Effects of Consuming Xylitol on Gut Microbiota and Lipid Metabolism in Mice
Abstract
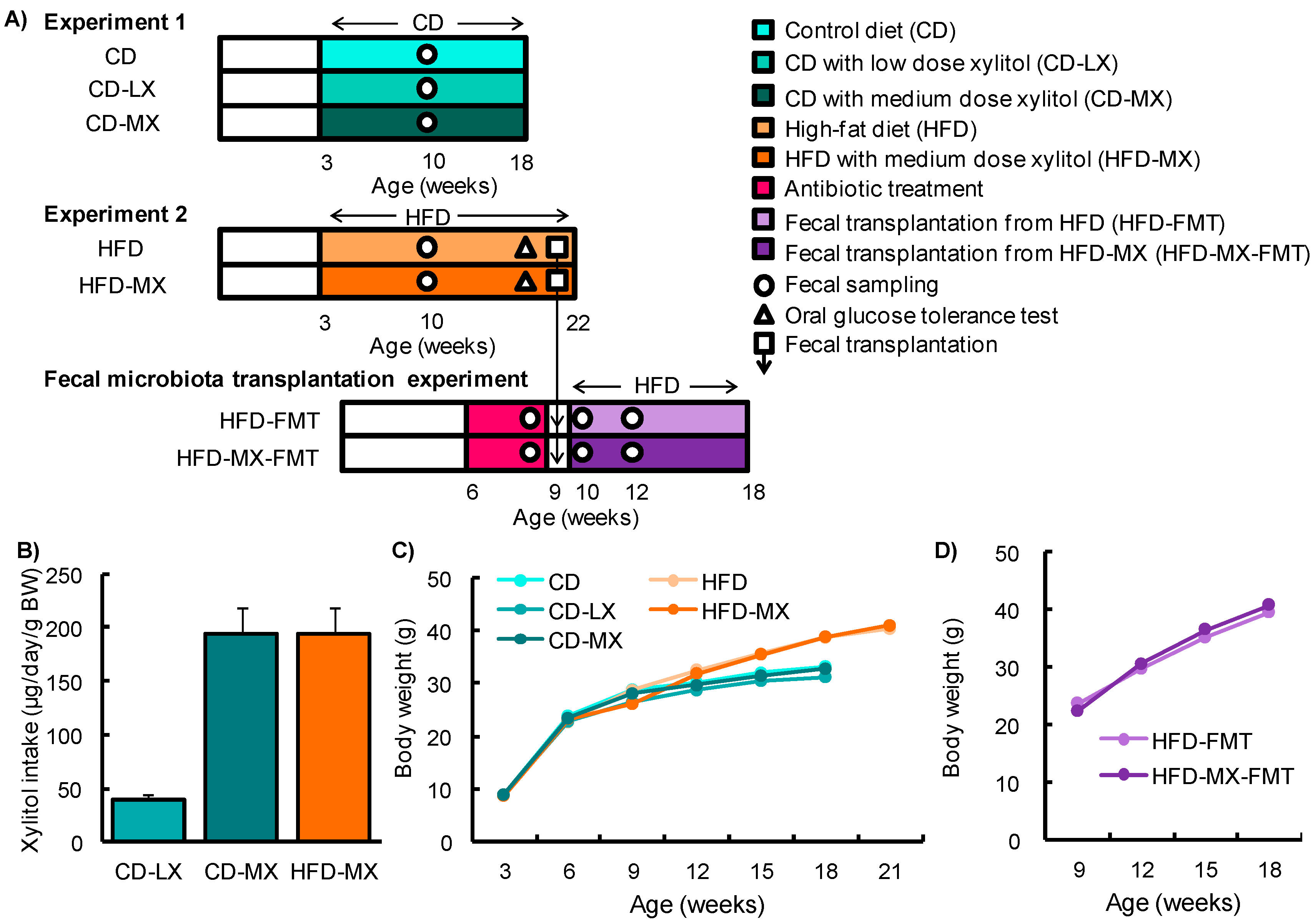
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Oral Glucose Tolerance Test
2.3. Extraction of Genomic DNA and Quantitative PCR
2.4. PCR-DGGE Analysis
2.5. Determination of Bacterial Strain by Sequence Analysis
2.6. Plasma and Hepatic Lipid Concentrations
2.7. RNA Preparation and Quantitative Reverse Transcriptase PCR
2.8. Metabolome Analysis of Cecum Luminal Content by Capillary Electrophoresis Electrospray Ionization Time-of-Flight Mass Spectrometry
2.9. Statistical Analyses
3. Results
4. Discussion and Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chassaing, B.; Koren, O.; Goodrich, J.K.; Poole, A.C.; Srinivasan, S.; Ley, R.E.; Gewirtz, A.T. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 2015, 519, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Kovatcheva-Datchary, P.; Nilsson, A.; Akrami, R.; Lee, Y.S.; De Vadder, F.; Arora, T.; Hallen, A.; Martens, E.; Bjorck, I.; Backhed, F. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of prevotella. Cell Metab. 2015, 22, 971–982. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.A.; Bennett, B.J. Diet and gut microbial function in metabolic and cardiovascular disease risk. Curr. Dia. Rep. 2016, 16, 93. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, B.O.; Backhed, F. Signals from the gut microbiota to distant organs in physiology and disease. Nat. Med. 2016, 22, 1079–1089. [Google Scholar] [CrossRef] [PubMed]
- Suez, J.; Korem, T.; Zeevi, D.; Zilberman-Schapira, G.; Thaiss, C.A.; Maza, O.; Israeli, D.; Zmora, N.; Gilad, S.; Weinberger, A.; et al. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature 2014, 514, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Soderling, E.M.; Ekman, T.C.; Taipale, T.J. Growth inhibition of streptococcus mutans with low xylitol concentrations. Curr. Microbiol. 2008, 56, 382–385. [Google Scholar] [CrossRef] [PubMed]
- Uebanso, T.; Taketani, Y.; Yamamoto, H.; Amo, K.; Ominami, H.; Arai, H.; Takei, Y.; Masuda, M.; Tanimura, A.; Harada, N.; et al. Paradoxical regulation of human FGF21 by both fasting and feeding signals: Is FGF21 a nutritional adaptation factor? PLoS ONE 2011, 6, e22976. [Google Scholar] [CrossRef] [PubMed]
- Amo, K.; Arai, H.; Uebanso, T.; Fukaya, M.; Koganei, M.; Sasaki, H.; Yamamoto, H.; Taketani, Y.; Takeda, E. Effects of xylitol on metabolic parameters and visceral fat accumulation. J. Clin. Biochem. Nutr. 2011, 49, 1–7. [Google Scholar] [PubMed]
- Rahman, M.A.; Islam, M.S. Xylitol improves pancreatic islets morphology to ameliorate type 2 diabetes in rats: A dose response study. J. Food Sci. 2014, 79, H1436–H1442. [Google Scholar] [CrossRef] [PubMed]
