A Dipteran’s Novel Sucker Punch: Evolution of Arthropod Atypical Venom with a Neurotoxic Component in Robber Flies (Asilidae, Diptera)
Abstract
:1. Introduction
2. Results
2.1. Assembled Transcripts and Numbers of Assigned Coding Regions
2.2. Transcriptomic and Proteomic Profiles Differ in Thoracic Glands
2.3. Similar Transcript Diversity but Different Expression Levels in Thoracic Gland and Body Tissue Transcriptomes
2.4. Anatomy of the Venom Delivery System
2.5. Both Species Exhibit Similar Venom Proteomes
2.6. Neurotoxic Activity of U-Asilidin1-Mar1a
3. Discussion
3.1. Highly Expressed Novel Proteins
3.2. Asilid Venoms Contain Putative ICK Neurotoxins
3.3. Missing “Usual Suspect” Enzymes in Putative Asilid Venom?
3.4. Scenario for Envenomation of Prey by Asilids
3.5. Implications in the Context of Fly and Insect Venom Evolution
4. Conclusions
5. Materials and Methods
5.1. Specimen Collection and Determination
5.2. Specimen Dissection and Sample Preservation
5.3. RNA Extraction, Transcriptome Sequencing and Assembly
5.4. Assessing Coding Regions and Expression Levels for Transcripts
5.5. Identification of Venom Protein Classes and Putative Toxins via Transcriptomics
5.6. Sample and Data Processing for Proteome Analyses
5.7. Chemical Synthesis of U-Asilidin1-Mar1a
5.8. HPLC Purification and Activity Tests of U-Asilidin1-Mar1a 1a with Bioassay
5.9. Morphological Work and Data Processing
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fry, B.G.; Roelants, K.; Champagne, D.E.; Scheib, H.; Tyndall, J.D.A.; King, G.F.; Nevalainen, T.J.; Norman, J.A.; Lewis, R.J.; Norton, R.S.; et al. The Toxicogenomic Multiverse: Convergent Recruitment of Proteins into Animal Venoms. Annu. Rev. Genom. Hum. Genet. 2009, 10, 483–511. [Google Scholar] [CrossRef] [PubMed]
- Casewell, N.R.; Wüster, W.; Vonk, F.J.; Harrison, R.A.; Fry, B.G. Complex cocktails: The evolutionary novelty of venoms. Trends Ecol. Evol. 2013, 28, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Von Reumont, B.M.; Campbell, L.I.; Richter, S.; Hering, L.; Sykes, D.; Hetmank, J.; Jenner, R.A.; Bleidorn, C. A Polychaete’s Powerful Punch: Venom Gland Transcriptomics of Glycera Reveals a Complex Cocktail of Toxin Homologs. Genome Biol. Evol. 2014, 6, 2406–2423. [Google Scholar] [CrossRef] [PubMed]
- Von Reumont, B.; Campbell, L.; Jenner, R. Quo Vadis Venomics? A Roadmap to Neglected Venomous Invertebrates. Toxins 2014, 6, 3488–3551. [Google Scholar] [CrossRef] [PubMed]
- Hargreaves, A.D.; Swain, M.T.; Hegarty, M.J.; Logan, D.W.; Mulley, J.F. Restriction and Recruitment—Gene Duplication and the Origin and Evolution of Snake Venom Toxins. Genome Biol. Evol. 2014, 6, 2088–2095. [Google Scholar] [CrossRef] [PubMed]
- Martinson, E.O.; Mrinalini; Kelkar, Y.D.; Chang, C.H.; Werren, J.H. The Evolution of Venom by Co-option of Single-Copy Genes. Curr. Biol. 2017, 27, R647–R649. [Google Scholar] [CrossRef] [PubMed]
- Wong, E.S.W.; Papenfuss, A.T.; Whittington, C.M.; Warren, W.C.; Belov, K. A Limited Role for Gene Duplications in the Evolution of Platypus Venom. Mol. Biol. Evol. 2011, 29, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Vonk, F.J.; Casewell, N.R.; Henkel, C.V.; Heimberg, A.M.; Jansen, H.J.; McCleary, R.J.R.; Kerkkamp, H.M.E.; Vos, R.A.; Guerreiro, I.; Calvete, J.J.; et al. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc. Natl. Acad. Sci. USA 2013, 110, 20651–20656. [Google Scholar] [CrossRef] [PubMed]
