-R.; Kim, O.Y. Involvement of miR-Let7A in inflammatory response and cell survival/apoptosis regulated by resveratrol in THP-1 macrophage. Nutr. Res. Pract. 2016, 10, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Jia, Z.; Li, A.; Jenkins, G.; Yang, X.; Hu, J.; Guo, W. Resveratrol repressed viability of U251 cells by miR-21 inhibiting of NF-κB pathway. Mol. Cell. Biochem. 2013, 382, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Bigagli, E.; Cinci, L.; Paccosi, S.; Parenti, A.; D’Ambrosio, M.; Luceri, C. Nutritionally relevant concentrations of resveratrol and hydroxytyrosol mitigate oxidative burst of human granulocytes and monocytes and the production of pro-inflammatory mediators in LPS-stimulated RAW 264.7 macrophages. Int. Immunopharmacol. 2017, 43, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Tomé-Carneiro, J.; Larrosa, M.; Yánez-Gascón, M.J.; Dávalos, A.; Gil-Zamorano, J.; Gonzálvez, M.; García-Almagro, F.J.; Ros, J.A.R.; Tomás-Barberán, F.A.; Espín, J.C. One-year supplementation with a grape extract containing resveratrol modulates inflammatory-related microRNAs and cytokines expression in peripheral blood mononuclear cells of type 2 diabetes and hypertensive patients with coronary artery disease. Pharmacol. Res. 2013. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Amar, J.; Iglesias, M.A.; Poggi, M.; Knauf, C.; Bastelica, D.; Neyrinck, A.M.; Fava, F.; Tuohy, K.M.; Chabo, C. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007, 1761–1772. [Google Scholar] [CrossRef] [PubMed]
- Martinez, K.B.; Leone, V.; Chang, E.B. Western diets, gut dysbiosis, and metabolic diseases: Are they linked? Gut Microbes 2017, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Núñez, B.; Pruimboom, L.; Dijck-Brouwer, D.A.J.; Muskiet, F.A.J. Lifestyle and nutritional imbalances associated with western diseases: Causes and consequences of chronic systemic low-grade inflammation in an evolutionary context. J. Nutr. Biochem. 2013, 24, 1183–1201. [Google Scholar] [CrossRef] [PubMed]
- O’Keefe, J.H.; Gheewala, N.M.; O’Keefe, J.O. Dietary strategies for improving post-prandial glucose, lipids, inflammation, and cardiovascular health. J. Am. Coll. Cardiol. 2008, 51, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qian, Y.; Fang, Q.; Zhong, P.; Li, W.; Wang, L.; Fu, W.; Zhang, Y.; Xu, Z.; Li, X. Saturated palmitic acid induces myocardial inflammatory injuries through direct binding to TLR4 accessory protein MD2. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.W.; Kwon, M.-J.; Choi, A.M.K.; Kim, H.-P.; Nakahira, K.; Hwang, D.H. Fatty acids modulate Toll-like receptor 4 activation through regulation of receptor dimerization and recruitment into lipid rafts in a reactive oxygen species-dependent manner. J. Biol. Chem. 2009, 284, 27384–27392. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Sohn, K.H.; Rhee, S.H.; Hwang, D. Saturated fatty acids, but not unsaturated fatty acids, induce the expression of cyclooxygenase-2 mediated through Toll-like receptor 4. J. Biol. Chem. 2001, 276, 16683–16689. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.-M.; Jeong, H.-J.; Park, S.-Y.; Lee, W. Saturated fatty acid-induced miR-195 impairs insulin signaling and glycogen metabolism in HepG2 cells. FEBS Lett. 2014, 588, 3939–3946. