Functional Characterization of the alb1 Orthologue Gene in the Ochratoxigenic Fungus Aspergillus carbonarius (AC49 strain)
Abstract
:1. Introduction
2. Results
2.1. A. carbonarius alb1 Gene Description and Phylogenetic Analysis
2.2. Generation of A. carbonarius Δalb1 Mutants
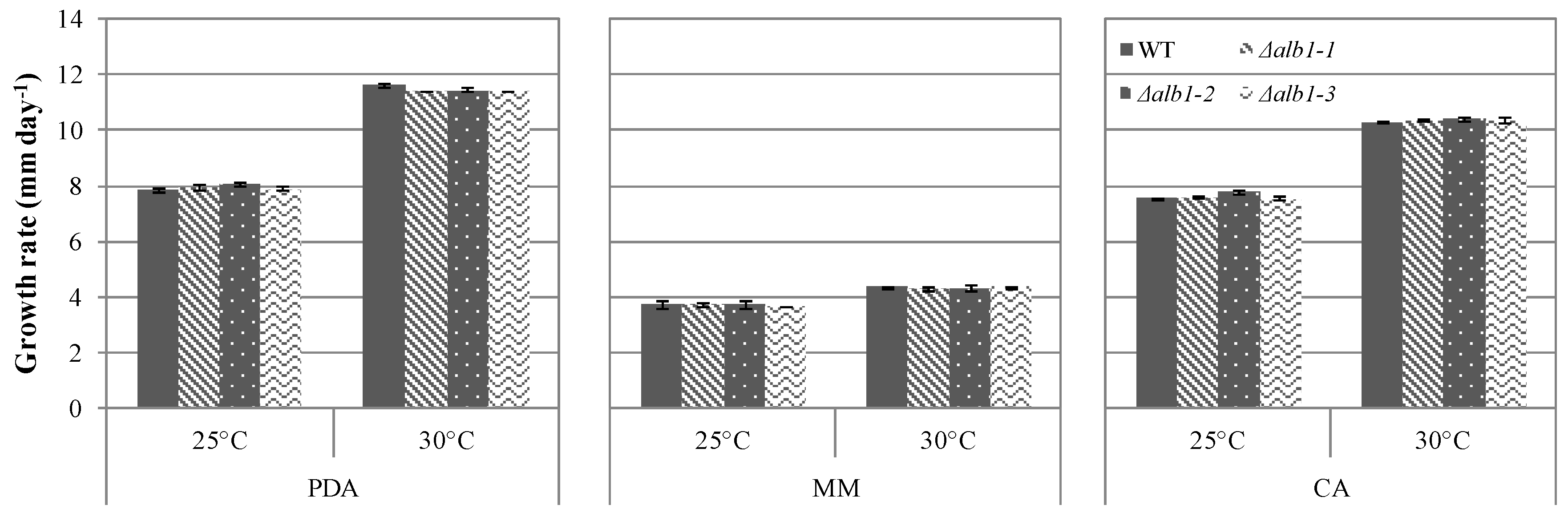
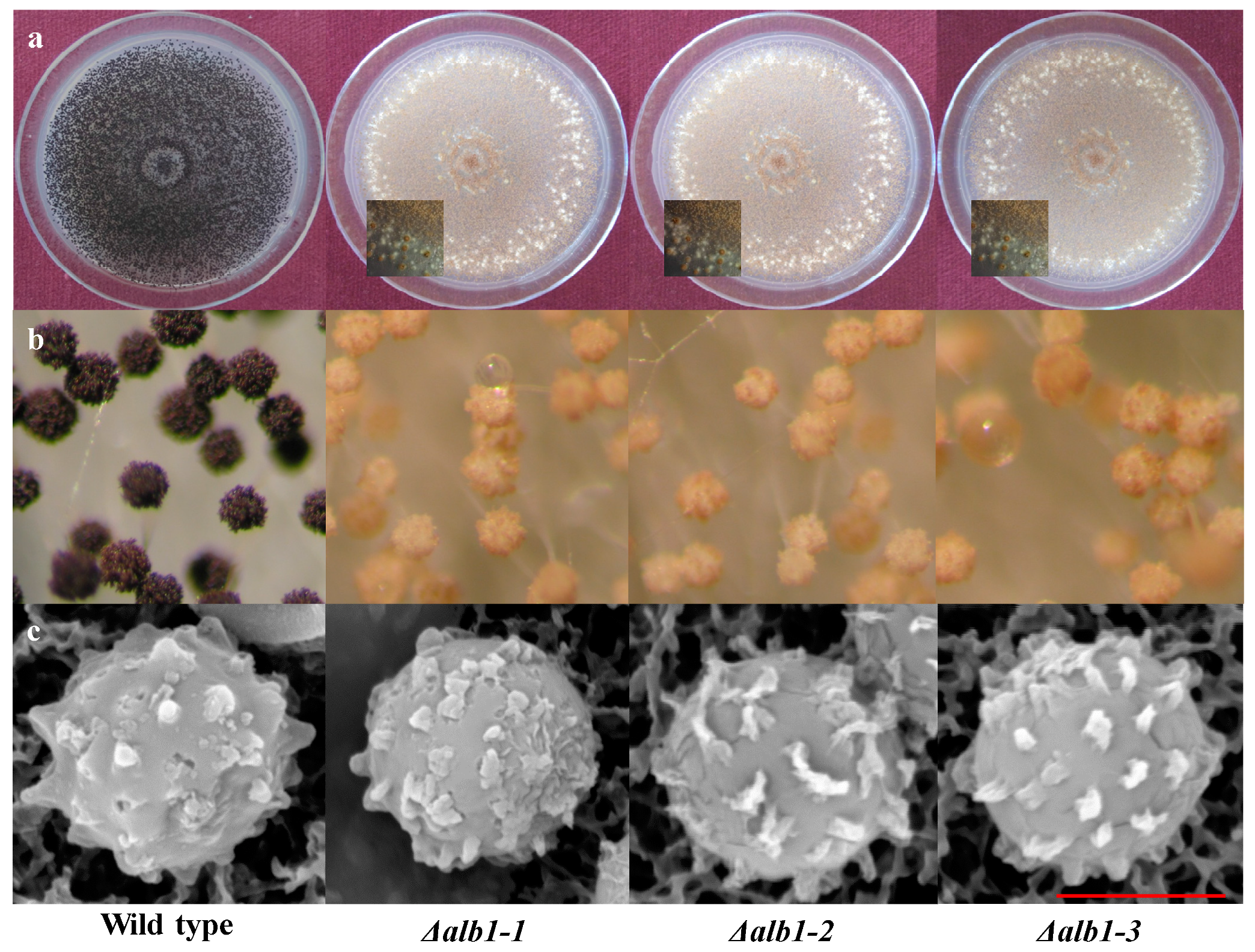
2.3. Phenotypic Characterization
2.4. OTA Production and Partitioning
2.5. Artificial Inoculation on Grape Berries
3. Discussion
4. Materials and Methods
4.1. Strains and Growth Conditions
4.2. Analysis of the A. carbonarius alb1 Gene
4.3. Generation of A. carbonarius Δalb1 Mutants
4.4. Morphological Studies and OTA Production
4.5. OTA Partitioning in WT and Δalb1 Strains
4.6. Artificial Inoculation on Grape Berries
4.7. OTA Extraction and Quantification
4.8. Scanning Electron Microscopy (SEM) Studies
4.9. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bellì, N.; Marın, S.; Sanchis, V.; Ramos, A.J. Influence of water activity and temperature on growth of isolates of Aspergillus section Nigri obtained from grapes. Int. J. Food Microbiol. 2004, 96, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Cabañes, F.J.; Accensi, F.; Bragulat, M.R.; Abarca, M.L.; Castellá, G.; Minguez Pons, A. What is the source of Ochratoxin A in wine? Int. J. Food Microbiol. 2002, 79, 213–215. [Google Scholar] [CrossRef]
- Pollastro, S.; Dongiovanni, C.; Abbatecola, A.; Tauro, G.; Natale, P.; Pascale, M.; Visconti, A.; Faretra, F. Wine contamination by Ochratoxin A in South Italy: Causes and preventive actions. J. Plant Pathol. 2003, 85, 281. [Google Scholar]
- International Agency for Research on Cancer (IARC). Ochratoxin A. In IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, 1st ed.; IARC Press: Lyon, France, 1993; Volume 56, pp. 489–521. [Google Scholar]
- Ray, A.C.; Eakin, R.E. Studies on the biosynthesis of Aspergillin by Aspergillus niger. Appl. Microbiol. 1975, 30, 909–915. [Google Scholar] [PubMed]
- Pal, A.K.; Gajjar, D.U.; Vasavada, A.R. DOPA and DHN pathways orchestrate melanin synthesis in Aspergillus species. Med. Mycol. 2014, 52, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Babitskaya, V.G.; Shcherba, V.; Filimonova, T.V.; Grigorchuk, E.A. Melanin Pigments from the Fungi Paecilomycesvariotii and Aspergillus carbonarius. Appl. Biochem. Microbiol. 2000, 36, 128–133. [Google Scholar] [CrossRef]