- Tamura, M.; Hoshi, C.; Hori, S. Xylitol affects the intestinal microbiota and metabolism of daidzein in adult male mice. Int. J. Mol. Sci. 2013, 14, 23993–24007. [Google Scholar] [CrossRef] [PubMed]
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2006, 47, 241–259. [Google Scholar] [CrossRef] [PubMed]
- Reynier, M.O.; Montet, J.C.; Gerolami, A.; Marteau, C.; Crotte, C.; Montet, A.M.; Mathieu, S. Comparative effects of cholic, chenodeoxycholic, and ursodeoxycholic acids on micellar solubilization and intestinal absorption of cholesterol. J. Lipid Res. 1981, 22, 467–473. [Google Scholar] [PubMed]
- Jiang, C.; Xie, C.; Lv, Y.; Li, J.; Krausz, K.W.; Shi, J.; Brocker, C.N.; Desai, D.; Amin, S.G.; Bisson, W.H.; et al. Intestine-selective farnesoid x receptor inhibition improves obesity-related metabolic dysfunction. Nat. Commun. 2015, 6, 10166. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.; Gioiello, A.; Noriega, L.; Strehle, A.; Oury, J.; Rizzo, G.; Macchiarulo, A.; Yamamoto, H.; Mataki, C.; Pruzanski, M.; et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009, 10, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Kimura, I.; Ozawa, K.; Inoue, D.; Imamura, T.; Kimura, K.; Maeda, T.; Terasawa, K.; Kashihara, D.; Hirano, K.; Tani, T.; et al. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat. Commun. 2013, 4, 1829. [Google Scholar] [CrossRef] [PubMed]
- Campus, G.; Cagetti, M.G.; Sale, S.; Petruzzi, M.; Solinas, G.; Strohmenger, L.; Lingstrom, P. Six months of high-dose xylitol in high-risk caries subjects—A 2-year randomised, clinical trial. Clin. Oral Investig. 2013, 17, 785–791. [Google Scholar] [CrossRef] [PubMed]
- Soderling, E.; ElSalhy, M.; Honkala, E.; Fontana, M.; Flannagan, S.; Eckert, G.; Kokaras, A.; Paster, B.; Tolvanen, M.; Honkala, S. Effects of short-term xylitol gum chewing on the oral microbiome. Clin. Oral Investig. 2015, 19, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Nohara, K.; Ajami, N.J.; Michalek, R.D.; Tian, X.; Wong, M.; Losee-Olson, S.H.; Petrosino, J.F.; Yoo, S.H.; Shimomura, K.; et al. Transmissible microbial and metabolomic remodeling by soluble dietary fiber improves metabolic homeostasis. Sci. Rep. 2015, 5, 10604. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Jackson, J.A.; Vilgalys, R.; Jackson, R.B. Assessment of soil microbial community structure by use of taxon-specific quantitative pcr assays. Appl. Environ. Microbiol. 2005, 71, 4117–4120. [Google Scholar] [CrossRef] [PubMed]
