Bacteriological and Immunological Profiling of Meconium and Fecal Samples from Preterm Infants: A Two-Year Follow-Up Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Sampling
2.2. Human Intestinal Tract Chip (HITChip) Analysis
2.3. Immunological Analysis
2.4. Statistical Analysis
3. Results
3.1. Characteristics of the Infants
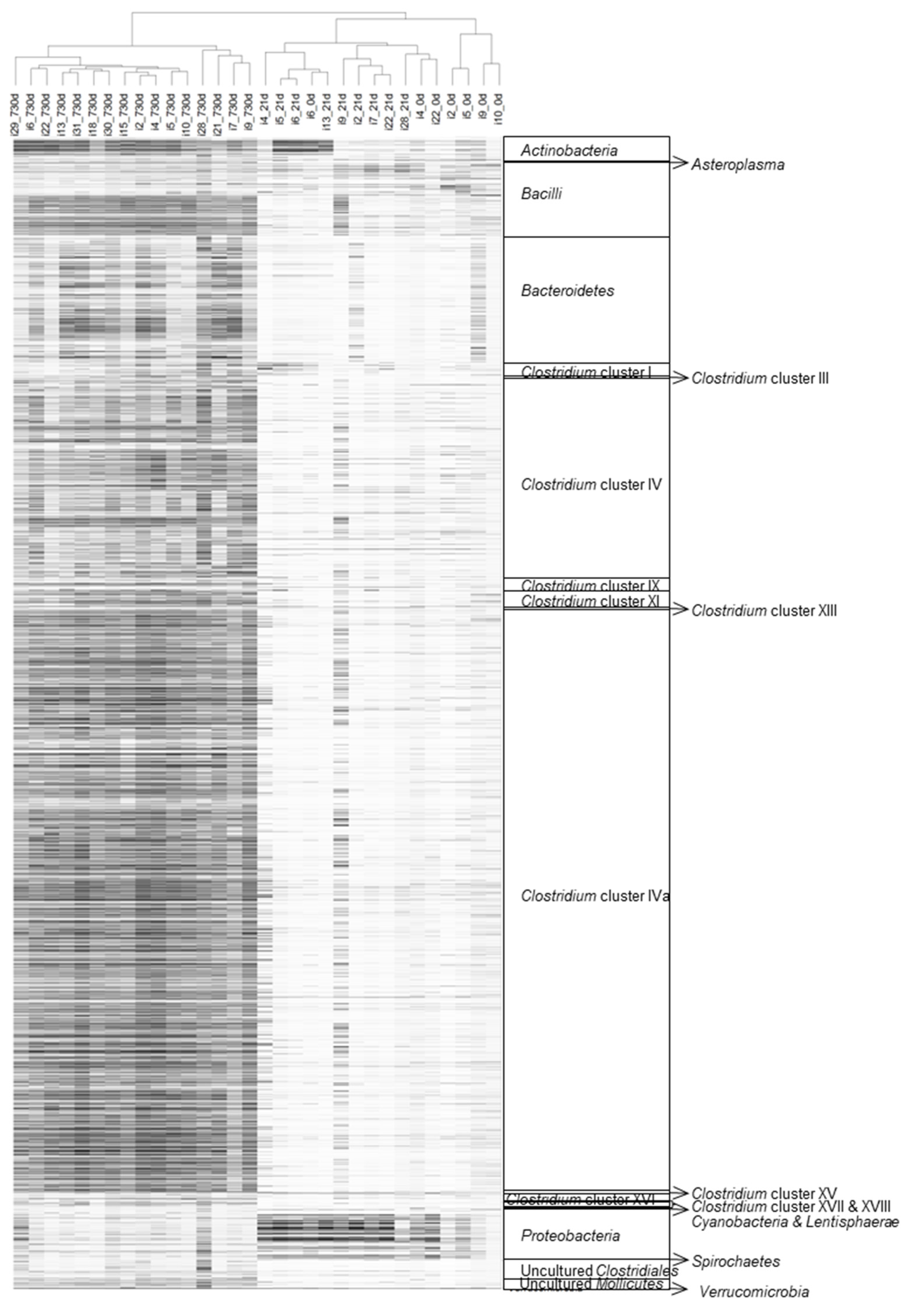
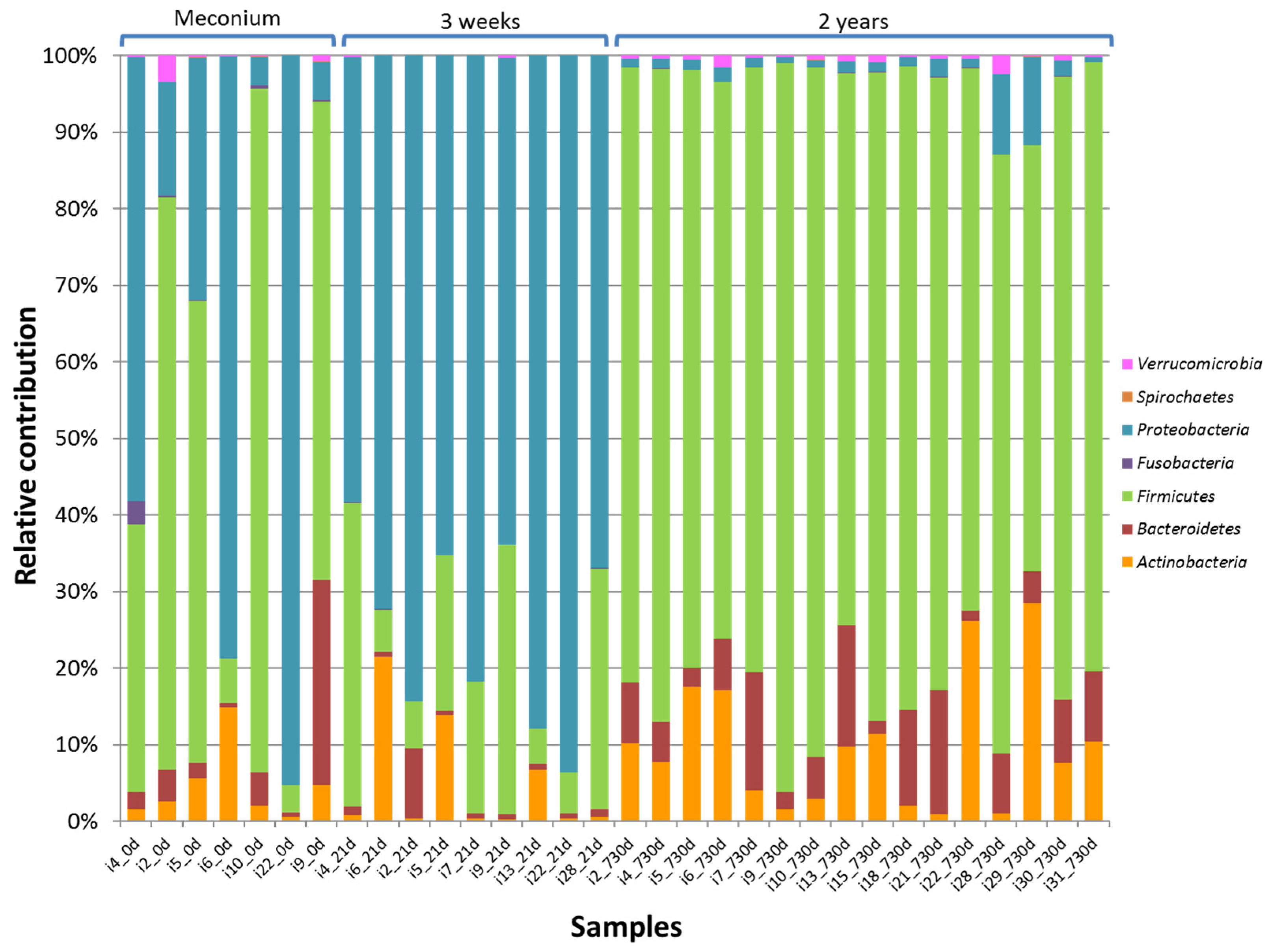
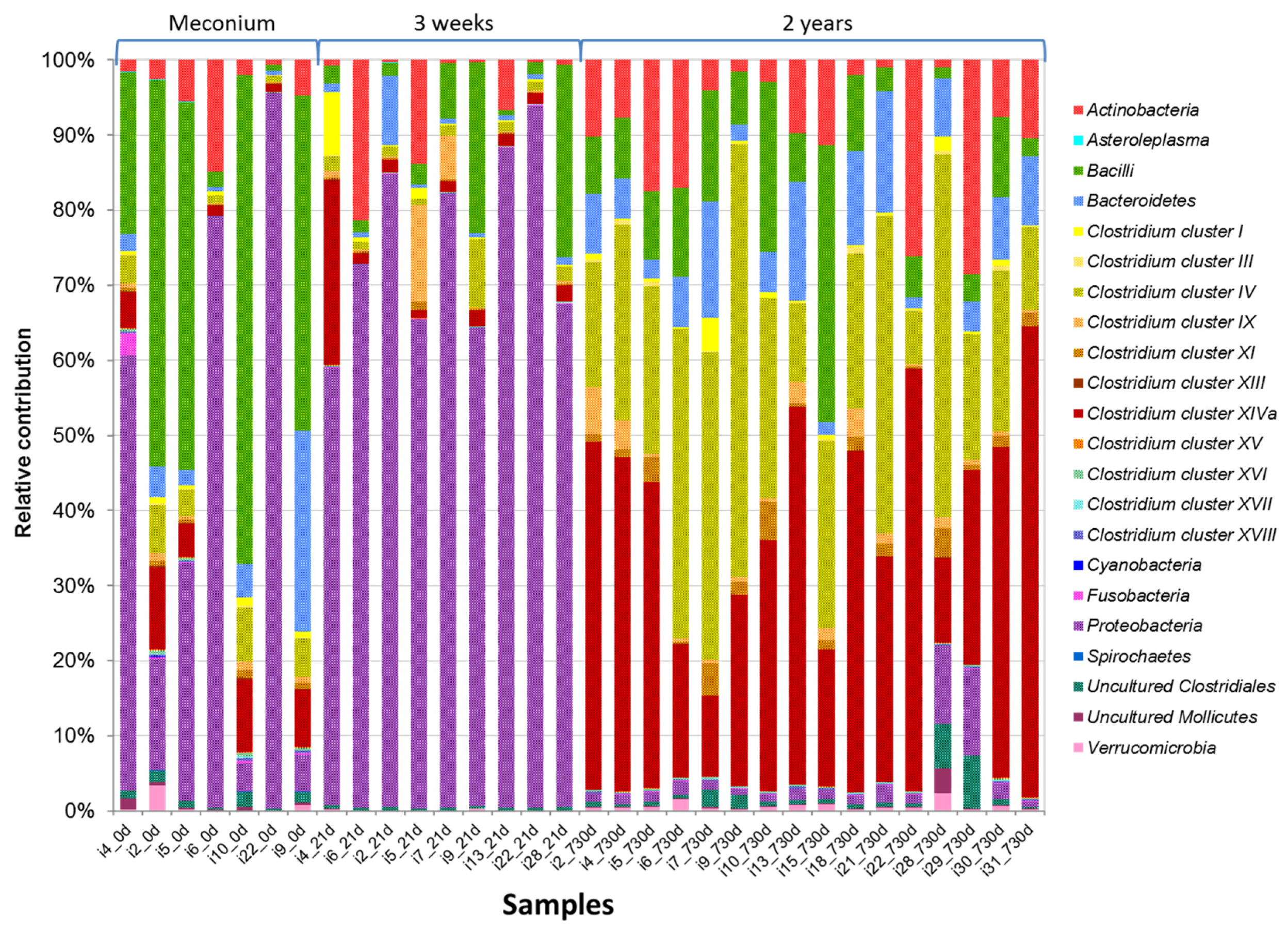
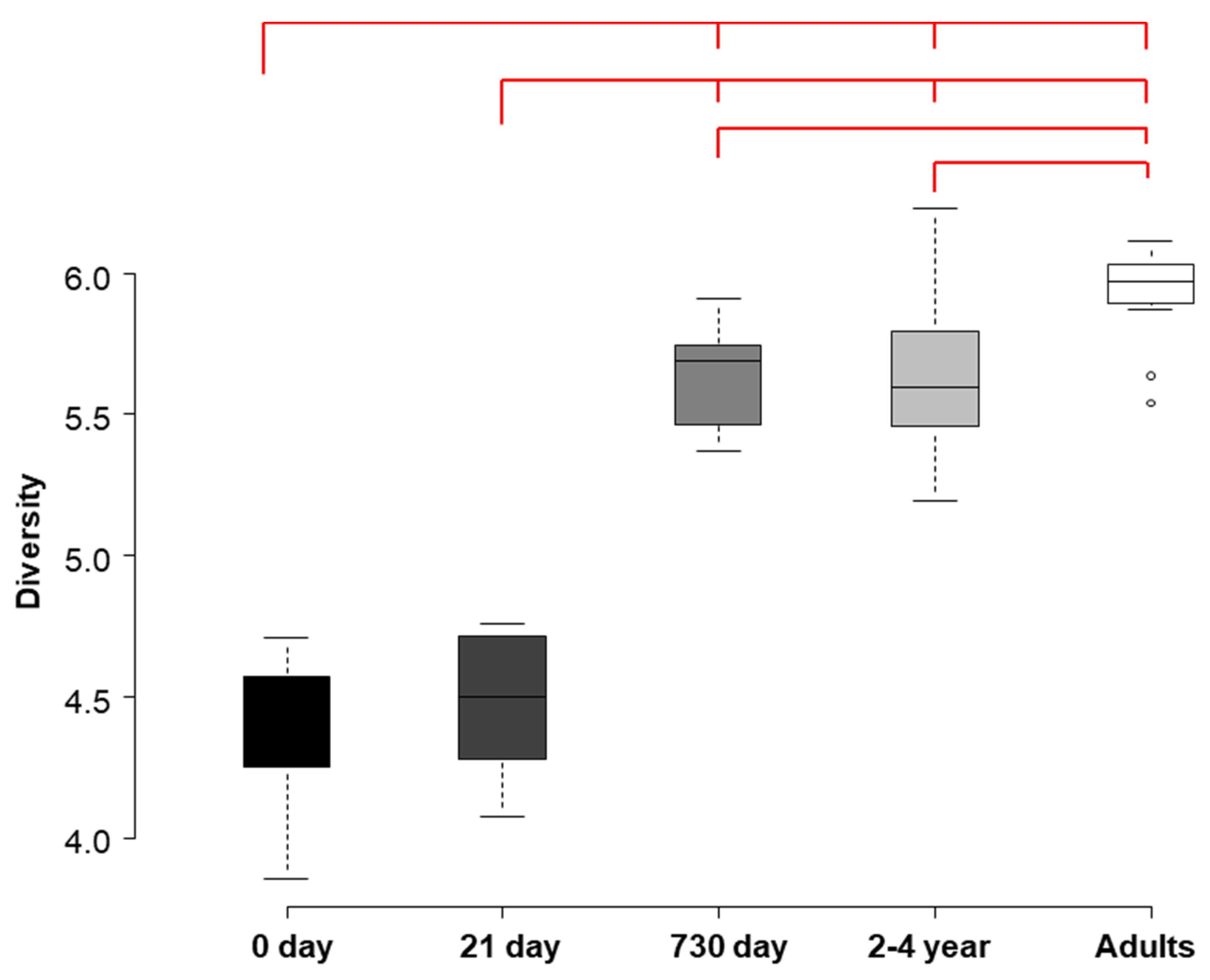
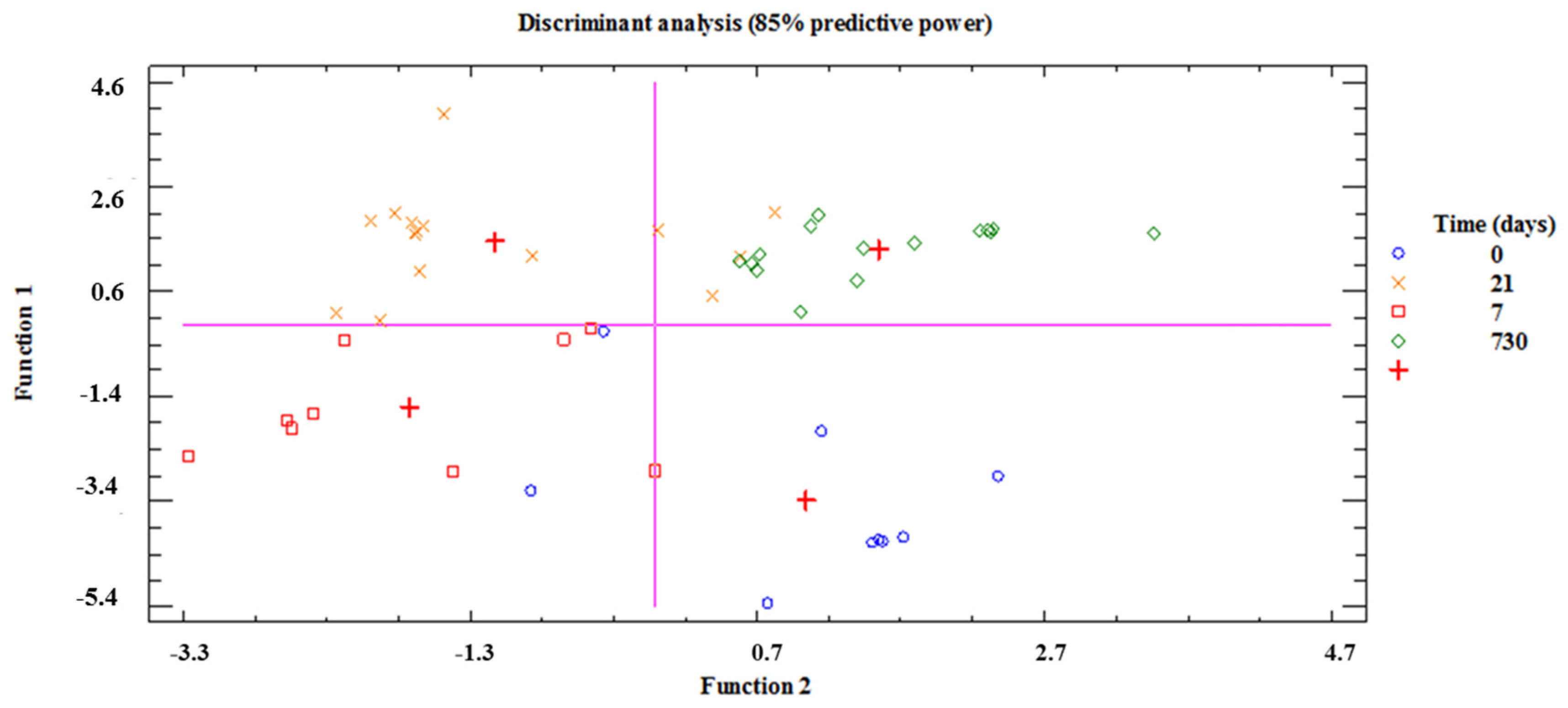
3.2. HITChip Analysis
3.3. Immunological Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author contributions
Conflicts of interest
References
- O’Toole, P.W.; Claesson, M.J. Gut microbiota: Changes throughout the lifespan from infancy to elderly. Int. Dairy J. 2010, 20, 281–291. [Google Scholar] [CrossRef]
- Woodmansey, E.J. Intestinal bacteria and ageing. J. Appl. Microbiol. 2007, 102, 1178–1186. [Google Scholar] [CrossRef] [PubMed]
- Harmsen, H.J.; Wildeboer-Veloo, A.C.; Raangs, G.C.; Wagendorp, A.A.; Klijn, N.; Bindels, J.G.; Welling, G.W. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J. Pediatr. Gastroenterol. Nutr. 2000, 30, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Mackie, R.I.; Sghir, A.; Gaskins, H.R. Developmental microbial ecology of the neonatal gastrointestinal tract. Am. J. Clin. Nutr. 1999, 69, 1035S–1045S. [Google Scholar] [PubMed]
- Fanaro, S.; Chierici, R.; Guerrini, P.; Vigi, V. Intestinal microflora in early infancy: Composition and development. Acta Paediatr. 2003, 92, 48–55. [Google Scholar] [CrossRef]
- Matamoros, S.; Gras-Leguen, C.; Le Vacon, F.; Potel, G.; de La Cochetiere, M.-F. Development of intestinal microbiota in infants and its impact on health. Trends Microbiol. 2013, 21, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, J.M.; Murphy, K.; Stanton, C.; Ross, R.P.; Kober, O.I.; Juge, N.; Avershina, E.; Rudi, K.; Narbad, A.; Jenmalm, M.C.; et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb. Ecol. Health Dis. 2015, 26, 26050. [Google Scholar] [CrossRef] [PubMed]
- Koenig, J.E.; Spor, A.; Scalfone, N.; Fricker, A.D.; Stombaugh, J.; Knight, R.; Angenent, L.T.; Ley, R.E. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4578–4585. [Google Scholar] [CrossRef] [PubMed]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Madan, J.C.; Salari, R.C.; Saxena, D.; Davidson, L.; O’Toole, G.A.; Moore, J.H.; Sogin, M.L.; Foster, J.A.; Edwards, W.H.; Palumbo, P.; et al. Gut microbial colonisation in premature neonates predicts neonatal sepsis. Arch. Dis. Child. Fetal Neonatal Ed. 2012, 97, F456–F462. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, L.; Moles, L.; Gueimonde, M.; Rodriguez, J.M. Perinatal Microbiomes’ Influence on Preterm Birth and Preterms’ Health: Influencing Factors and Modulation Strategies. J. Pediatr. Gastroenterol. Nutr. 2016, 63, e193–e203. [Google Scholar] [CrossRef] [PubMed]
- Penders, J.; Thijs, C.; Vink, C.; Stelma, F.F.; Snijders, B.; Kummeling, I.; van den Brandt, P.A.; Stobberingh, E.E. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 2006, 118, 511–521. [Google Scholar] [CrossRef] [PubMed]