- Tsai, H.F.; Wheeler, M.H.; Chang, Y.C.; Kwon-Chung, K.J. A developmentally regulated gene cluster involved in conidial pigment biosynthesis in Aspergillus fumigatus. J. Bacteriol. 1998, 181, 6469–6477. [Google Scholar]
- Tsai, H.F.; Fujii, I.; Watanabe, A.; Wheeler, M.H.; Chang, Y.C.; Yasuoka, Y.; Ebizuka, Y.; Kwon-Chung, K.J. Pentaketide melanin biosynthesis in Aspergillus fumigatus requires chain-length shortening of a heptaketide precursor. J. Biol. Chem. 2001, 276, 29292–29298. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, A.; Fujii, I.; Tsai, H.F.; Chang, Y.C.; Kwon-Chung, K.J.; Ebizuka, Y. Aspergillus fumigatus alb1 encodes Naphthopyrone synthase when expressed in Aspergillus oryzae. FEMS Microbiol. Lett. 2000, 192, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Fujii, I.; Yasuoka, Y.; Tsai, H.F.; Chang, Y.C.; Kwon-Chung, K.J.; Ebizuka, Y. Hydrolytic polyketide shortening by ayg1p, a novel enzyme involved in fungal melanin biosynthesis. J. Biol. Chem. 2004, 279, 44613–44620. [Google Scholar] [CrossRef] [PubMed]
- Pihet, M.; Vandeputte, P.; Tronchin, G.; Renier, G.; Saulnier, P.; Georgeault, S.; Mallet, R.; Chabasse, D.; Symoens, F.; Bouchara, J.P. Melanin is an essential component for the integrity of the cell wall of Aspergillus fumigatus conidia. BMC Microbiol. 2009, 9, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugareva, V.; Härtl, A.; Brock, M.; Hübner, K.; Rohde, M.; Heinekamp, T.; Brakhage, A.A. Characterisation of the laccase-encoding gene abr2 of the dihydroxynaphthalene-like melanin gene cluster of Aspergillus fumigatus. Arch. Microbiol. 2006, 186, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, T.R.; Park, J.; Arentshorst, M.; van Welzen, A.M.; Lamers, G.; vanKuyk, P.A.; Damveld, R.A.; van den Hondel, C.A.M.; Nielsen, K.F.; Frisvard, J.C.; et al. The molecular and genetic basis of conidial pigmentation in Aspergillus niger. Fungal Genet. Biol. 2011, 48, 544–553. [Google Scholar] [CrossRef] [PubMed]
- Keller, N.P.; Turner, G.; Bennett, J.W. Fungal secondary metabolism—From biochemistry to genomics. Nat. Rev. Microbiol. 2005, 3, 937–947. [Google Scholar] [CrossRef] [PubMed]