- Tannock, G.W.; Munro, K.; Harmsen, H.J.; Welling, G.W.; Smart, J.; Gopal, P.K. Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosusDR20. Appl. Environ. Microbiol. 2000, 66, 2578–2588. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Xia, X.; Tang, R.; Zhou, J.; Zhao, H.; Wang, K. Development of a real-time pcr method for firmicutes and bacteroidetes in faeces and its application to quantify intestinal population of obese and lean pigs. Lett. Appl. Microbiol. 2008, 47, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Bekele, A.Z.; Koike, S.; Kobayashi, Y. Genetic diversity and diet specificity of ruminal prevotella revealed by 16s rrna gene-based analysis. FEMS Microbiol. Lett. 2010, 305, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Uebanso, T.; Ohnishi, A.; Kitayama, R.; Yoshimoto, A.; Nakahashi, M.; Shimohata, T.; Mawatari, K.; Takahashi, A. Effects of low-dose non-caloric sweetener consumption on gut microbiota in mice. Nutrients 2017, 9, 560. [Google Scholar] [CrossRef] [PubMed]
- Uebanso, T.; Taketani, Y.; Fukaya, M.; Sato, K.; Takei, Y.; Sato, T.; Sawada, N.; Amo, K.; Harada, N.; Arai, H.; et al. Hypocaloric high-protein diet improves fatty liver and hypertriglyceridemia in sucrose-fed obese rats via two pathways. Am. J. Physiol. Endocrinol. Metab. 2009, 297, E76–E84. [Google Scholar] [CrossRef] [PubMed]
- Kami, K.; Fujimori, T.; Sato, H.; Sato, M.; Yamamoto, H.; Ohashi, Y.; Sugiyama, N.; Ishihama, Y.; Onozuka, H.; Ochiai, A.; et al. Metabolomic profiling of lung and prostate tumor tissues by capillary electrophoresis time-of-flight mass spectrometry. Metabolomics 2013, 9, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, Y.; Hirayama, A.; Ishikawa, T.; Nakamura, S.; Shimizu, K.; Ueno, Y.; Tomita, M.; Soga, T. Depiction of metabolome changes in histidine-starved Escherichia coli by CE-TOFMS. Mol. Biosyst. 2008, 4, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, H.K.; Gudmundsdottir, V.; Nielsen, H.B.; Hyotylainen, T.; Nielsen, T.; Jensen, B.A.; Forslund, K.; Hildebrand, F.; Prifti, E.; Falony, G.; et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 2016, 535, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Neyrinck, A.M.; Maton, N.; Delzenne, N.M. Oligofructose promotes satiety in rats fed a high-fat diet: Involvement of glucagon-like peptide-1. Obes. Res. 2005, 13, 1000–1007. [Google Scholar] [CrossRef] [PubMed]
- Rabot, S.; Membrez, M.; Bruneau, A.; Gerard, P.; Harach, T.; Moser, M.; Raymond, F.; Mansourian, R.; Chou, C.J. Germ-free C57BL/6J mice are resistant to high-fat-diet-induced insulin resistance and have altered cholesterol metabolism. FASEB J. 2010, 24, 4948–4959. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Chang, D.H.; Ahn, S.; Kim, B.C. Whole genome sequencing of “faecalibaculum rodentium” ALO17, isolated from C57bl/6J laboratory mouse feces. Gut Pathog. 2016, 8, 3. [Google Scholar] [CrossRef] [PubMed]
- Morotomi, M.; Nagai, F.; Sakon, H.; Tanaka, R. Dialister succinatiphilus sp. nov. and Barnesiella intestinihominis sp. Nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2008, 58, 2716–2720. [Google Scholar] [CrossRef] [PubMed]