- Torrazza, M.R.; Neu, J. The developing intestinal microbiome and its relationship to health and disease in the neonate. J. Perinatol. 2011, 31 (Suppl. S1), S29–S34. [Google Scholar] [CrossRef] [PubMed]
- Magne, F.; Abély, M.; Boyer, F.; Morville, P.; Pochart, P.; Suau, A. Low species diversity and high interindividual variability in faeces of preterm infants as revealed by sequences of 16S rRNA genes and PCR-temporal temperature gradient gel electrophoresis profiles. FEMS Microbiol. Ecol. 2006, 57, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Gewolb, I.; Schwalbe, R.; Taciak, V.; Harrison, T.; Panigrahi, P. Stool microflora in extremely low birthweight infants. Arch. Dis. Child. Fetal Neonatal Ed. 1999, 80, F167–F173. [Google Scholar] [CrossRef] [PubMed]
- Jacquot, A.; Neveu, D.; Aujoulat, F.; Mercier, G.; Marchandin, H.; Jumas-Bilak, E.; Picaud, J.-C. Dynamics and clinical evolution of bacterial gut microflora in extremely premature patients. J. Pediatr. 2011, 158, 390–396. [Google Scholar] [CrossRef] [PubMed]
- LaTuga, M.S.; Ellis, J.C.; Cotton, C.M.; Goldberg, R.N.; Wynn, J.L.; Jackson, R.B.; Seed, P.C. Beyond Bacteria: A Study of the Enteric Microbial Consortium in Extremely Low Birth Weight Infants. PLoS ONE 2011, 6, e27858. [Google Scholar] [CrossRef] [PubMed]
- Arboleya, S.; Binetti, A.; Salazar, N.; Fernández, N.; Solís, G.; Hernández-Barranco, A.; Margolles, A.; de los Reyes-Gavilán, C.G.; Gueimonde, M. Establishment and development of intestinal microbiota in preterm neonates. FEMS Microbiol. Ecol. 2012, 79, 763–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arboleya, S.; Solís, G.; Fernández, N.; de los Reyes-Gavilán, C.G.; Gueimonde, M. Facultative to strict anaerobes ratio in the preterm infant microbiota: A target for intervention? Gut Microbes 2012, 3, 583–598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butel, M.J.; Suau, A.; Campeotto, F.; Magne, F.; Aires, J.; Ferraris, L.; Kalach, N.; Leroux, B.; Dupont, C. Conditions of bifidobacterial colonization in preterm infants: A prospective analysis. J. Pediatr. Gastroenterol. Nutr. 2007, 44, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Mshvildadze, M.; Neu, J.; Shuster, J.; Theriaque, D.; Li, N.; Mai, V. Intestinal microbial ecology in premature infants assessed with non-culture-based techniques. J. Pediatr. 2010, 156, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Young, C.; Neu, J. Molecular Modulation of Intestinal Epithelial Barrier: Contribution of Microbiota. Available online: https://www.hindawi.com/journals/bmri/2010/305879/ (accessed on 30 August 2017).
- Di Mauro, A.; Neu, J.; Riezzo, G.; Raimondi, F.; Martinelli, D.; Francavilla, R.; Indrio, F. Gastrointestinal function development and microbiota. Ital. J. Pediatr. 2013, 39, 15. [Google Scholar] [CrossRef] [PubMed]
- Neu, J. Developmental aspects of maternal-fetal, and infant gut microbiota and implications for long-term health. Matern. Health Neonatol. Perinatol. 2015, 1, 6. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.A.; Saghafian-Hedengren, S.; Haileselassie, Y.; Roos, S.; Troye-Blomberg, M.; Nilsson, C.; Sverremark-Ekström, E. Early-life gut bacteria associate with IL-4-, IL-10- and IFN-γ production at two years of age. PLoS ONE 2012, 7, e49315. [Google Scholar] [CrossRef] [PubMed]