- Gerin, D.; De Miccolis Angelini, R.M.; Pollastro, S.; Faretra, F. RNA-Seq Reveals OTA-related gene transcriptional changes in Aspergillus carbonarius. PLoS ONE 2016, 11, e0147089. [Google Scholar] [CrossRef] [PubMed]
- Frandsen, R.J.; Andersson, J.A.; Kristensen, M.B.; Giese, H. Efficient four fragment cloning for the Construction of vectors for targeted gene replacement in filamentous fungi. BMC Mol. Biol. 2008, 9, 70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frisvad, J.C.; Petersen, L.M.; Lyhne, E.K.; Larsen, T.O. Formation of Sclerotia and production of Indoloterpenes by Aspergillus niger and other species in Section Nigri. PLoS ONE 2014, 9, e94857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abarca, M.L.; Accensi, F.; Bragulat, M.R.; Castella, G.; Cabanes, F.J. Aspergillus carbonarius as the main source of ochratoxin A contamination in dried vine fruits from the Spanish market. J. Food Prot. 2003, 66, 504–506. [Google Scholar] [CrossRef] [PubMed]
- Simões, M.F.; Santos, C.; Lima, N. Structural diversity of Aspergillus (section Nigri) spores. Microsc. Microanal. 2013, 19, 1151–1158. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, K.; Jahn, B.; Gehringer, H.; Schmidt, A.; Wanner, G.; Brakhage, A.A. Identification of a polyketide synthase gene (pksP) of Aspergillus fumigatus involved in conidial pigment biosynthesis and virulence. Med. Microbiol. Immunol. 1998, 187, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Krijgsheld, P.; Bleichrodt, R.V.; Van Veluw, G.J.; Wang, F.; Müller, W.H.; Dijksterhuis, J.; Wösten, H.A.B. Development in Aspergillus. Stud. Mycol. 2013, 74, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, K.F.; Mogensen, J.M.; Johansen, M.; Larsen, T.O.; Frisvad, J.C. Review of secondary metabolites and mycotoxins from the Aspergillus niger group. Anal. Bioanal. Chem. 2009, 395, 1225–1242. [Google Scholar] [CrossRef] [PubMed]
- Adams, T.H.; Wieser, J.K.; Yu, J.H. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 1998, 62, 35–54. [Google Scholar] [PubMed]