- Ubeda, C.; Bucci, V.; Caballero, S.; Djukovic, A.; Toussaint, N.C.; Equinda, M.; Lipuma, L.; Ling, L.; Gobourne, A.; No, D.; et al. Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infect. Immun. 2013, 81, 965–973. [Google Scholar] [CrossRef] [PubMed]
- Arrieta, M.C.; Stiemsma, L.T.; Dimitriu, P.A.; Thorson, L.; Russell, S.; Yurist-Doutsch, S.; Kuzeljevic, B.; Gold, M.J.; Britton, H.M.; Lefebvre, D.L.; et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 2015, 7, 307ra152. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Onodera, K.; Akiba, Y. Effect of dietary xylitol on growth and inflammatory responses in immune stimulated chickens. Br. Poult. Sci. 1999, 40, 552–554. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.L.; Wi, G.R.; Kim, H.J. Ameliorating effect of dietary xylitol on human respiratory syncytial virus (hRSV) infection. Biol. Pharm. Bull. 2016, 39, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Geidenstam, N.; Al-Majdoub, M.; Ekman, M.; Spegel, P.; Ridderstrale, M. Metabolite profiling of obese individuals before and after a one year weight loss program. Int. J. Obes. 2017. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.J.; Gerasimidis, K.; Edwards, C.A.; Shaikh, M.G. Role of gut microbiota in the aetiology of obesity: Proposed mechanisms and review of the literature. J. Obes. 2016, 2016, 7353642. [Google Scholar] [CrossRef] [PubMed]
- Woods, H.F.; Krebs, H.A. Xylitol metabolism in the isolated perfused rat liver. Biochem. J. 1973, 134, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Kabashima, T.; Kawaguchi, T.; Wadzinski, B.E.; Uyeda, K. Xylulose 5-phosphate mediates glucose-induced lipogenesis by xylulose 5-phosphate-activated protein phosphatase in rat liver. Proc. Natl. Acad. Sci. USA 2003, 100, 5107–5112. [Google Scholar] [CrossRef] [PubMed]
- Uyeda, K.; Yamashita, H.; Kawaguchi, T. Carbohydrate responsive element-binding protein (ChREBP): A key regulator of glucose metabolism and fat storage. Biochem. Pharmacol. 2002, 63, 2075–2080. [Google Scholar] [CrossRef]



| Primer Name | Sequence (5′–3′) | Reference |
|---|---|---|
| Eub338F | ACTCCTACGGGAGGCAGCAG | [20] |
| Eub518R | ATTACCGCGGCTGCTGG | |
| HDA1-GC-F | CGCCCGGGGCGCGCCCCGGGCGGGGCGGGGGCACGGGGGGACTCCTACGGGAGGCAGCAGT | [21] |
| HDA2-R | GTATTACCGCGGCTGCTGGCAC | |
| Bact934F | GGARCATGTGGTTTAATTCGATGAT | [22] |
| Bact1060R | AGCTGACGACAACCATGCAG | |
| Firm934F | GGAGYATGTGGTTTAATTCGAAGCA | |
| Firm1060R | AGCTGACGACAACCATGCAC | |
| Prevotella-F | CATGACGTTACCCGCAGAAGAAG | [23] |
| Prevotella-R | TCCTGCACGCTACTTGGCTG | |
| mChREBP-F | TCAGCACTTCCACAAGCATC | NM_021455.4 |
| mChREBP-R | GCATTAGCAACAGTGCAGGA | |