- Undheim, E.A.B.; Mobli, M.; King, G.F. Toxin structures as evolutionary tools: Using conserved 3D folds to study the evolution of rapidly evolving peptides. BioEssays 2016, 38, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Thomas, G.W.C.; Hahn, M.W.; Hahn, Y. The effects of increasing the number of taxa on inferences of molecular convergence. Genome Biol. Evol. 2017, 9, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Casewell, N.R.; Visser, J.C.; Baumann, K.; Dobson, J.; Han, H.; Kuruppu, S.; Morgan, M.; Romilio, A.; Weisbecker, V.; Mardon, K.; et al. The Evolution of Fangs, Venom, and Mimicry Systems in Blenny Fishes. Curr. Biol. 2017, 27, 1184–1191. [Google Scholar] [CrossRef] [PubMed]
- Undheim, E.; Fry, B.; King, G. Centipede Venom: Recent Discoveries and Current State of Knowledge. Toxins 2015, 7, 679–704. [Google Scholar] [CrossRef] [PubMed]
- Von Reumont, B.M.; Blanke, A.; Richter, S.; Alvarez, F.; Bleidorn, C.; Jenner, R.A. The First Venomous Crustacean Revealed by Transcriptomics and Functional Morphology: Remipede Venom Glands Express a Unique Toxin Cocktail Dominated by Enzymes and a Neurotoxin. Mol. Biol. Evol. 2014, 31, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.A.; Madio, B.; Jin, J.; Undheim, E.A.B.; Fry, B.G.; King, G.F. Melt with This Kiss: Paralyzing and Liquefying Venom of The Assassin Bug Pristhesancus plagipennis (Hemiptera: Reduviidae). Mol. Cell. Proteom. 2017, 16, 552–566. [Google Scholar] [CrossRef] [PubMed]
- Yeates, D.K.; Wiegmann, B.M.; Courtney, G.W.; Meier, R.; Lambkin, C.; Pape, T. Phylogeny and systematics of Diptera: Two decades of progress and prospects. Zootaxa 2007, 1668, 265–590. [Google Scholar]
- Wiegmann, B.M.; Trautwein, M.D.; Winkler, I.S.; Barr, N.B.; Kim, J.-W.; Lambkin, C.; Bertone, M.A.; Cassel, B.K.; Bayless, K.M.; Heimberg, A.M.; et al. Episodic radiations in the fly tree of life. Proc. Natl. Acad. Sci. USA 2011, 108, 5690–5695. [Google Scholar] [CrossRef] [PubMed]
- Dikow, T. A phylogenetic hypothesis for Asilidae based on a total evidence analysis of morphological and DNA sequence data (Insecta: Diptera: Brachycera: Asiloidea). Org. Divers. Evol. 2009, 9, 165–188. [Google Scholar] [CrossRef]
- Kahan, D. The toxic effect of the bite and the proteolytic activity of the saliva and stomach contents of the robber flies (Diptera Asilidae). Isr. J. Zool. 1964, 13, 47–57. [Google Scholar] [CrossRef]
- Melin, D.E. Contributions to the Knowledge of the Biology, Metamorphosis and Distribution of the Swedish Asilids in Relation to the Whole Family of Asilids; Zoologiska Bidrag Från Uppsala; Almqvist & Wiksells: Uppsala, Sweden, 1923. [Google Scholar]
- Hobby, B.M. Rhodesian Asilidae (Diptera) and Their Prey Collected by Mr C. F. M. Swynnerton. J. Anim. Ecol. 1935, 4, 90. [Google Scholar] [CrossRef]
- Dennis, D.S.; Lavigne, R.J. Hymenoptera as prey of robber flies (Diptera: Asilidae) with new prey records. J. Entomol. Res. Soc. 2007, 9, 23–42. [Google Scholar]
- Dennis, D.S.; Lavigne, R.J.; Dennis, J.G. Hemiptera (heteroptera/homoptera) as prey of robber flies (Diptera: Asilidae) with unpublished records. J. Entomol. Res. Soc. 2010, 12, 27–42. [Google Scholar]
- Dennis, D.S.; Lavigne, R.J.; Dennis, J.G. Spiders (Araneae) as prey of robber flies (Diptera: Asilidae). J. Entomol. Res. Soc. 2012, 14, 65–76. [Google Scholar]
- Joern, A.; Rudd, N.T. Impact of predation by the robber fly Proctacanthus milbertii (Diptera: Asilidae) on grasshopper (Orthoptera: Acrididae) populations. Oecologia 1982, 55, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Rabinovich, M.; Corley, J.C. An important new predator of honey bees the robber fly Mallophora ruficauda Wiedemann (Diptera, Asilidae) in Argentina. Am. Bee J. 1997, 137, 303–306. [Google Scholar]
- Owsley, W.B. The Comparative Morphology of Internal Structures of the Asilidae (Diptera). Ann. Entomol. Soc. Am. 1946, 39, 33–68. [Google Scholar] [CrossRef]