- Maynard, C.L.; Elson, C.O.; Hatton, R.D.; Weaver, C.T. Reciprocal interactions of the intestinal microbiota and immune system. Nature 2012, 489, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Quirós, A.; Arranz, E.; Solis, G.; Villar, A.; Ramos, A.; Coto, D. Cord blood interleukin-10 levels are increased in preterm newborns. Eur. J. Pediatr. 2000, 159, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Wan, A.K.L.; Seow, W.K.; Purdie, D.M.; Bird, P.S.; Walsh, L.J.; Tudehope, D.I. Immunoglobulins in saliva of preterm and full-term infants. Oral Microbiol. Immunol. 2003, 18, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Matoba, N.; Yu, Y.; Mestan, K.; Pearson, C.; Ortiz, K.; Porta, N.; Thorsen, P.; Skogstrand, K.; Hougaard, D.M.; Zuckerman, B.; et al. Differential patterns of 27 cord blood immune biomarkers across gestational age. Pediatrics 2009, 123, 1320–1328. [Google Scholar] [CrossRef] [PubMed]
- Sjögren, Y.M.; Tomicic, S.; Lundberg, A.; Böttcher, M.F.; Björkstén, B.; Sverremark-Ekström, E.; Jenmalm, M.C. Influence of early gut microbiota on the maturation of childhood mucosal and systemic immune responses. Clin. Exp. Allergy 2009, 39, 1842–1851. [Google Scholar] [CrossRef] [PubMed]
- Lusyati, S.; Hulzebos, C.V.; Zandvoort, J.; Sukandar, H.; Sauer, P.J.J. Cytokines patterns in newborn infants with late onset sepsis. J. Neonatal Perinat. Med. 2013, 6, 153–163. [Google Scholar] [CrossRef]
- Mohan, R.; Koebnick, C.; Schildt, J.; Mueller, M.; Radke, M.; Blaut, M. Effects of Bifidobacterium lactis Bb12 supplementation on body weight, fecal pH, acetate, lactate, calprotectin, and IgA in preterm infants. Pediatr. Res. 2008, 64, 418–422. [Google Scholar] [CrossRef] [PubMed]
- Moles, L.; Manzano, S.; Fernández, L.; Montilla, A.; Corzo, N.; Ares, S.; Rodríguez, J.M.; Espinosa-Martos, I. Bacteriological, biochemical, and immunological properties of colostrum and mature milk from mothers of extremely preterm infants. J. Pediatr. Gastroenterol. Nutr. 2015, 60, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Topf, H.-G.; Gröschl, M.; Fröhlich, T.; Tzschoppe, A.; Wenzl, T.G.; Köhler, H. Influence of gestational age, cesarean section, and type of feeding on fecal human beta-defensin 2 and tumor necrosis factor-alpha. J. Pediatr. Gastroenterol. Nutr. 2010, 51, 103–105. [Google Scholar] [CrossRef] [PubMed]
- Westerbeek, E.A.M.; Mørch, E.; Lafeber, H.N.; Fetter, W.P.F.; Twisk, J.W.R.; Van Elburg, R.M. Effect of neutral and acidic oligosaccharides on fecal IL-8 and fecal calprotectin in preterm infants. Pediatr. Res. 2011, 69, 255–258. [Google Scholar] [CrossRef] [PubMed]
- Moles, L.; Gómez, M.; Heilig, H.; Bustos, G.; Fuentes, S.; de Vos, W.; Fernández, L.; Rodríguez, J.M.; Jiménez, E. Bacterial Diversity in Meconium of Preterm Neonates and Evolution of Their Fecal Microbiota during the First Month of Life. PLoS ONE 2013, 8, e66986. [Google Scholar] [CrossRef] [PubMed]
- Rajilić-Stojanović, M.; Heilig, H.G.H.J.; Molenaar, D.; Kajander, K.; Surakka, A.; Smidt, H.; de Vos, W.M. Development and application of the human intestinal tract chip, a phylogenetic microarray: Analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environ. Microbiol. 2009, 11, 1736–1751. [Google Scholar] [CrossRef] [PubMed]