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.-M.; Min, K.-H.; Lee, W. Induction of miR-96 by dietary saturated fatty acids exacerbates hepatic insulin resistance through the suppression of INSR and IRS-1. PLoS ONE 2016, 11, e0169039. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.-M.; Min, K.-H.; Lee, W. MiR-1271 upregulated by saturated fatty acid palmitate provokes impaired insulin signaling by repressing INSR and IRS-1 expression in HepG2 cells. Biochem. Biophys. Res. Commun. 2016, 478, 1786–1791. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.-M.; Jeong, H.-J.; Park, S.-Y.; Lee, W. Induction of miR-29a by saturated fatty acids impairs insulin signaling and glucose uptake through translational repression of IRS-1 in myocytes. FEBS Lett. 2014, 588, 2170–2176. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Sasano, T.; Sugiyama, K.; Kurokawa, J.; Tamura, N.; Soejima, Y.; Sawabe, M.; Isobe, M.; Furukawa, T. High-fat diet increases vulnerability to atrial arrhythmia by conduction disturbance via miR-27b. J. Mol. Cell. Cardiol. 2016, 90, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Talukdar, S.; Bae, E.J.; Imamura, T.; Morinaga, H.; Fan, W.; Li, P.; Lu, W.J.; Watkins, S.M.; Olefsky, J.M. GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell 2010, 142, 687–698. [Google Scholar] [CrossRef]
- Calder, P.C. Omega-3 polyunsaturated fatty acids and inflammatory processes: Nutrition or pharmacology? Br. J. Clin. Pharmacol. 2013, 75, 645–662. [Google Scholar] [CrossRef] [PubMed]
- Roessler, C.; Kuhlmann, K.; Hellwing, C.; Leimert, A.; Schumann, J. Impact of polyunsaturated fatty acids on miRNA profiles of monocytes/macrophages and endothelial cells—A pilot study. Int. J. Mol. Sci. 2017, 18, 284. [Google Scholar] [CrossRef] [PubMed]
- Visioli, F.; Giordano, E.; Nicod, N.M.; Dávalos, A. Molecular targets of omega 3 and conjugated linoleic fatty acids—“micromanaging” cellular response. Front. Physiol. 2012, 3, 42. [Google Scholar] [CrossRef] [PubMed]
- Krishnamoorthy, S.; Recchiuti, A.; Chiang, N.; Fredman, G.; Serhan, C.N. Resolvin D1 receptor stereoselectivity and regulation of inflammation and proresolving microRNAs. Am. J. Pathol. 2012, 180, 2018–2027. [Google Scholar] [CrossRef] [PubMed]
- Fredman, G.; Li, Y.; Dalli, J.; Chiang, N.; Serhan, C.N. Self-limited versus delayed resolution of acute inflammation: Temporal regulation of pro-resolving mediators and microRNA. Sci. Rep. 2012, 2, 639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Filgueiras, L.R.; Wang, S.; Serezani, A.P.M.; Peters-Golden, M.; Jancar, S.; Serezani, C.H. Leukotriene B4 enhances the generation of proinflammatory microRNAs to promote MyD88-dependent macrophage activation. J. Immunol. 2014, 192, 2349–2356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zárate, R.; El Jaber-Vazdekis, N.; Tejera, N.; Pérez, J.A.; Rodríguez, C. Significance of long chain polyunsaturated fatty acids in human health. Clin. Transl. Med. 2017, 6, 25. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Dalli, J.; Chiang, N.; Baron, R.M.; Quintana, C.; Serhan, C.N. Plasticity of leukocytic exudates in resolving acute inflammation is regulated by MicroRNA and proresolving mediators. Immunity 2013, 14, 885–898. [Google Scholar] [CrossRef] [PubMed]