- Wardill, T.J.; Fabian, S.T.; Pettigrew, A.C.; Stavenga, D.G.; Nordström, K.; Gonzalez-Bellido, P.T. A Novel Interception Strategy in a Miniature Robber Fly with Extreme Visual Acuity. Curr. Biol. 2017, 27, 854–859. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, F.G.S. The Relation between the Feeding-habits and the Structure of the Month-parts in the Asilidae (Diptera). Proc. Zool. Soc. Lond. 1925, 95, 599–638. [Google Scholar] [CrossRef]
- Musso, J.J.; Garnier, R.; Legier, F. Comparison of toxicity of venom of some asilids (Diptera Brachycera) on locusts. Ann. Soc. Entomol. Fr. 1978, 14, 177–184. [Google Scholar]
- King, G.F. Venoms as a platform for human drugs: Translating toxins into therapeutics. Expert Opin. Biol. Ther. 2011, 11, 1469–1484. [Google Scholar] [CrossRef] [PubMed]
- King, G.F.; Hardy, M.C. Spider-Venom Peptides: Structure, Pharmacology, and Potential for Control of Insect Pests. Annu. Rev. Entomol. 2013, 58, 475–496. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, S.; Otto, C.; Kurtz, S.; Sharma, C.M.; Khaitovich, P.; Vogel, J.; Stadler, P.F.; Hackermüller, J. Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLoS Comput. Biol. 2009, 5, e1000502. [Google Scholar] [CrossRef] [PubMed]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Otto, C.; Stadler, P.F.; Hoffmann, S. Lacking alignments? The next-generation sequencing mapper segemehl revisited. Bioinformatics 2014, 30, 1837–1843. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Undheim, E.A.B.; Jones, A.; Clauser, K.R.; Holland, J.W.; Pineda, S.S.; King, G.F.; Fry, B.G. Clawing through Evolution: Toxin Diversification and Convergence in the Ancient Lineage Chilopoda (Centipedes). Mol. Biol. Evol. 2014, 31, 2124–2148. [Google Scholar] [CrossRef] [PubMed]
- Magalhães, G.S.; Lopes-Ferreira, M.; Junqueira-de-Azevedo, I.L.M.; Spencer, P.J.; Araújo, M.S.; Portaro, F.C.V.; Ma, L.; Valente, R.H.; Juliano, L.; Fox, J.W.; et al. Natterins, a new class of proteins with kininogenase activity characterized from Thalassophryne nattereri fish venom. Biochimie 2005, 87, 687–699. [Google Scholar] [CrossRef] [PubMed]
- Tamura, S.; Yamakawa, M.; Shiomi, K. Purification, characterization and cDNA cloning of two natterin-like toxins from the skin secretion of oriental catfish Plotosus lineatus. Toxicon 2011, 58, 430–438. [Google Scholar] [CrossRef] [PubMed]
- Lopes-Ferreira, M.; Grund, L.Z.; Lima, C. Thalassophryne nattereri fish venom: From the envenoming to the understanding of the immune system. J. Venom. Anim. Toxins Incl. Trop. Dis. 2014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pallaghy, P.K.; Norton, R.S.; Nielsen, K.J.; Craik, D.J. A common structural motif incorporating a cystine knot and a triple-stranded β-sheet in toxic and inhibitory polypeptides. Protein Sci. 1994, 3, 1833–1839. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Smith, R.; Fletcher, J.I.; Wilson, H.; Wood, C.J.; Howden, M.E.H.; King, G.F. Structure-function studies of ω-atracotoxin, a potent antagonist of insect voltage-gated calcium channels. Eur. J. Biochem. 1999, 264, 488–494. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, J.I.; Smith, R.; O’Donoghue, S.I.; Nilges, M.; Connor, M.; Howden, M.E.; Christie, M.J.; King, G.F. The structure of a novel insecticidal neurotoxin, omega-atracotoxin-HV1, from the venom of an Australian funnel web spider. Nat. Struct. Biol. 1997, 4, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Madio, B.; Undheim, E.A.B.; King, G.F. Revisiting venom of the sea anemone Stichodactyla haddoni: Omics techniques reveal the complete toxin arsenal of a well-studied sea anemone genus. J. Proteom. 2017, 166, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Von Reumont, B.; Undheim, E.; Jauss, R.-T.; Jenner, R. Venomics of Remipede Crustaceans Reveals Novel Peptide Diversity and Illuminates the Venom’s Biological Role. Toxins 2017, 9, 234. [Google Scholar] [CrossRef] [PubMed]