- Marshall, M.A.; Timberlake, W.E. Aspergillus nidulanswet A activates spore-specific gene expression. Mol. Cell. Biol. 1991, 11, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.H. Regulation of development in Aspergillus nidulans and Aspergillus fumigatus. Mycobiology 2010, 38, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zhou, B.; Yin, C.; Guo, Y.; Lin, Y.; Pan, L.; Wang, B. Characterization of natural antisense transcript, sclerotia development and secondary metabolism by strand-specific RNA sequencing of Aspergillus flavus. PLoS ONE 2014, 9, e97814. [Google Scholar] [CrossRef] [PubMed]
- Jin, F.J.; Takahashi, T.; Matsushima, K.I.; Hara, S.; Shinohara, Y.; Maruyama, J.I. SclR, a basic helix-loop-helix transcription factor, regulates hyphal morphology and promotes sclerotial formation in Aspergillus oryzae. Eukaryot. Cell 2011, 10, 945–955. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, J. DHN melanin biosynthesis in the plant pathogenic fungus Botrytis cinerea is based on two developmentally regulated key enzyme (PKS)-encoding genes. Mol. Microbiol. 2016, 99, 729–748. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.K.; Scharfenstein, L.L.; Li, R.W.; Arroyo-Manzanares, N.; De Saeger, S.; Di Mavungu, J.D. Aspergillus flavusasw, a gene homolog of Aspergillus nidulansoef, regulates sclerotial development and biosynthesis of sclerotium-associated secondary metabolites. Fungal Genet. Biol. 2017, 104, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Zhi, Q.Q.; Li, J.Y.; Liu, Q.Y.; He, Z.M. A cytosine Methyltransferase ortholog dmtA is involved in the sensitivity of Aspergillus flavus to environmental stresses. Fungal Biol. 2017, 121, 501–514. [Google Scholar] [CrossRef] [PubMed]
- Calvo, A.M.; Cary, J.W. Association of fungal secondary metabolism and sclerotial biology. Front. Microbiol. 2015, 6, 62. [Google Scholar] [CrossRef] [PubMed]
- Hicks, J.K.; Yu, J.H.; Keller, N.P.; Adams, T.H. Aspergillus sporulation and mycotoxin production both require inactivation of the FadA Gα protein-dependent signaling pathway. EMBO J. 1997, 16, 4916–4923. [Google Scholar] [CrossRef] [PubMed]
- Wicklow, D.T.; Dowd, P.F.; Alfatafta, A.A.; Gloer, J.B. Ochratoxin A: An antiinsectan metabolite from the sclerotia of Aspergillus carbonarius NRRL 369. Can. J. Microbiol. 1996, 42, 1100–1103. [Google Scholar] [CrossRef] [PubMed]
- Atoui, A.; Mitchell, D.; Mathieu, F.; Magan, N.; Lebrihi, A. Partitioning of ochratoxin A in mycelium and conidia of Aspergillus carbonarius and the impact on toxin contamination of grapes and wine. J. Appl. Microbiol. 2007, 103, 961–968. [Google Scholar] [CrossRef] [PubMed]