- Recchiuti, A.; Krishnamoorthy, S.; Fredman, G.; Chiang, N.; Serhan, C.N. MicroRNAs in resolution of acute inflammation: Identification of novel resolvin D1-miRNA circuits. FASEB J. 2011, 25, 544–560. [Google Scholar] [CrossRef] [PubMed]
- Codagnone, M.; Recchiuti, A.; Lanuti, P.; Pierdomenico, A.M.; Cianci, E.; Patruno, S.; Mari, V.C.; Simiele, F.; Di Tomo, P.; Pandolfi, A.; et al. Lipoxin A4 stimulates endothelial miR-126-5p expression and its transfer via microvesicles. FASEB J. 2017, 31, 1856–1866. [Google Scholar] [CrossRef] [PubMed]
- Chiu, S.-C.; Chiang, E.-P.I.; Tsai, S.-Y.; Wang, F.-Y.; Pai, M.-H.; Syu, J.-N.; Cheng, C.-C.; Rodriguez, R.L.; Tang, F.-Y. Eicosapentaenoic acid induces neovasculogenesis in human endothelial progenitor cells by modulating c-kit protein and PI3-K/Akt/eNOS signaling pathways. J. Nutr. Biochem. 2014, 25, 934–945. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Ge, Y.; Zhang, J.; Xue, M.; Li, Q.; Lin, D.; Ma, W. PUFA diets alter the microRNA expression profiles in an inflammation rat model. Mol. Med. Rep. 2015, 11, 4149–4157. [Google Scholar] [CrossRef] [PubMed]
- Siriwardhana, N.; Kalupahana, N.S.; Cekanova, M.; LeMieux, M.; Greer, B.; Moustaid-Moussa, N. Modulation of adipose tissue inflammation by bioactive food compounds. J. Nutr. Biochem. 2013, 24, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Hatcher, H.; Planalp, R.; Cho, J.; Torti, F.M.; Torti, S.V. Curcumin: From ancient medicine to current clinical trials. Cell. Mol. Life Sci. 2008, 65, 1631–1652. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Liu, F.; Ding, L.; You, M.; Yue, H.; Zhou, Y.; Hou, Y. Anti-inflammatory effects of curcumin are associated with down regulating microRNA-155 in LPS-treated macrophages and mice. Pharm. Biol. 2017, 55, 1263–1273. [Google Scholar] [CrossRef] [PubMed]
- Al-Ansari, M.M.; Aboussekhra, A. miR-146b-5p mediates p16-dependent repression of IL-6 and suppresses paracrine procarcinogenic effects of breast stromal fibroblasts. Oncotarget 2015, 6, 30006. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Song, Z.; Shao, W.; Du, W.W.; Zhao, L.R.; Zeng, K.; Yang, B.B.; Jin, T. Curcumin represses mouse 3T3-L1 cell adipogenic differentiation via inhibiting miR-17-5p and stimulating the Wnt signalling pathway effector Tcf7l2. Cell Death Dis. 2017, 8, e2559. [Google Scholar] [CrossRef] [PubMed]
- Marunaka, Y.; Marunaka, R.; Sun, H.; Yamamoto, T.; Kanamura, N.; Inui, T.; Taruno, A. Actions of quercetin, a polyphenol, on blood pressure. Molecules 2017, 22, 209. [Google Scholar] [CrossRef] [PubMed]
- Boesch-Saadatmandi, C.; Wagner, A.E.; Wolffram, S.; Rimbach, G. Effect of quercetin on inflammatory gene expression in mice liver in vivo—Role of redox factor 1, miRNA-122 and miRNA-125b. Pharmacol. Res. 2012, 65, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Boesch-Saadatmandi, C.; Loboda, A.; Wagner, A.E.; Stachurska, A.; Jozkowicz, A.; Dulak, J.; Döring, F.; Wolffram, S.; Rimbach, G. Effect of quercetin and its metabolites isorhamnetin and quercetin-3-glucuronide on inflammatory gene expression: Role of miR-155. J. Nutr. Biochem. 2011, 22, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Graham, H.N. Green tea composition, consumption, and polyphenol chemistry. Prev. Med. 1992, 21, 334–350. [Google Scholar] [CrossRef]