| 18sF | AAACGGCTACCACATCCAAG | NR_003278.3 |
| 18sR | GGCCTCGAAAGAGTCCTGTA | |
| mPklr-F | TTGTGCTGACAAAGACTGGC | NM_013631 |
| mPklr-R | CCACGAAGCTTTCCACTTTC | |
| mFasn-F | TGCCTTCGGTTCAGTCTCTT | NM_007988.3 |
| mFasn-R | GGGCAACTTAAAGGTGGACA | |
| mScd1-F | CGAGGGTTGGTTGTTGATCT | NM_009127.4 |
| mScd1-R | GCCCATGTCTCTGGTGTTTT | |
| m II-6-F | CTGATGCTGGTGACAACCAC | NM_031168.2 |
| m II-6-R | TCCACGATTTCCCAGAGAAC | |
| mTnf-F | AGCCTGTAGCCCACGTCGTA | NM_013693.3 |
| mTnf-R | TCTTTGAGATCCATGCCGTTG |
| Diet | |||||
|---|---|---|---|---|---|
| CD (n = 5) | CD-LX (n = 5) | CD-MX (n = 5) | HFD (n = 5) | HFD-MX (n = 6) | |
| Final body weight, g | 33.4 ± 0.3 | 31.2 ± 0.5 | 32.5 ± 0.8 | 38.5 ± 1.3 | 40.5 ± 1.3 |
| Visceral fat, g/kg body weight | 15.2 ± 1.2 | 15.8 ± 1.4 | 19.4 ± 3.8 | 43.9 ± 4.5 | 49.1 ± 2.8 |
| Cecum weight, g/kg body weight | 17.7 ± 0.9 | 16.8 ± 1.3 | 16.3 ± 0.9 | 8.9 ± 0.8 | 8.8 ± 1.4 |
| Hepatic parameters | |||||
| Liver, g/kg body weight | 48.6 ± 0.7 | 45.1 ± 1.3 | 43.0 ± 2.3 | 43.5 ± 3.8 | 47.6 ± 3.4 |
| Total cholesterol, mmol/liver | 7.5 ± 0.8 | 7.2 ± 0.4 | 7.1 ± 0.5 | 21.3 ± 2.8 | 33.7 ± 7.3 |
| Triglycerides, mmol/liver | 8.8 ± 0.8 | 11.4 ± 1.3 | 12.0 ± 1.6 | 56.0 ± 12.3 | 74.2 ± 9.2 |
| Plasma parameters | |||||
| Total cholesterol, mmol/L | 2.0 ± 0.1 | 2.3 ± 0.1 | 2.0 ± 0.2 | 3.6 ± 0.7 | 4.2 ± 0.3 |
| Triglycerides, mmol/L | 1.2 ± 0.1 | 1.5 ± 0.1 | 1.4 ± 0.3 | 1.0 ± 0.1 | 0.8 ± 0.1 |
| Diet | ||
|---|---|---|
| HFD-FMT (n = 4) | HFD-MX-FMT (n = 3) | |
| Final body weight, g | 39.5 ± 2.9 | 40.8 ± 1.4 |
| Visceral fat, g/kg body weight | 55.1 ± 6.3 | 60.6 ± 8.8 |
| Cecum weight, g/kg body weight | 6.6 ± 0.8 | 8.5 ± 1.2 |
| Hepatic parameters | ||
| Liver, g/kg body weight | 44.1 ± 1.8 | 45.1 ± 6.5 |
| Total cholesterol, mmol/liver | 82.3 ± 11.1 | 101.4 ± 16.7 |
| Triglycerides, mmol/liver | 23.8 ± 4.6 | 31.5 ± 3.6 |
| Plasma parameters | ||
| Total cholesterol, mmol/L | 4.9 ± 0.1 | 5.6 ± 0.3 * |
| Triglycerides, mmol/L | 1.1 ± 0.2 | 1.0 ± 0.2 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uebanso, T.; Kano, S.; Yoshimoto, A.; Naito, C.; Shimohata, T.; Mawatari, K.; Takahashi, A. Effects of Consuming Xylitol on Gut Microbiota and Lipid Metabolism in Mice. Nutrients 2017, 9, 756. https://doi.org/10.3390/nu9070756
Uebanso T, Kano S, Yoshimoto A, Naito C, Shimohata T, Mawatari K, Takahashi A. Effects of Consuming Xylitol on Gut Microbiota and Lipid Metabolism in Mice. Nutrients. 2017; 9(7):756. https://doi.org/10.3390/nu9070756
Chicago/Turabian StyleUebanso, Takashi, Saki Kano, Ayumi Yoshimoto, Chisato Naito, Takaaki Shimohata, Kazuaki Mawatari, and Akira Takahashi. 2017. "Effects of Consuming Xylitol on Gut Microbiota and Lipid Metabolism in Mice" Nutrients 9, no. 7: 756. https://doi.org/10.3390/nu9070756