- De Goffau, M.C.; Fuentes, S.; van den Bogert, B.; Honkanen, H.; de Vos, W.M.; Welling, G.W.; Hyöty, H.; Harmsen, H.J. Aberrant gut microbiota composition at the onset of type 1 diabetes in young children. Diabetologia 2014, 57, 1569–1577. [Google Scholar] [CrossRef] [PubMed]
- Braak, C.J.F.; Smilauer, P. Canoco Reference Manual and User’s Guide: Software for Ordination; Version 5.0; Microcomputer Power: Ithaca, NY, USA, 2012. [Google Scholar]
- Moles, L.; Gómez, M.; Jiménez, E.; Fernández, L.; Bustos, G.; Chaves, F.; Cantón, R.; Rodríguez, J.M.; Del Campo, R. Preterm infant gut colonization in the neonatal ICU and complete restoration 2 years later. Clin. Microbiol. Infect. 2015, 21, 936.e1–936.e10. [Google Scholar] [CrossRef] [PubMed]
- Gómez, M.; Moles, L.; Melgar, A.; Ureta, N.; Bustos, G.; Fernández, L.; Rodríguez, J.M.; Jiménez, E. Early Gut Colonization of Preterm Infants: Effect of Enteral Feeding Tubes. J. Pediatr. Gastroenterol. Nutr. 2016, 62, 893–900. [Google Scholar] [CrossRef] [PubMed]
- Palmer, C.; Bik, E.M.; DiGiulio, D.B.; Relman, D.A.; Brown, P.O. Development of the human infant intestinal microbiota. PLoS Biol. 2007, 5, e177. [Google Scholar] [CrossRef] [PubMed]
- Hao, W.-L.; Lee, Y.-K. Microflora of the gastrointestinal tract: A review. Public Health Microbiol. Methods Protoc. 2004, 491–502. [Google Scholar]
- Wilks, M. Bacteria and early human development. Early Hum. Dev. 2007, 83, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Bennet, R.; Eriksson, M.; Nord, C.E. The fecal microflora of 1–3-month-old infants during treatment with eight oral antibiotics. Infection 2002, 30, 158–160. [Google Scholar] [CrossRef] [PubMed]
- Sholtens, P.A.M.J.; Alliet, P.; Raes, M.; Alles, M.S.; Kroes, H.; Boehm, G.; Knipples, L.M.J.; Knol, J.; Vandenpla, Y. Faecal Secretory Immunoglobulin A Is Increased in Healthy Infants Who Receive a Formula with Short-Chain Galacto-Oligosaccharides and Long-Chain Fructo-Oligosaccharides. J. Nutr. 2008, 138, 1141–1147. [Google Scholar]
- Sierra, C.; Bernal, M.J.; Blasco, J.; Martínez, R.; Dalmau, J.; Ortuño, I.; Espín, B.; Vasallo, M.I.; Gil, D.; Vidal, M.L.; et al. Prebiotic effect during the first year of life in healthy infants fed formula containing GOS as the only prebiotic: A multicentre, randomised, double-blind and placebo-controlled trial. Eur. J. Nutr. 2015, 54, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Ha, S.; Tsuji, M.; Fagarasan, S. Intestinal IgA synthesis: A primitive form of adaptive immunity that regulates microbial communities in the gut. Semin. Immunol. 2007, 19, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Brandtzaeg, P. The mucosal immune system and its integration with the mammary glands. J. Pediatr. 2010, 156, S8–S15. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, R. Cytokines in human milk. J. Pediatr. 2010, 156, S36–S40. [Google Scholar] [CrossRef] [PubMed]
- Tomicić, S.; Johansson, G.; Voor, T.; Björkstén, B.; Böttcher, M.F.; Jenmalm, M.C. Breast milk cytokine and IgA composition differ in Estonian and Swedish mothers-relationship to microbial pressure and infant allergy. Pediatr. Res. 2010, 68, 330–334. [Google Scholar] [CrossRef] [PubMed]