- Manach, C.; Williamson, G.; Morand, C.; Scalbert, A.; Rémésy, C. Bioavailability and bioefficacy of polyphenols in humans. I. Review of 97 bioavailability studies. Am. J. Clin. Nutr. 2005, 81, 230S–242S. [Google Scholar] [PubMed]
- Arola-Arnal, A.; Blade, C. Proanthocyanidins modulate microRNA expression in human HepG2 cells. PLoS ONE 2011, 6, e25982. [Google Scholar] [CrossRef] [PubMed]
- Yamada, S.; Tsukamoto, S.; Huang, Y.; Makio, A.; Kumazoe, M.; Yamashita, S.; Tachibana, H. Epigallocatechin-3-O-gallate up-regulates microRNA-let-7b expression by activating 67-kDa laminin receptor signaling in melanoma cells. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Rayman, M.P. The importance of selenium to human health. Lancet 2000, 356, 233–241. [Google Scholar] [CrossRef] [Green Version]
- Flores-Mateo, G.; Navas-Acien, A.; Pastor-Barriuso, R.; Guallar, E. Selenium and coronary heart disease: A meta-analysis. Am. J. Clin. Nutr. 2006, 84, 762–773. [Google Scholar] [PubMed]
- Bleys, J.; Navas-Acien, A.; Guallar, E. Serum selenium levels and all-cause, cancer, and cardiovascular mortality among US adults. Arch. Intern. Med. 2008, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Liu, Z.; Yang, G.; Gao, D.; Niu, X. MicroRNA expression profiles in rats with selenium deficiency and the possible role of the Wnt/β-catenin signaling pathway in cardiac dysfunction. Int. J. Mol. Med. 2015, 35, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Alehagen, U.; Johansson, P.; Aaseth, J.; Alexander, J.; Wågsäter, D. Significant changes in circulating microRNA by dietary supplementation of selenium and coenzyme Q10 in healthy elderly males. A subgroup analysis of a prospective randomized double-blind placebo-controlled trial among elderly Swedish citizens. PLoS ONE 2017, 12, e0174880. [Google Scholar] [CrossRef] [PubMed]
- Chasapis, C.T.; Loutsidou, A.C.; Spiliopoulou, C.A.; Stefanidou, M.E. Zinc and human health: An update. Arch. Toxicol. 2012, 86, 521–534. [Google Scholar] [CrossRef] [PubMed]
- Alder, H.; Taccioli, C.; Chen, H.; Jiang, Y.; Smalley, K.J.; Fadda, P.; Ozer, H.G.; Huebner, K.; Farber, J.L.; Croce, C.M. Dysregulation of miR-31 and miR-21 induced by zinc deficiency promotes esophageal cancer. Carcinogenesis 2012. [Google Scholar] [CrossRef] [PubMed]
- Ryu, M.-S.; Langkamp-Henken, B.; Chang, S.-M.; Shankar, M.N.; Cousins, R.J. Genomic analysis, cytokine expression, and microRNA profiling reveal biomarkers of human dietary zinc depletion and homeostasis. Proc. Natl. Acad. Sci. USA 2011, 108, 20970–20975. [Google Scholar] [CrossRef] [PubMed]
- Zeljic, K.; Supic, G.; Magic, Z. New insights into vitamin D anticancer properties: Focus on miRNA modulation. Mol. Genet. Genomics 2017, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Dambal, S.; Giangrecom, A.A.; Acosta, A.M.; Fairchild, A.; Richards, Z.; Deaton, R.; Wagner, D.; Vieth, R.; Gann, P.H.; Kajdacsy-Balla, A.; et al. microRNAs and DICER1 are regulated by 1,25-dihydroxyvitamin D in prostate stroma. J. Steroid Biochem. Mol. Biol. 2017, 167, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Kempinska-Podhorodecka, A.; Milkiewicz, M.; Wasik, U.; Ligocka, J.; Zawadzki, M.; Krawczyk, M.; Milkiewicz, P. Decreased expression of vitamin D receptor affects an immune response in primary biliary cholangitis via the VDR-miRNA155-SOCS1 pathway. Int. J. Mol. Sci. 2017, 18, 289. [Google Scholar] [CrossRef] [PubMed]