- Musso, J.J. Observations sur le comortement alimentaires l’anatomie e l’histologie des galndes salivaires de deux asilides mediterraneens. Ann. Soc. Entomol. Fr. 1968, 4, 245–255. [Google Scholar]
- Cohen, A.C. Extra-Oral Digestion in Predaceous Terrestrial Arthropoda. Annu. Rev. Entomol. 1995, 40, 85–103. [Google Scholar] [CrossRef]
- Cohen, A.C. Solid-to-Liquid Feeding: The Inside(s) Story of Extra-Oral Digestion in Predaceous Arthropoda. Am. Entomol. 1998, 44, 103–117. [Google Scholar] [CrossRef]
- Chisenhall, D.M.; Londono, B.L.; Christofferson, R.C.; Mores, C.N. Dengue-2 Alters Salivary Gland Protein Expression in Infected Aedes Aegypti Mosquitoes. Am. J. Trop. Med. Hyg. 2010, 83, 389. [Google Scholar]
- Almeras, L.; Fontaine, A.; Belghazi, M.; Bourdon, S.; Boucomont-Chapeaublanc, E.; Orlandi-Pradines, E.; Baragatti, M.; Corre-Catelin, N.; Reiter, P.; Pradines, B.; et al. Salivary Gland Protein Repertoire from Aedes aegypti Mosquitoes. Vector-Borne Zoonotic Dis. 2010, 10, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Wongkamchai, S.; Khongtak, P.; Leemingsawat, S.; Komalamisra, N.; Junsong, N.; Kulthanan, K.; Wisuthsarewong, W.; Boitano, J.J. Comparative identification of protein profiles and major allergens of saliva, salivary gland and whole body extracts of mosquito species in Thailand. Asian Pac. J. Allergy Immunol. 2010, 28, 162–169. [Google Scholar] [PubMed]
- Ma, D.; Li, Y.; Dong, J.; An, S.; Wang, Y.; Liu, C.; Yang, X.; Yang, H.; Xu, X.; Lin, D.; et al. Purification and characterization of two new allergens from the salivary glands of the horsefly, Tabanus yao. Allergy 2010, 66, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Volfova, V.; Tothova, V.; Volf, P. Hyaluronidase activity in the salivary glands of tabanid flies. Insect Biochem. Mol. Biol. 2016, 73, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Yang, H.; Ma, D.; Wu, J.; Wang, Y.; Song, Y.; Wang, X.; Lu, Y.; Yang, J.; Lai, R. Toward an Understanding of the Molecular Mechanism for Successful Blood Feeding by Coupling Proteomics Analysis with Pharmacological Testing of Horsefly Salivary Glands. Mol. Cell. Proteom. 2008, 7, 582–590. [Google Scholar] [CrossRef] [PubMed]
- Takác, P.; Nunn, M.A.; Mészáros, J.; Pechánová, O.; Vrbjar, N.; Vlasáková, P.; Kozánek, M.; Kazimírová, M.; Hart, G.; Nuttall, P.A.; et al. Vasotab, a vasoactive peptide from horse fly Hybomitra bimaculata (Diptera, Tabanidae) salivary glands. J. Exp. Biol. 2006, 209, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Misof, B.; Liu, S.; Meusemann, K.; Peters, R.S.; Flouri, T.; Beutel, R.G.; Niehuis, O.; Petersen, M. Phylogenomics resolves the timing and pattern of insect evolution. Science 2014, 346, 763–767. [Google Scholar] [CrossRef] [PubMed]
- Sunagar, K.; Moran, Y. The Rise and Fall of an Evolutionary Innovation: Contrasting Strategies of Venom Evolution in Ancient and Young Animals. PLoS Genet. 2015, 11, e1005596. [Google Scholar] [CrossRef] [PubMed]
- Geller-Grimm, F. Photographic Atlas and Identification Key to the Robber Flies of Germany (Diptera: Asilidae). Available online: http://www.robberflies.info/keyger/start.html (accessed on 1 October 2017).
- A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/ (accessed on 1 October 2017).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- TransDecoder (Find Coding Regions within Transcripts). Available online: https://github.com/TransDecoder (accessed on 1 October 2017).
- UniProt: A hub for protein information. Nucleic Acids Res. 2014, 43, D204–D212. [CrossRef]
- HMMER: Biosequence Analysis Using Profile Hidden Markov Models. Available online: http://hmmer.org/ (accessed on 1 October 2017).