- Castellote, C.; Casillas, R.; Ramírez-Santana, C.; Pérez-Cano, F.J.; Castell, M.; Moretones, M.G.; López-Sabater, M.C.; Franch, A. Premature delivery influences the immunological composition of colostrum and transitional and mature human milk. J. Nutr. 2011, 141, 1181–1187. [Google Scholar] [CrossRef] [PubMed]
- Mai, V.; Torrazza, R.M.; Ukhanova, M.; Wang, X.; Sun, Y.; Li, N.; Shuster, J.; Sharma, R.; Hudak, M.L.; Neu, J. Distortions in development of intestinal microbiota associated with late onset sepsis in preterm infants. PLoS ONE 2013, 8, e52876. [Google Scholar] [CrossRef] [PubMed]
- Rogosch, T.; Kerzel, S.; Hoss, K.; Hoersch, G.; Zemlin, C.; Heckmann, M.; Berek, C.; Schroeder, H.W.; Maier, R.F.; Zemlin, M. IgA response in preterm neonates shows little evidence of antigen-driven selection. J. Immunol. 2012, 189, 5449–5456. [Google Scholar] [CrossRef] [PubMed]

| Infant | Gestational Age (week) | Delivery Mode | Gender | Birth Weight (g) |
|---|---|---|---|---|
| 2 | 30 | Caesarean section | Male | 1550 |
| 4 | 27 | Caesarean section | Female | 1080 |
| 5 | 30 | Caesarean section | Male | 2030 |
| 6 | 30 | Vaginal | Male | 1760 |
| 7 | 24 | Caesarean section | Female | 600 |
| 9 | 27 | Vaginal | Male | 1540 |
| 10 | 26 | Caesarean section | Female | 790 |
| 13 | 32 | Vaginal | Female | 1310 |
| 15 | 30 | Vaginal | Female | 1350 |
| 18 | 24 | Vaginal | Male | 740 |
| 21 | 28 | Caesarean section | Male | 1100 |
| 22 | 31 | Vaginal | Female | 1430 |
| 28 | 27 | Vaginal | Female | 1040 |
| 29 | 29 | Caesarean section | Male | 680 |
| 30 | 30 | Vaginal | Female | 1370 |
| 31 | 28 | Caesarean section | Female | 1150 |
| Infant | Hospital Stay (days) | Antibiotic Therapy (days) | Mechanical Ventilation (days) | Parenteral Nutrition (days) | Nasogastric Feeding Tube (days) | Meconium Expulsion (h) |
|---|---|---|---|---|---|---|
| 2 | 42 | 3 | 0 | 5 | 38 | 59 |
| 4 | 60 | 4 | 0.5 | 3 | 48 | 24 |
| 5 | 27 | 3 | 2 | 0 | 26 | 11 |
| 6 | 27 | 0 | 0 | 0 | 26 | 7 |
| 7 | 113 | 5 | 9 | 8 | 107 | 116 |
| 9 | 68 | 4 | 2 | 5 | 58 | 96 |
| 10 | 84 | 7 | 0.5 | 6 | 70 | 0 |
| 13 | 28 | 3 | 0 | 0 | 21 | 12 |
| 15 | 44 | 3 | 1 | 7 | 38 | 11 |
| 18 | 116 | 27 | 35 | 13 | 112 | 144 |
| 21 | 73 | 7 | 10 | 14 | 62 | 48 |
| 22 | 37 | 3 | 0 | 4 | 35 | 3 |
| 28 | 68 | 7 | 0 | 7 | 62 | 12 |
| 29 | 70 | 15 | 8 | 9 | 60 | 144 |
| 30 | 41 | 4 | 0 | 3 | 40 | |
| 31 | 52 | 3 | 0 | 6 | 47 | 9 |
| Mean (95% CI) | 58.64 (41.35; 75.93) | 4.85 (2.66; 7.05) | 52.40 (36.03; 68.82) | 46.4 (17.22;75.58) | ||
| Median (IQR) | 3.5 (3.0–7.0) | 0.25 (0.00–1.75) |
| Phylum/Order | Genus-Like Phylogenetic Group ¥ | 21 Days Mean (95% CI) | 730 Days Mean (95% CI) | p-Value * | Fold Change # |
|---|---|---|---|---|---|
| Actinobacteria | Collinsella | 0.04 (0.03; 0.04) | 0.25 (0.18; 0.32) | 0.009 | ↑6.77 |
| Bacteroidetes | Allistipes et rel. | 0.09 (0.08; 0.11) | 0.38 (0.17; 0.59) | 0.009 | ↑4.06 |
| Bacteroides ovatus et rel. | 0.06 (0.05; 0.07) | 0.38 (0.05; 0.71) | 0.009 | ↑6.69 | |
| Bacteroides plebeius et rel. | 0.04 (0.03; 0.04) | 0.12 (0.08; 0.16) | 0.009 | ↑3.23 | |
| Bacteroides splachnicus et rel. | 0.09(0.08; 0.11) | 0.26 (0.11; 0.41) | 0.016 | ↑2.73 | |
| Bacteroides stercoris et rel. | 0.04 (0.03; 0.04) | 0.11 (0.05; 0.16) | 0.009 | ↑2.97 | |
| Bacteroides vulgatus et rel. | 0.08 (0.05; 0.09) | 3.12 (1.54; 4.70) | 0.009 | ↑39.50 | |