- Karkeni, E.; Bonnet, L.; Marcotorchino, J.; Tourniaire, F.; Astier, J.; Ye, J.; Landrier, J.F. Vitamin D limits inflammation-linked microRNA expression in adipocytes in vitro and in vivo: A new mechanism for the regulation of inflammation by vitamin D. Epigenetics 2017. [Google Scholar] [CrossRef] [PubMed]
- Panizo, S.; Carrillo-Lopez, N.; Naves-Diaz, M.; Solache-Berrocal, G.; Martinez-Arias, L.; Rodrigues-Diez, R.R.; Fernandez-Vazquez, A.; Martinez-Salgado, C.; Ruiz-Ortega, M.; Dusso, A.; et al. Regulation of miR-29b and miR-30c by vitamin D receptor activators contributes to attenuate uraemia-induced cardiac fibrosis. Nephrol. Dial. Transplant. 2017. [Google Scholar] [CrossRef] [PubMed]
- Mansouri, L.; Lundwall, K.; Moshfegh, A.; Jacobson, S.H.; Lundahl, J.; Spaak, J. Vitamin D receptor activation reduces inflammatory cytokines and plasma MicroRNAs in moderate chronic kidney disease—A randomized trial. BMC Nephrol. 2017, 18, 161. [Google Scholar] [CrossRef] [PubMed]
- Giangreco, A.A.; Vaishnav, A.; Wagner, D.; Finelli, A.; Fleshner, N.; Van der Kwast, T.; Vieth, R.; Nonn, L. Tumor suppressor microRNAs, miR-100 and-125b, are regulated by 1, 25-dihydroxyvitamin D in primary prostate cells and in patient tissue. Cancer Prev. Res. 2013, 6, 483–494. [Google Scholar] [CrossRef] [PubMed]
- Perri, M.; Caroleo, M.C.; Liu, N.; Gallelli, L.; De Sarro, G.; Kagechika, H.; Cione, E. 9-cis Retinoic acid modulates myotrophin expression and its miR in physiological and pathophysiological cell models. Exp. Cell Res. 2017, 354, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Gao, Y.; Yu, M.; Wu, H.; Ai, Z.; Wu, Y.; Liu, H.; Du, J.; Guo, Z.; Zhang, Y. Retinoic acid induces embryonic stem cell differentiation by altering both encoding RNA and microRNA expression. PLoS ONE 2015, 10, e0132566. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Wall, D.; Curran, C.; Newell, J.; Kerin, M.J.; Dwyer, R.M. MicroRNA-10a is reduced in breast cancer and regulated in part through retinoic acid. BMC Cancer 2015, 15, 345. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Kanno, T.; Nakayamada, S.; Hirahara, K.; Sciume, G.; Muljo, S.A.; Kuchen, S.; Casellas, R.; Wei, L.; Kanno, Y.; et al. TGF-beta and retinoic acid induce the microRNA miR-10a, which targets Bcl-6 and constrains the plasticity of helper T cells. Nat. Immunol. 2012, 13, 587–595. [Google Scholar] [CrossRef] [PubMed]
- Tsang, W.P.; Kwok, T.T. Epigallocatechin gallate up-regulation of miR-16 and induction of apoptosis in human cancer cells. J. Nutr. Biochem. 2010, 21, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Hou, D.; Chen, X.; Li, D.; Zhu, L.; Zhang, Y.; Li, J.; Bian, Z.; Liang, X.; Cai, X. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: Evidence of cross-kingdom regulation by microRNA. Cell Res. 2012, 22, 107. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Dai, G.-H.; Ren, Z.-M.; Tong, Y.-L.; Yang, F.; Zhu, Y.-Q. Identification of dietetically absorbed rapeseed (Brassica campestris L.) bee pollen microRNAs in serum of mice. Biomed. Res. Int. 2016. [Google Scholar] [CrossRef]