- Calvo, A.M.; Wilson, R.A.; Bok, J.W.; Keller, N.P. Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 2002, 66, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucl. Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Michielse, C.B.; Hooykaas, P.J.; van den Hondel, C.A.; Ram, A.F. Agrobacterium-mediated transformation of the filamentous fungus Aspergillus awamori. Nat. Protoc. 2008, 3, 1671–1678. [Google Scholar] [CrossRef] [PubMed]
- Crespo-Sempere, A.; Marin, S.; Sanchis, V.; Ramos, A.J. VeA and LaeA transcriptional factors regulate ochratoxin A biosynthesis in Aspergillus carbonarius. Int. J. Food Microbiol. 2013, 166, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Ramakers, C.; Ruijter, J.M.; Deprez, R.H.L.; Moorman, A.F. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucl. Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, K.E.; Srb, A.M. Heterokaryosis and parasexuality in the fungus Ascochyta imperfecta. Am. J. Bot. 1965, 52, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Pollastro, S.; De Miccolis Angelini, R.M.; Natale, P.; Pastore, C.; Pascale, M.; Faretra, F. Observations on ochratoxin A production in Aspergillus carbonarius and Aspergillus niger. In Proceedings of the International Workshop: Ochratoxin A in Grapes and Wine: Prevention and Control, Marsala, Italy, 20–21 October 2005. [Google Scholar]
- Visconti, A.; Pascale, M.; Centonze, G. Determination of ochratoxin A in wine by means of immunoaffinity column clean-up and high-performance liquid chromatography. J. Chromatogr. 1999, 864, 89–101. [Google Scholar] [CrossRef]



| Primer Name | Primer Sequence (5′-3′) | Target DNA (Organism) |
|---|---|---|
| Amplification of promoter and terminator | ||
| ALB1_O1 | GGTCTTAAUGGAATACCTGGACGCTGTTG | Promoter (A. carbonarius WT) |
| ALB1_O2 | GGCATTAAUCGGATCGATTGGGTTGCATT | |
| ALB1_A3 | GGACTTAAUCCAGTGAATGACCGAATGCA | Terminator (A. carbonarius WT) |
| ALB1_A4 | GGGTTTAAUAGACTTCGTACGCCACAGAA | |
| Screening of transformants | ||
| RF-5 | GTTTGCAGGGCCATAGAC | Promoter (E. coli DH5α) |
| RF-2 | TCTCCTTGCATGCACCATTCCTTG | |
| RF-1 | AAATTTTGTGCTCACCGCCTGGAC | Terminator (E. coli DH5α) |
| RF-6 | ACGCCAGGGTTTTCCCAGTC | |
| ALB1_1F | GAACTCACGGCCCTCAAAGA | Promoter (A. carbonarius Δalb1) |