| Parabacteroides distasonis et rel. | 0.06 (0.05; 0.07) | 0.32 (0.16; 0.48) | 0.009 | ↑5.20 | |
| Prevotella melaninogenica et rel. | 0.11 (0.09; 0.13) | 0.88 (−0.16; 1.92) | 0.033 | ↑8.20 | |
| Prevotella oralis et rel. | 0.04 (0.03; 0.04) | 0.24 (−0.02; 0.50) | 0.016 | ↑6.55 | |
| Prevotella tannerae et rel. | 0.02 (0.02; 0.03) | 0.10 (0.07; 0.13) | 0.009 | ↑4.20 | |
| Tannerella et rel. | 0.05 (0.04; 0.06) | 0.13 (0.11; 0.16) | 0.009 | ↑2.63 | |
| Bacilli | Enterococcus | 3.49 (−1.05; 8.03) | 0.10 (0.06; 0.13) | 0.009 | ↓36.18 |
| Granulicatella | 1.00 (0.19; 1.80) | 0.02 (0.01; 0.02) | 0.009 | ↓61.31 | |
| Lactococcus | 0.02 (0.01; 0.02) | 0.73 (−0.02; 1.48) | 0.056 | ↑41.05 | |
| Staphylococcus | 0.19 (−0.05; 0.43) | 0.02 (nd) | 0.033 | ↓10.86 | |
| Streptococcus intermedius et rel. | 0.04 (0.02; 0.05) | 0.15 (0.11; 0.20) | 0.033 | ↑3.96 | |
| Clostridium cluster III | Clostridium stercorarium et rel. | 0.03 (0.03; 0.04) | 0.28 (0.15; 0.41) | 0.009 | ↑9.26 |
| Clostridium cluster IV | Clostridium cellulosi et rel. | 0.13 (0.11; 16) | 3.49 (1.20; 5.79) | 0.009 | ↑26.48 |
| Clostridium leptum et rel. | 0.18 (0.09; 0.28) | 2.16 (0.74; 3.57) | 0.009 | ↑11.81 | |
| Clostridium orbiscindens et rel. | 1.03 (−0.68; 2.73) | 5.59 (3.90; 7.29) | 0.042 | ↑5.44 | |
| Faecalibacterium prausnitzii et rel. | 0.17 (0.14; 0.20) | 5.02 (1.81; 8.22) | 0.009 | ↑29.35 | |
| Oscillospira guillermondii et rel. | 0.15 (0.13; 0.18) | 3.10 (−0.13; 6.32) | 0.009 | ↑20.04 | |
| Papillibacter cinnamivorans et rel. | 0.06 (0.05; 0.07) | 0.50 (0.34; 0.66) | 0.009 | ↑8.56 | |
| Ruminococcus bromii et rel. | 0.02 (0.02; 0.03) | 1.31 (0.20; 2.43) | 0.009 | ↑52.65 | |
| Ruminococcus callidus et rel. | 0.08 (0.07; 0.09) | 0.86 (−0.06; 1.78) | 0.009 | ↑10.57 | |
| Sporobacter termitidis et rel. | 0.16 (0.14; 0.19) | 1.89 (0.96; 2.81) | 0.009 | ↑11.56 | |
| Subdoligranulum variable at rel. | 0.11 (0.09; 0.13) | 2.98 (1.48; 4.48) | 0.009 | ↑27.51 | |
| Clostridium cluster IX | Dialister | 0.05 (0.02; 0.07) | 0.75 (−0.03; 1.54) | 0.009 | ↑16.10 |
| Clostridium cluster XI | Anaerovorax odorimutans et rel. | 0.05 (0.04; 0.06) | 0.41 (0.27; 0.55) | 0.009 | ↑7.79 |
| Clostridium cluster XIVa | Anaerostipes caccae et rel. | 0.09 (0.05; 0.12) | 3.94 (2.10; 5.77) | 0.009 | ↑45.14 |
| Bryantella formatexigens et rel. | 0.14 (0.08; 0.21) | 0.69 (0.50; 0.89) | 0.009 | ↑4.82 | |
| Butyrivibrio crossotus et rel. | 0.16 (0.09; 0.23) | 1.41 (1.14; 1.68) | 0.009 | ↑8.89 | |
| Clostridium colinum et rel. | 0.05 (0.04; 0.06) | 0.14 (0.06; 0.23) | 0.009 | ↑2.77 | |
| Clostridium sphenoides et rel. | 0.12 (0.09; 0.14) | 0.85 (0.56; 1.14) | 0.009 | ↑7.38 | |
| Clostridium symbiosum et rel. | 0.22 (0.13; 0.32) | 1.81 (1.38; 2.24) | 0.009 | ↑8.09 | |
| Coprococcus eutactus et rel. | 0.08 (0.06; 0.09) | 4.96 (2.87; 7.04) | 0.009 | ↑64.93 | |
| Dorea formicigenerans et rel. | 0.16 (0.08; 0.23) | 2.63 (1.86; 3.40) | 0.009 | ↑16.72 | |
| Eubacterium hallii et rel. | 0.04 (0.02; 0.06) | 2.23 (1.40; 3.05) | 0.009 | ↑52.41 | |
| Eubacterium rectale et rel. | 0.06 (0.04; 0.08) | 0.42 (0.26; 0.58) | 0.009 | ↑7.09 | |
| Eubacterium ventriosum et rel. | 0.05 (0.02; 0.07) | 0.28 (0.13; 0.42) | 0.009 | ↑6.00 | |
| Lachnobacillus bovis et rel. | 0.07 (0.05; 0.07) | 0.31 (0.26; 0.36) | 0.009 | ↑4.43 | |