- Baier, S.R.; Nguyen, C.; Xie, F.; Wood, J.R.; Zempleni, J. MicroRNAs are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow milk and affect gene expression in peripheral blood mononuclear cells, HEK-293 kidney cell cultures, and mouse livers. J. Nutr. 2014, 144, 1495–1500. [Google Scholar] [CrossRef] [PubMed]
- Snow, J.W.; Hale, A.E.; Isaacs, S.K.; Baggish, A.L.; Chan, S.Y. Ineffective delivery of diet-derived microRNAs to recipient animal organisms. RNA Biol. 2013, 10, 1107–1116. [Google Scholar] [CrossRef] [PubMed]
- Micó, V.; Martín, R.; Lasunción, M.A.; Ordovás, J.M.; Daimiel, L. Unsuccessful detection of plant microRNAs in beer, extra virgin olive oil and human plasma after an acute ingestion of extra virgin olive oil. Plant Foods Hum. Nutr. 2016, 71, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Philip, A.; Ferro, V.A.; Tate, R.J. Determination of the potential bioavailability of plant microRNAs using a simulated human digestion process. Mol. Nutr. Food Res. 2015, 59, 1962–1972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, M.; Sang, X.; Hong, Z. Beyond nutrients: Food-derived microRNAs provide cross-kingdom regulation. Bioessays 2012, 34, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Benmoussa, A.; Lee, C.H.C.; Laffont, B.; Savard, P.; Laugier, J.; Boilard, E.; Gilbert, C.; Fliss, I.; Provost, P. Commercial dairy cow milk microRNAs resist digestion under simulated gastrointestinal tract conditions. J. Nutr. 2016, 146, 2206–2215. [Google Scholar] [CrossRef] [PubMed]
- Denzler, R.; Stoffel, M. Uptake and function studies of maternal milk-derived microRNAs. J. Biol. Chem. 2015, 290, 23680–23691. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Y.; Liu, Y.; Liu, H.; Wang, H.; Jin, W.; Zhang, Y.; Zhang, C.; Xu, D. Role of plant MicroRNA in cross-species regulatory networks of humans. BMC Syst. Biol. 2016, 10. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.E.; Piegholdt, S.; Ferraro, M.; Pallauf, K.; Rimbach, G. Food derived microRNAs. Food Funct. 2015, 6, 714–718. [Google Scholar] [CrossRef] [PubMed]
- Igaz, I.; Igaz, P. Hypothetic interindividual and interspecies relevance of microRNAs released in body fluids. EXS 2015, 106, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Lukasik, A.; Zielenkiewicz, P. Plant MicroRNAs—Novel players in natural medicine? Int. J. Mol. Sci. 2016, 18. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, K.; Yamamoto, Y.; Matsuoka, R.; Ochiya, T. Maintaining good miRNAs in the body keeps the doctor away? Perspectives on the relationship between food-derived natural products and microRNAs in relation to exosomes/extracellular vesicles. Mol. Nutr. Food Res. 2017. [Google Scholar] [CrossRef] [PubMed]
| Nutrient | microRNA | Regulation and Tissue | Model | Targets | Reference |
|---|---|---|---|---|---|
| EPA/DHA | miR-146a-5p, -7b-5p, -155-5p, -125a-3p | ↑ cells | Macrophage cell line (RAW264.7) and epithelial cells (TIME) | - | Roessler et al. [106] |
| EPA | miR-221 | ↓ cells | Human endothelial progenitor cells (hEPC) | - | Chiu et al. [115] |
| Oleic acid | miR-21 | ↑ cells | Hepatocytes | PTEN | Vinciguerra et al. [16] |
| Resveratrol | miR-20a-5p | ↓ plasma | Human (♀) | Ikk | Li et al. [89] |
| miR-663 | ↑ cells | Human monocytes (THP-1) | JunB; JunD | Tili et al. [15] | |
| miR-155 | ↓ cells | Human monocytes (THP-1) | - | Tili et al. [15] | |
| miR-Let7A | ↑ cells | Human monocytes (THP-1) | - | Song et al. [88] | |