| hph_1F | ACGAGGTCGCCAACATCTTCTTCT | |
| ALB1_2R | ATTCACCCCGGTTTCCTCAC | Terminator (A. carbonarius Δalb1) |
| hph_PRO4 | GCACCAAGCAGCAGATGATA | |
| HMBF1 | CTGTCGAGAAGTTTCTGATCG | Hygromicin (A. carbonarius Δalb1) |
| HMBR1 | CTGATAGAGTTGGTCAAGACC | |
| ALB1_3F | CTTGGTAGGATCCGCGAGAC | Alb1 (A. carbonarius Δalb1) |
| ALB1_4R | CGGCATCGAAAGCGCAAATA | |
| Determination of T-DNA copy number | ||
| ALB1_7F | ATTTCCGAACGGGGTAACTC | Alb1 (A. carbonarius Δalb1) |
| ALB1_8R | CAAGGTCTCTTGCAATGCTG | |
| nrps_1F | GAGCAGCTACCGGAGCTATT | nrps (A. carbonarius Δalb1) |
| nrps_2R | GCATCGCATGAGTGAGTTGT | |
| Strain | Sclerotia (Number ± Standard Error) | |||||
|---|---|---|---|---|---|---|
| PDA | MM | CA | ||||
| 25 °C | 30 °C | 25 °C | 30 °C | 25 °C | 30 °C | |
| WT | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| Δalb1-1 | 9 ± 1 | 81 ± 1 | 4 ± 0 | 26 ± 2 | 53 ± 2 | 61 ± 4 |
| Δalb1-2 | 23 ± 3 | 82 ± 1 | 11 ± 1 | 25 ± 1 | 56 ± 5 | 66 ± 10 |
| Δalb1-3 | 20 ± 2 | 108 ± 1 | 10 ± 1 | 26 ± 2 | 44 ± 10 | 49 ± 4 |
| Strain | OTA (ng/mm2 ± Standard Error) | ||
|---|---|---|---|
| MM * | PDA | CA | |
| 25 °C | |||
| WT | 11.59 ± 0.67 B | 87.89 ± 2.39 B | 296.00 ± 22.41 B |
| Δalb1-1 | 90.15 ± 12.73 A (677.8) ** | 124.78 ± 1.19 A (41.9) | 492.26 ± 34.18 A (66.3) |
| Δalb1-2 | 85.81 ± 7.00 A (640.4) | 116.46 ± 3.39 A (32.5) | 503.45 ± 13.59 A (70.1) |
| Δalb1-3 | 99.21 ± 6.71 A (755.9) | 119.69 ± 5.75 A (36.2) | 550.03 ± 41.31 A (85.5) |
| 30 °C | |||
| WT | 12.03 ± 0.55 B | 28.80 ± 1.14 B | 47.11 ± 2.92 B |
| Δalb1-1 | 44.41 ± 5.28 A (269.2) | 50.34 ± 1.33 A (70.8) | 126.68 ± 5.40 A (168.9) |
| Δalb1-2 | 43.97 ± 0.98 A (265.5) | 48.35 ± 1.82 A (67.9) | 117.97 ± 4.90 A (150.4) |
| Δalb1-3 | 39.37 ± 1.19 A (227.2) | 43.08 ± 2.64 A (49.6) | 112.08 ± 12.43 A (137.9) |
| Strain | Mycelium (ng mg−1) | Conidia (pg 10−2 Conidia) | Medium (ng mg−1) |
|---|---|---|---|
| 4 DAI | |||
| WT | 3.3 ± 0.8 (a) | 21.1 ± 1.8 (a A) | 3.8 ± 0.6 (cC) |
| Δalb1-1 | 3.0 ± 0.5 (a) | 8.9 ± 1.7 (bc B) | 9.5 ± 2.1 (ab AB) |
| Δalb1-2 | 3.1 ± 0.8 (a) | 9.1 ± 1.2 (b AB) | 12.8 ± 2.1 (a A) |
| Δalb1-3 | 2.6 ± 0.2 (a) | 6.1 ± 1.0 (c B) | 6.6 ± 0.8 (bc BC) |
| 6 DAI | |||
| WT | 0.4 ± 0.1 (b A) | 23.5 ± 3.9 (a A) | 5.1 ± 1.1 (bB) |
| Δalb1-1 | 0.9 ± 0.3 (ab A) | 12.1 ± 2.5 (ab A) | 12.0 ± 1.5 (aAB) |