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016. [Google Scholar] [CrossRef] [PubMed]
- Brekhman, V.; Malik, A.; Haas, B.; Sher, N.; Lotan, T. Transcriptome profiling of the dynamic life cycle of the scypohozoan jellyfish Aurelia aurita. BMC Genom. 2015, 16, 74. [Google Scholar] [CrossRef] [PubMed]
- Lewis Ames, C.; Ryan, J.F.; Bely, A.E.; Cartwright, P.; Collins, A.G. A new transcriptome and transcriptome profiling of adult and larval tissue in the box jellyfish Alatina alata: An emerging model for studying venom, vision and sex. BMC Genom. 2016, 17. [Google Scholar] [CrossRef]
- Jungo, F.; Bougueleret, L.; Xenarios, I.; Poux, S. The UniProtKB/Swiss-Prot Tox-Prot program: A central hub of integrated venom protein data. Toxicon 2012. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.-S.; Tang, S.; Qi, Y.-K.; Wang, Z.-P.; Liu, L. Chemical synthesis of proteins using peptide hydrazides as thioester surrogates. Nat. Protoc. 2013, 8, 2483–2495. [Google Scholar] [CrossRef] [PubMed]
- Yushkevich, P.A.; Piven, J.; Hazlett, H.C.; Smith, R.G.; Ho, S.; Gee, J.C.; Gerig, G. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage 2006. [Google Scholar] [CrossRef] [PubMed]
- Blender. Available online: blender.org (accessed on 1 October 2017).

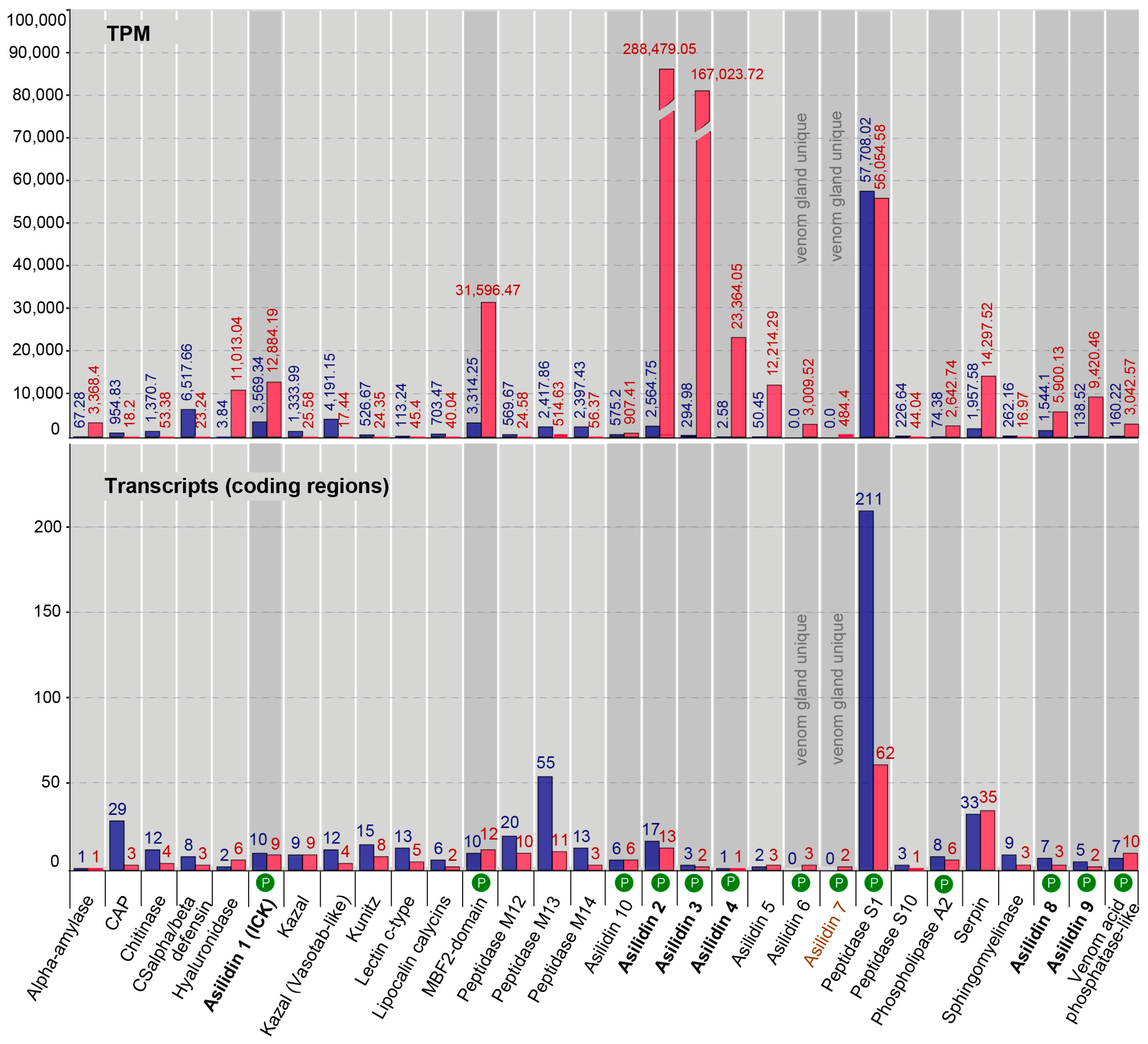
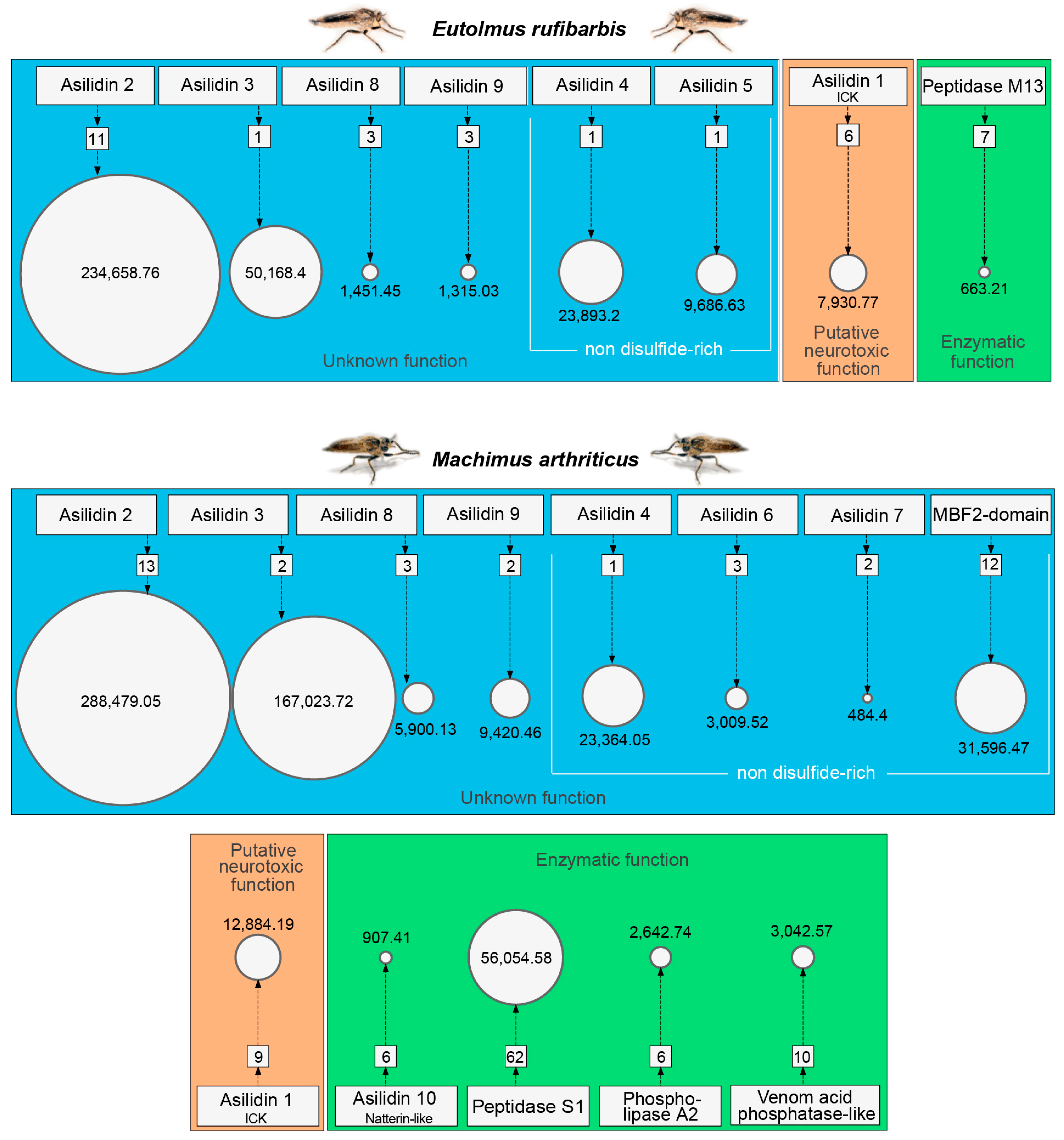

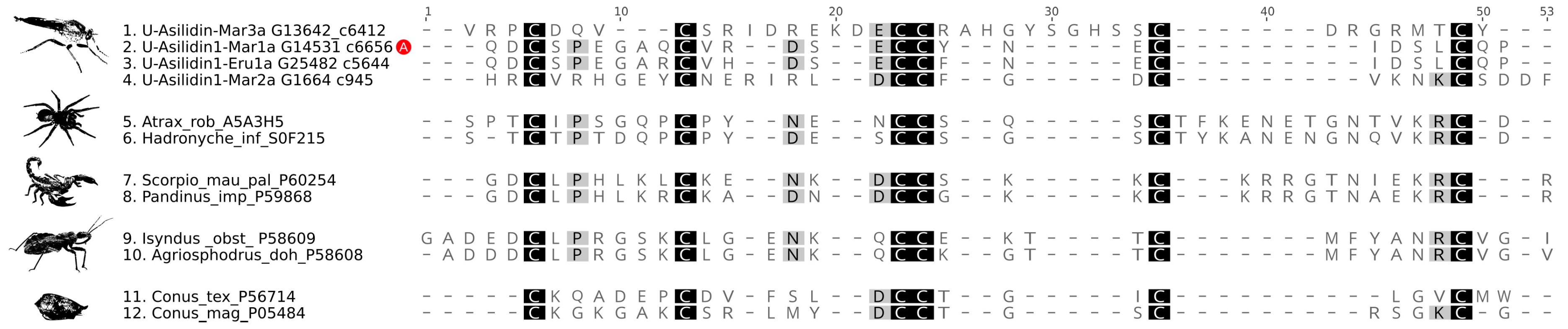


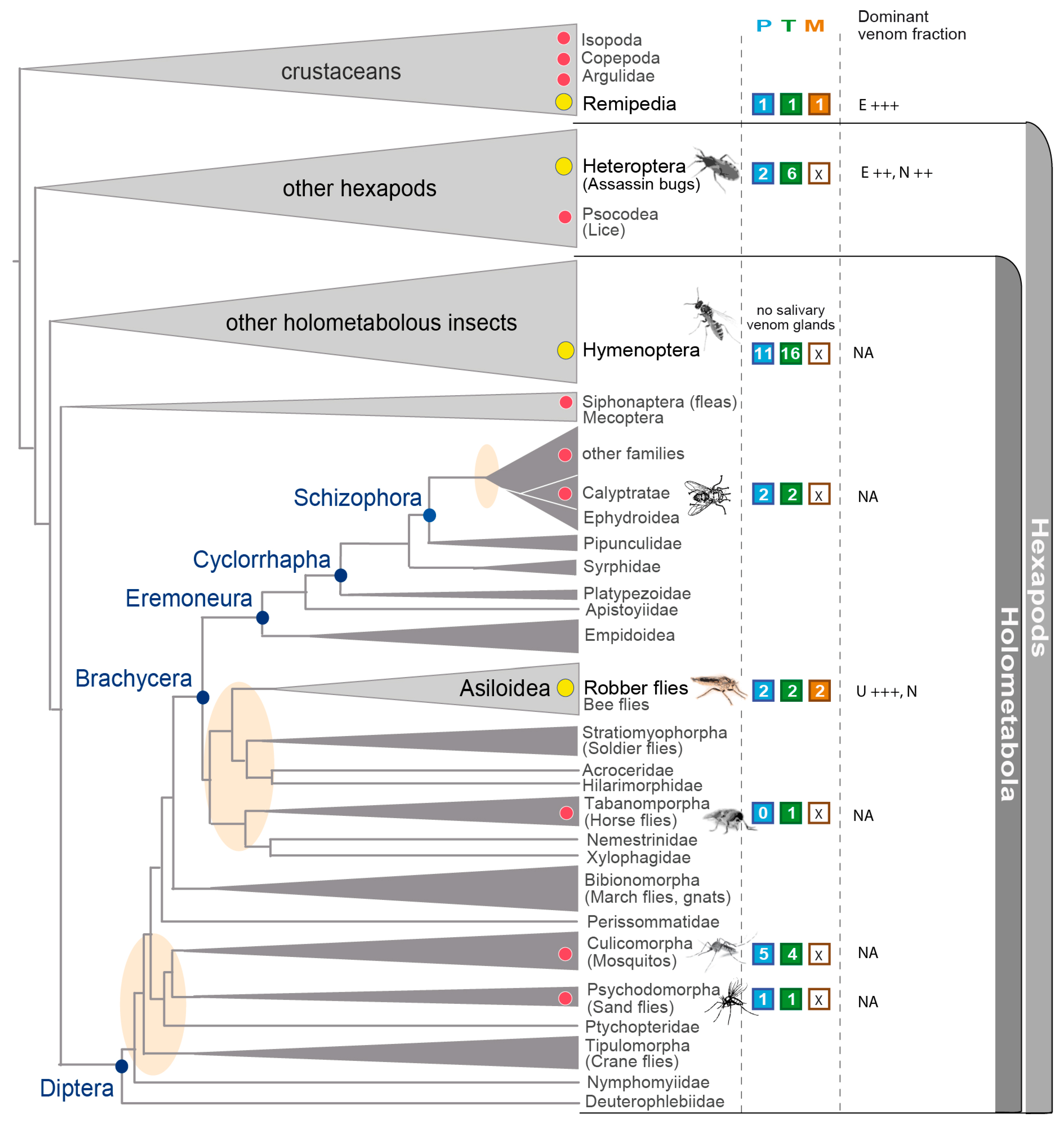

| Species | Tissue | Library Size (Raw Reads) | Library Size (Processed Reads) | Contigs of Cleaned Assembly | Extracted Coding Regions | Coding Regions (TPM ≥ 1, Kallisto) | Coding Regions (TPM ≥ 1, Segemehl) |
|---|---|---|---|---|---|---|---|
| Eutolmus rufibarbis | Thoracic gland | 108,632,880 | 87,187,856 | 56,640 | 33,218 | 16,049 | 15,523 |
| Body tissue | 64,751,420 | 50,968,759 | 70,281 | 42,919 | 32,920 | 28,827 | |
| Machimus arthriticus | Thoracic gland | 106,668,752 | 83,421,201 | 69,849 | 41,816 | 17,798 | 15,346 |
| Body tissue | 64,651,716 | 44,208,096 | 67,504 | 42,784 | 34,478 | 30,629 |