| miR-21 | ↓ cells | Human glioblastoma cells (U251) | - | Li et al. [89] | |
| miR-146 | ↓ cells | Macrophage cell line (RAW264.7) | Nfr2 | Bigagli et al. [90] | |
| miR-21, miR-181b, miR-663, miR-30c2 | ↑ plasma | Human (♀) | - | Tomé-Carneiro et al. [91] | |
| miR-155, miR-34a | ↓ plasma | Human (♀) | - | Tomé-Carneiro et al. [91] | |
| Curcumin | miR-155-5p | ↓ cells | Macrophage cell line RAW264.7 | SOCS1; SHIP1 | Ma et al. [119] |
| ↓ liver and kidney | Male C57BL/6 mice | ||||
| miR181b | ↑ cells | Breast cancer cell (MDA-MB-231) | CXCL-1; CXCL-2 | Kronski et al. [20] | |
| miR-17-5p | ↑ cells | 3T3-L1 | Tcf7l2 | Tian et al. [121] | |
| Epigallocatechin gallate (EGCG) | miR-let-7b | ↑ cells | Human melanoma cells (Mewo and A375) | HMGA2; PKA; PP2A | Yamada et al. [128] |
| miR-16 | ↑ cells | Human hepatocellular carcinoma cells (HepG2) | Bcl-2 | Tsang et al. [148] | |
| miR-140-3p | ↑ cells | Human chondrocytes from osteoarthritic cartilage | ADAMTS5 | Rasheed et al. [19] | |
| Quercetin | miR-122 | ↑ liver | Female C57BL/6 mice | AOAH | Boesch-Saadatmandi et al. [123] |
| miR-125b | ↑ liver | Female C57BL/6 mice | TNF-α | Boesch-Saadatmandi et al. [123] | |
| miR-155 | ↓ cells | Macrophage cell line RAW264.7 | TNF-α | Boesch-Saadatmandi et al. [124] | |
| Selenium | miR-185, miR-625, miR-203, miR-429 | ↓ cells | Human intestinal cell line (Caco-2) | - | Maciel-Dominguez et al. [18] |
| miR-374, miR-16, miR-199a-5p, miR-195, miR-30e* | ↑ heart | Rats with selenium deficiency (miRNAs extracted from harvested heart) | - | Xing et al. [132] | |
| miR-3571, miR-675, miR-450a* | ↓ heart | - | Xing et al. [132] | ||
| Zinc | miR-31, miR-21 | ↑ esophagus and tongue | Rats with zinc deficiency | PPP2R2A; PDCD4 | Alder et al. [135] |
| miR-204, miR-296-5p | ↓ serum | Young male humans on dietary zinc depletion regimen | - | Ryu et al. [136] | |
| Vitamin D | miR-100 | ↑ cells | Primary prostatic epithelial cells | PLK1 | Giangreco et al. [143] |
| miR-125b | ↑ cells | E2F3 | Giangreco et al. [143] | ||
| miR 432-5p, miR 495-3p, and miR 576-5p | ↓ plasma | Patients with moderate chronic kidney disease | - | Mansouri et al. [142] | |
| Vitamin A | miR-200b, miR-200c | ↓ cells | Mouse embryonic stem cells | Oct4; Nanog | Zhang et al. [145] |
| miR-10a | ↑ cells | Breast cancer cell lines (T47D and SK-BR-3) | - | Khan et al. [146] | |
| ↑ cells | Naturally occurring Treg cells from Foxp3EGFP mouse | Bcl-6; Ncor2 | Takahashi et al. [147] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quintanilha, B.J.; Reis, B.Z.; Duarte, G.B.S.; Cozzolino, S.M.F.; Rogero, M.M. Nutrimiromics: Role of microRNAs and Nutrition in Modulating Inflammation and Chronic Diseases. Nutrients 2017, 9, 1168. https://doi.org/10.3390/nu9111168
Quintanilha BJ, Reis BZ, Duarte GBS, Cozzolino SMF, Rogero MM. Nutrimiromics: Role of microRNAs and Nutrition in Modulating Inflammation and Chronic Diseases. Nutrients. 2017; 9(11):1168. https://doi.org/10.3390/nu9111168
Chicago/Turabian StyleQuintanilha, Bruna J., Bruna Z. Reis, Graziela B. Silva Duarte, Silvia M. F. Cozzolino, and Marcelo M. Rogero. 2017. "Nutrimiromics: Role of microRNAs and Nutrition in Modulating Inflammation and Chronic Diseases" Nutrients 9, no. 11: 1168. https://doi.org/10.3390/nu9111168