| Lachnospira pectinoschiza et rel. | 0.10 (0.07; 0.13) | 0.40 (0.28; 0.53) | 0.009 | ↑4.08 | |
| Roseburia intestinalis et rel. | 0.03 (0.01; 0.05) | 0.24 (0.15; 0.34) | 0.009 | ↑7.86 | |
| Ruminococcus lactaris et rel. | 0.03 (0.02; 0.03) | 0.42 (−0.18; 1.02) | 0.009 | ↑16.30 | |
| Ruminococcus obeum et rel. | 0.26 (0.21; 0.31) | 11.42 (8.18; 14.66) | 0.009 | ↑43.96 | |
| Uncultured Clostridiales | Uncultured Clostridiales I | 0.17 (0.14; 0.20) | 1.11 (0.12; 2.10) | 0.009 | ↑6.48 |
| Uncultured Clostridiales II | 0.18 (0.16; 0.21) | 0.36 (0.21; 0.51) | 0.009 | ↑1.97 | |
| Uncultured Mollicutes | Uncultured Mollicutes | 0.08 (0.07; 0.10) | 0.32 (−0.07; 0.70) | 0.022 | ↑3.94 |
| Proteobacteria | Burkholderia | 0.01 (nd) | 0.10 (−0.04; 0.24) | 0.016 | ↑10.98 |
| Enterobacter aerogenes et rel. | 14.82 (8.07; 21.57) | 0.21 (0.10; 0.32) | 0.009 | ↓69.90 | |
| Escherichia coli et rel. | 37.06 (24.37; 49.76) | 0.82 (−0.25; 1.88) | 0.009 | ↓45.41 | |
| Klebisiella pneumoniae et rel. | 15.75 (10.18; 21.32) | 0.11 (0.05; 0.18) | 0.009 | ↓138.34 | |
| Oxalobacter formigenes et rel. | 0.02 (0.02; 0.03) | 0.17 (0.02; 0.32) | 0.009 | ↑7.35 | |
| Proteus et rel. | 0.29 (0.17; 0.42) | 0.09 (0.05; 0.12) | 0.009 | ↓3.46 | |
| Serratia | 5.18 (1.88; 8.49) | 0.15 (−0.04; 0.33) | 0.009 | ↓35.00 | |
| Sutterella wadsworthia et rel. | 0.08 (0.07; 0.10) | 0.23 (0.07; 0.39) | 0.009 | ↑2.79 | |
| Vibrio | 0.11 (0.08; 0.15) | 0.04 (0.03; 0.04) | 0.009 | ↓2.94 | |
| Yersinia et rel. | 1.16 (0.63; 1.68) | 0.04 (0.03; 0.04) | 0.009 | ↓31.05 | |
| Verrucomicrobia | Akkermansia | 0.06 (0.00; 0.13) | 0.62 (0.34; 0.91) | 0.016 | ↑9.64 |
| Meconium (n = 9) | Feces (7 days) (n = 9) | Feces (21 days) (n = 15) | Feces (2 years) (n = 15) | p-Value ♯ | p-Value ǂ | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| n (%) 1 | Median (IQR) | n (%) | Median (IQR) | n (%) | Median (IQR) | n (%) | Median (IQR) | |||
| Innate immunity | ||||||||||
| IL-1β | 2 (22.22) | 1.24 (0.68–1.80) | 7 (77.78) | 0.13 (0.05–0.30) | 13 (86.67) | 0.05 (0.03–0.38) | 0 | - | 0.000 | 0.520 |
| IL-6 | 1 (11.11) | 0.03 | 2 (22.22) | 0.02 (0.02–0.03) | 3 (20.00) | 0.05 (0.05–0.71) | 4 (26.67) | 0.3 (0.15–0.47) | 0.094 | 0.255 |
| IL-12p70 | 1 (11.11) | 0.06 | 2 (22.22) | 0.11 (0.08–0.15) | 4 (26.67) | 0.07 (0.05–0.12) | 4 (26.67) | 0.84 (0.28–1.38) | 0.018 | 0.601 |
| IFN-γ * | 0 | - | 0 | - | 1 (6.67) | 3.27 | 3 (20.00) | 9.27 (−7.44; 25.98) | 0.000 | 0.521 |
| TNF-α | 1 (11.11) | 0.14 | 2 (22.22) | 0.15 (0.13–0.17) | 5 (33.33) | 0.18 (0.10–0.25) | 3 (20.00) | 0.27 (0.17–0.43) | 0.002 | 0.934 |
| Acquired immunity | ||||||||||
| IL-2 | 1 (11.11) | 0.92 | 1 (11.11) | 0.09 | 2 (13.33) | 0.04 (0.03–0.05) | 4 (26.67) | 0.04 (0.02–0.15) | 0.003 | 0.373 |
| IL-4 | 4 (44.44) | 0.01 (0.00–0.01) | 6 (66.67) | 0.01 (0.00–0.01) | 11 (73.33) | 0.01 (0.00–0.01) | 15 (100.00) | 0.01 (0.00–0.01) | 0.000 | 0.292 |
| IL-10 | 0 | - | 1 (11.11) | 0.03 | 1 (6.67) | 0.04 | 4 (26.67) | 0.03 (0.02–0.23) | 0.000 | 0.675 |
| IL-13 * | 1 (11.11) | 0.03 | 1 (11.11) | 0.05 | 2 (13.33) | 0.08 (−0.10; 0.26) | 1 (6.67) | 0.05 | 0.568 | 0.571 |
| IL-17 | 4 (44.44) | 3.09 (2.32–3.31) | 2 (22.22) | 0.08 (0.05–0.12) | 9 (60.00) | 0.07 (0.03–0.19) | 12 (80.00) | 0.08 (0.04–0.06) | 0.000 | 0.122 |