| Protein Family | Structural Fold | Scaffold | Number of Residues | Molecular Weight (Average) | Proteome Hits E. rufibarbis | Proteome Hits M. arthriticus |
|---|---|---|---|---|---|---|
| Asilidin1 | ICK | Cx3-6Cx5-9CCx3-10Cx4-6C | 51–65 | 6.2 kDa | 1 | 3 |
| Asilidin2 | unknown | Cx35Cx8-13Cx36-39C | 267–339 | 33.7 kDa | 5 | 8 |
| Asilidin3 | unknown | Cx7-8Cx22Cx14C | 88–104 | 11.36 kDa | 1 | 2 |
| Asilidin4 | unknown | no cysteine scaffold | 86 | 18.47 kDa | 1 | 1 |
| Asilidin5 | unknown | no cysteine scaffold | 273 | 30.2 kDa | 1 | 0 |
| Asilidin6 | unknown | no cysteine scaffold, two P rich domains | 115 | 13.12 kDa | 0 | 2 |
| Asilidin7 | unknown | no cysteine scaffold | 139 | 15.43 kDa | 0 | 1 |
| Asilidin8 | SVWC-domain | Cx16Cx4Cx10Cx7Cx14CCx5C | 101 | 11.29 kDa | 1 | 1 |
| Asilidin9 | SVWC-domain | Cx22Cx4Cx10Cx8Cx12CCx4C | 118–119 | 13.5 kDa | 1 | 1 |
| Asilidin10 | Natterin-like | no cysteine scaffold, P rich domain | 146–226 | 20.38 kDa | 0 | 2 