| Δalb1-2 | 1.6 ± 0.4 (a A) | 11.3 ± 2.8 (ab A) | 13.9 ± 2.2 (a A) |
| Δalb1-3 | 1.1 ± 0.5 (ab A) | 7.1 ± 2.0 (b A) | 12.2 ± 1.7 (a AB) |
| 8 DAI | |||
| WT | 1.0 ± 0.1 (b A) | 3.0 ± 0.5 (a) | 9.7 ± 2.4 (a) |
| Δalb1-1 | 1.3 ± 0.2 (b A) | 1.2 ± 0.2 (a) | 13.5 ± 1.5 (a) |
| Δalb1-2 | 2.9 ± 0.8 (a A) | 1.5 ± 0.2 (a) | 12.5 ± 1.6 (a) |
| Δalb1-3 | 2.2 ± 0.4 (ab A) | 1.3 ± 0.3 (a) | 14.5 ± 3.7 (a) |
| 10 DAI | |||
| WT | 1.0 ± 0.1 (b A) | 5.6 ± 0.5 (a A) | 8.0 ± 1.0 (b A) |
| Δalb1-1 | 2.6 ± 0.7 (ab A) | 2.7 ± 0.5 (b B) | 13.7 ± 2.5 (a A) |
| Δalb1-2 | 1.9 ± 0.3 (ab A) | 2.5 ± 0.4 (b B) | 12.2 ± 0.7 (ab A) |
| Δalb1-3 | 3.3 ± 0.9 (a A) | 2.0 ± 0.3 (b B) | 11.4 ± 1.0 (ab A) |
| Strain | 25 °C | 30 °C |
|---|---|---|
| Lesion diameter (mm) | ||
| WT | 33.2 ± 2.8 a | 47.0 ± 1.0 a |
| ΔAlb1-1 | 35.2 ± 3.0 a | 51.3 ± 3.8 a |
| ΔAlb1-2 | 33.3 ± 0.9 a | 52.7 ± 1.4 a |
| ΔAlb1-3 | 33.3 ± 1.6 a | 54.2 ± 5.8 a |
| OTA (ng g−1) | ||
| WT | 315.1 ± 14.8 a | 272.2 ± 26.3 bB |
| ΔAlb1-1 | 450.4 ± 67.5 a | 1380.7 ± 204.8 aAB |
| ΔAlb1-2 | 515.3 ± 77.3 a | 1548.9 ± 187.8 a A |
| ΔAlb1-3 | 482.8 ± 33.6 a | 1929.7 ± 388.3 a A |
| Sclerotia (No.) | ||
| WT | 0 ± 0 bC | 0 ± 0 bB |
| ΔAlb1-1 | 2 ± 0 abAB | 19 ± 2 aA |
| ΔAlb1-2 | 1 ± 0 bAB | 14 ± 2 aAB |
| ΔAlb1-3 | 3 ± 0 aA | 24 ± 1 aA |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gerin, D.; González-Candelas, L.; Ballester, A.-R.; Pollastro, S.; De Miccolis Angelini, R.M.; Faretra, F. Functional Characterization of the alb1 Orthologue Gene in the Ochratoxigenic Fungus Aspergillus carbonarius (AC49 strain). Toxins 2018, 10, 120. https://doi.org/10.3390/toxins10030120
Gerin D, González-Candelas L, Ballester A-R, Pollastro S, De Miccolis Angelini RM, Faretra F. Functional Characterization of the alb1 Orthologue Gene in the Ochratoxigenic Fungus Aspergillus carbonarius (AC49 strain). Toxins. 2018; 10(3):120. https://doi.org/10.3390/toxins10030120
Chicago/Turabian StyleGerin, Donato, Luis González-Candelas, Ana-Rosa Ballester, Stefania Pollastro, Rita Milvia De Miccolis Angelini, and Francesco Faretra. 2018. "Functional Characterization of the alb1 Orthologue Gene in the Ochratoxigenic Fungus Aspergillus carbonarius (AC49 strain)" Toxins 10, no. 3: 120. https://doi.org/10.3390/toxins10030120