| Chemokines | ||||||||||
| IL-8 * | 4 (44.44) | 0.07 (−0.08; 0.21) | 1 (11.11) | 0.05 | 3 (20.00) | 0.09 (−0.02; 0.20) | 0 | - | 0.000 | 0.865 |
| MCP-1 | 3 (33.33) | 0.03 (0.02–0.03) | 1 (11.11) | 0.03 | 4 (26.67) | 0.05 (0.03–0.06) | 9 (60.00) | 0.19 (0.06–0.88) | 0.000 | 0.080 |
| MIP-1β | 6 (66.67) | 0.62 (0.05–1.78) | 6 (66.67) | 0.05 (0.04–0.16) | 8 (53.33) | 0.07 (0.02–0.10) | 2 (13.33) | 0.05 (0.04–0.07) | 0.000 | 0.639 |
| GRO-α | 3 (33.33) | 0.18 (0.16–8.28) | 0 (0.00) | - | 6 (40.00) | 0.16 (0.11–0.19) | 3 (20.00) | 0.14 (0.13–0.22) | 0.000 | 0.537 |
| Hematopoyetic factors | ||||||||||
| IL-5 * | 0 | - | 0 | - | 1 (6.67) | 0.03 | 5 (33.33) | 0.25(0.03; 0.47) | 0.000 | 0.322 |
| IL-7 | 1 (11.11) | 0.03 | 1 (11.11) | 0.04 | 3 (20.00) | 0.03 (0.02–0.05) | 0 | - | 0.000 | 0.766 |
| G-CSF | 4 (44.44) | 1.64 (0.34–4.29) | 6 (66.67) | 0.06 (0.04–0.17) | 12 (80.00) | 0.28 (0.15–0.55) | 12 (80.00) | 0.14 (0.11–0.46) | 0.000 | 0.220 |
| GM-CSF | 9 (100.00) | 0.59 a (0.28–55.03) | 7 (77.78) | 0.23 ab (0.12–0.40) | 15 (100.00) | 0.23 ab (0.18–0.26) | 15 (100.00) | 0.67 ac (0.62–0.87) | 0.000 | 0.002 |
| Meconium (n = 9) | Feces (7 days) (n = 9) | Feces (21 days) (n = 15) | Feces (2 years) (n = 15) | p-Value ♯ | p-Value ǂ | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| n (%) 1 | Median (IQR) | n (%) | Median (IQR) | n (%) | Median (IQR) | n (%) | Median (IQR) | |||
| IgG1 | 4 (44.44) | 0.03 (0.02–0.06) | 2 (22.22) | 0.34 (0.18–0.50) | 3 (20.00) | 0.32 (0.17–0.76) | 1 (6.67) | 0.06 | 0.000 | 0.621 |
| IgG2 | 6 (66.67) | 1.54 (0.84–3.39) | 7 (77.78) | 0.99 (0.84–1.19) | 3 (20.00) | 0.35 (0.27–0.62) | 2 (13.33) | 0.34 (0.24–0.44) | 0.000 | 0.062 |
| IgG3 * | 0 | - | 1 (11.11) | 0.01 | 0 | - | 2 (13.33) | 0.01 (−0.01; 0.03) | 0.000 | 0.326 |
| IgG4 * | 2 (22.22) | 0.00 (−0.01; 0.01) | 3 (33.33) | 0.01 (−0.00; 0.02) | 1 (6.67) | 0.00 | 1 (6.67) | 0.01 | 0.000 | 0.510 |
| IgM | 1 (2.08) | 0.55 | 6 (12.50) | 2.27 (0.53–5.14) | 11 (22.92) | 0.54 (0.33–0.88) | 1 (6.67) | 0.08 | 0.000 | 0.368 |
| IgA | 5 (55.56) | 1.02 a (0.48–67.45) | 8 (88.89) | 26.62 ab (10.42–58.88) | 15 (100.00) | 6.24 ab (3.69–26.13) | 13 (86.67) | 1.13 ac (0.81–3.18) | 0.000 | 0.004 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gómez, M.; Moles, L.; Espinosa-Martos, I.; Bustos, G.; De Vos, W.M.; Fernández, L.; Rodríguez, J.M.; Fuentes, S.; Jiménez, E. Bacteriological and Immunological Profiling of Meconium and Fecal Samples from Preterm Infants: A Two-Year Follow-Up Study. Nutrients 2017, 9, 1293. https://doi.org/10.3390/nu9121293
Gómez M, Moles L, Espinosa-Martos I, Bustos G, De Vos WM, Fernández L, Rodríguez JM, Fuentes S, Jiménez E. Bacteriological and Immunological Profiling of Meconium and Fecal Samples from Preterm Infants: A Two-Year Follow-Up Study. Nutrients. 2017; 9(12):1293. https://doi.org/10.3390/nu9121293
Chicago/Turabian StyleGómez, Marta, Laura Moles, Irene Espinosa-Martos, Gerardo Bustos, Willem M. De Vos, Leónides Fernández, Juan M. Rodríguez, Susana Fuentes, and Esther Jiménez. 2017. "Bacteriological and Immunological Profiling of Meconium and Fecal Samples from Preterm Infants: A Two-Year Follow-Up Study" Nutrients 9, no. 12: 1293. https://doi.org/10.3390/nu9121293