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Drukewitz, S.H.; Fuhrmann, N.; Undheim, E.A.B.; Blanke, A.; Giribaldi, J.; Mary, R.; Laconde, G.; Dutertre, S.; Von Reumont, B.M. A Dipteran’s Novel Sucker Punch: Evolution of Arthropod Atypical Venom with a Neurotoxic Component in Robber Flies (Asilidae, Diptera). Toxins 2018, 10, 29. https://doi.org/10.3390/toxins10010029
Drukewitz SH, Fuhrmann N, Undheim EAB, Blanke A, Giribaldi J, Mary R, Laconde G, Dutertre S, Von Reumont BM. A Dipteran’s Novel Sucker Punch: Evolution of Arthropod Atypical Venom with a Neurotoxic Component in Robber Flies (Asilidae, Diptera). Toxins. 2018; 10(1):29. https://doi.org/10.3390/toxins10010029
Chicago/Turabian StyleDrukewitz, Stephan Holger, Nico Fuhrmann, Eivind A. B. Undheim, Alexander Blanke, Julien Giribaldi, Rosanna Mary, Guillaume Laconde, Sébastien Dutertre, and Björn Marcus Von Reumont. 2018. "A Dipteran’s Novel Sucker Punch: Evolution of Arthropod Atypical Venom with a Neurotoxic Component in Robber Flies (Asilidae, Diptera)" Toxins 10, no. 1: 29. https://doi.org/10.3390/toxins10010029





