Association with AflR in Endosomes Reveals New Functions for AflJ in Aflatoxin Biosynthesis
Abstract
:1. Introduction
2. Results
2.1. Complementation Experiments Reveal a Strong Species Specificity for AflJ Function
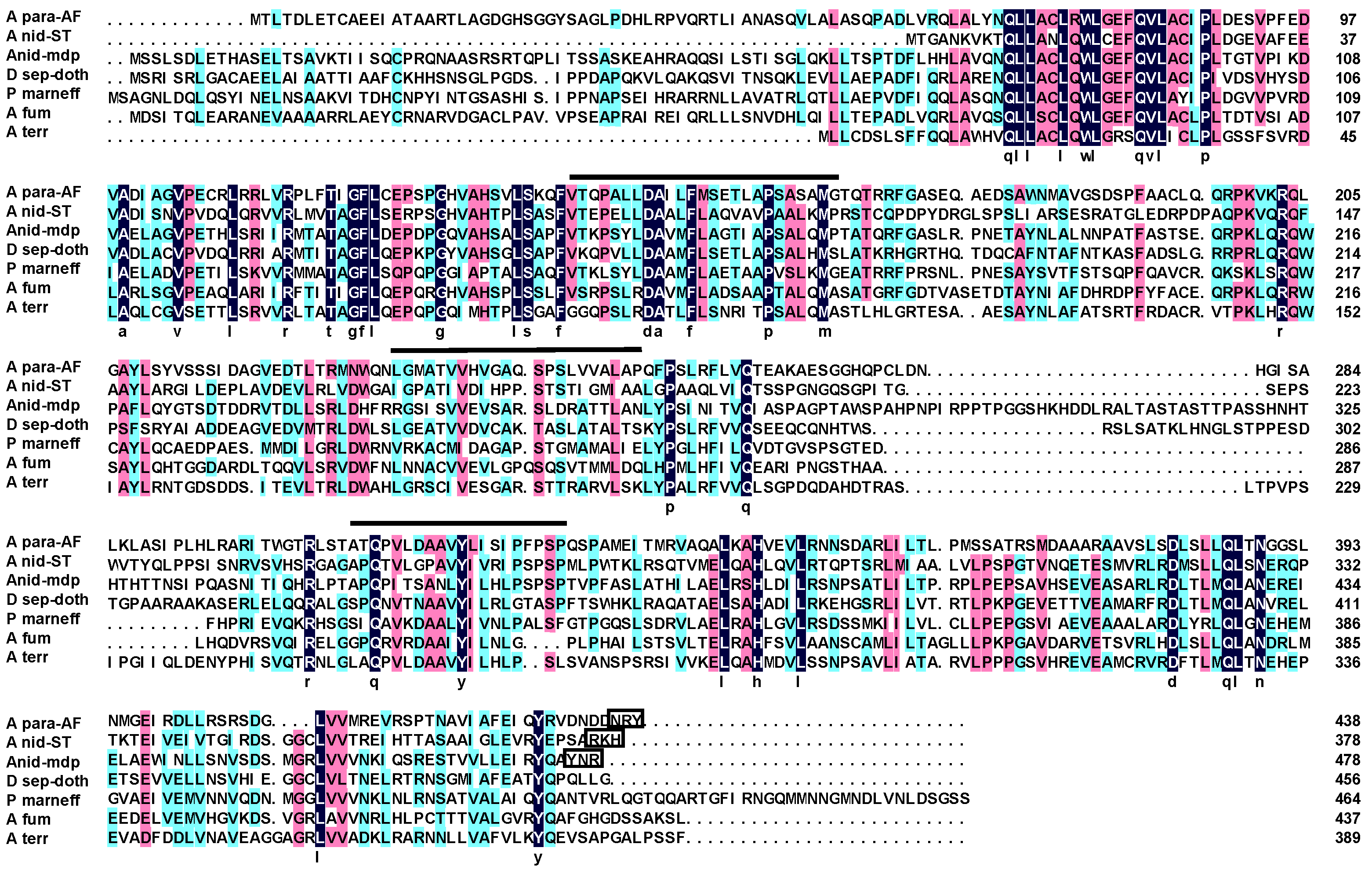
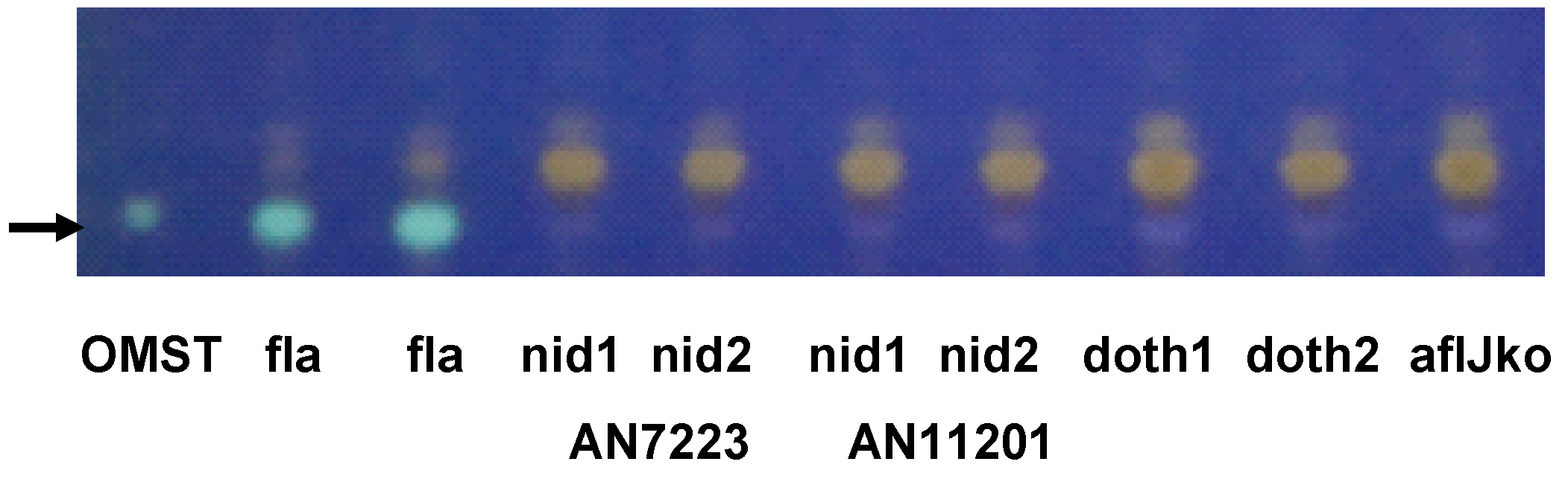
2.2. Aflatoxin Transcript Accumulation in Wild Type and ∆aflJ Transformed with aflJ from other Fungi
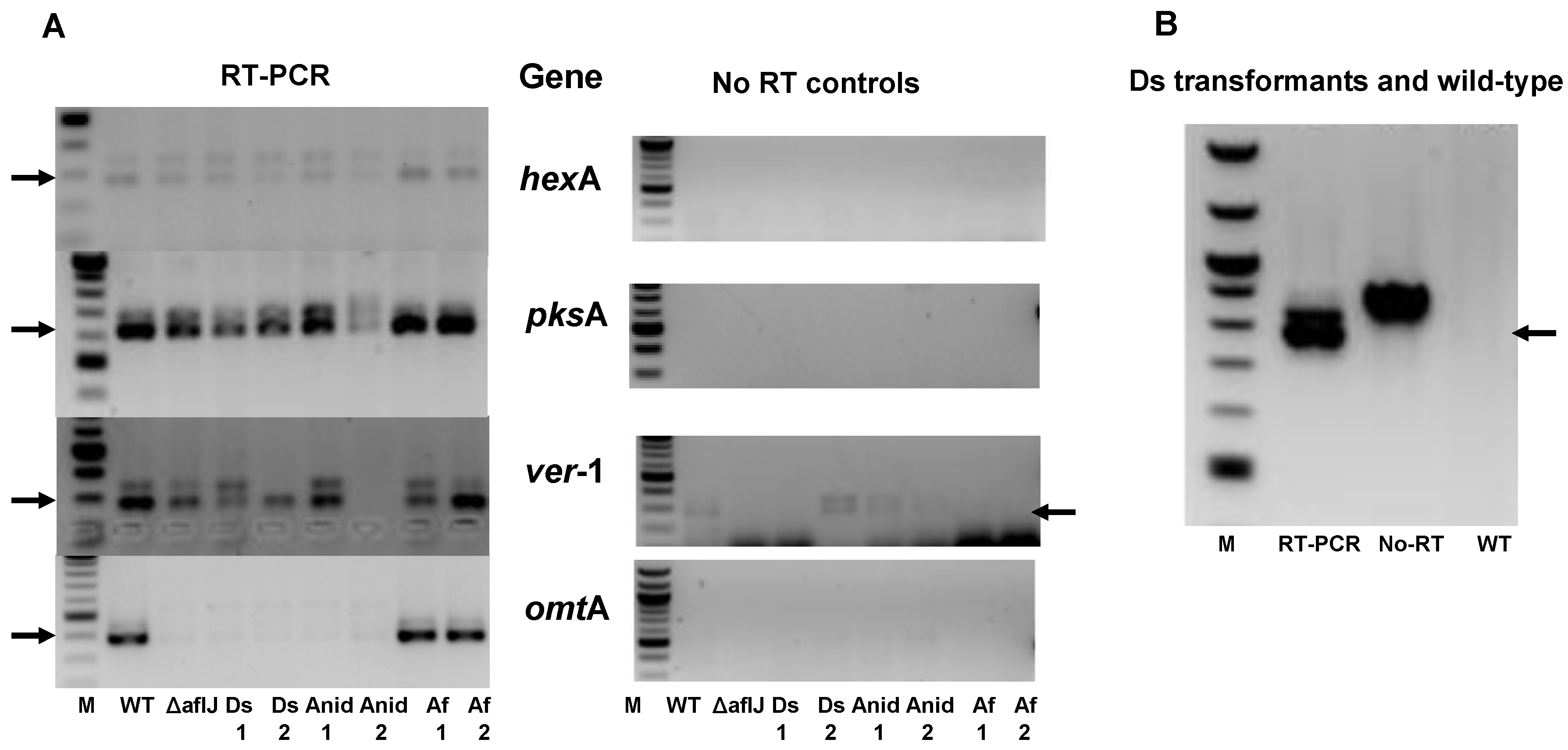
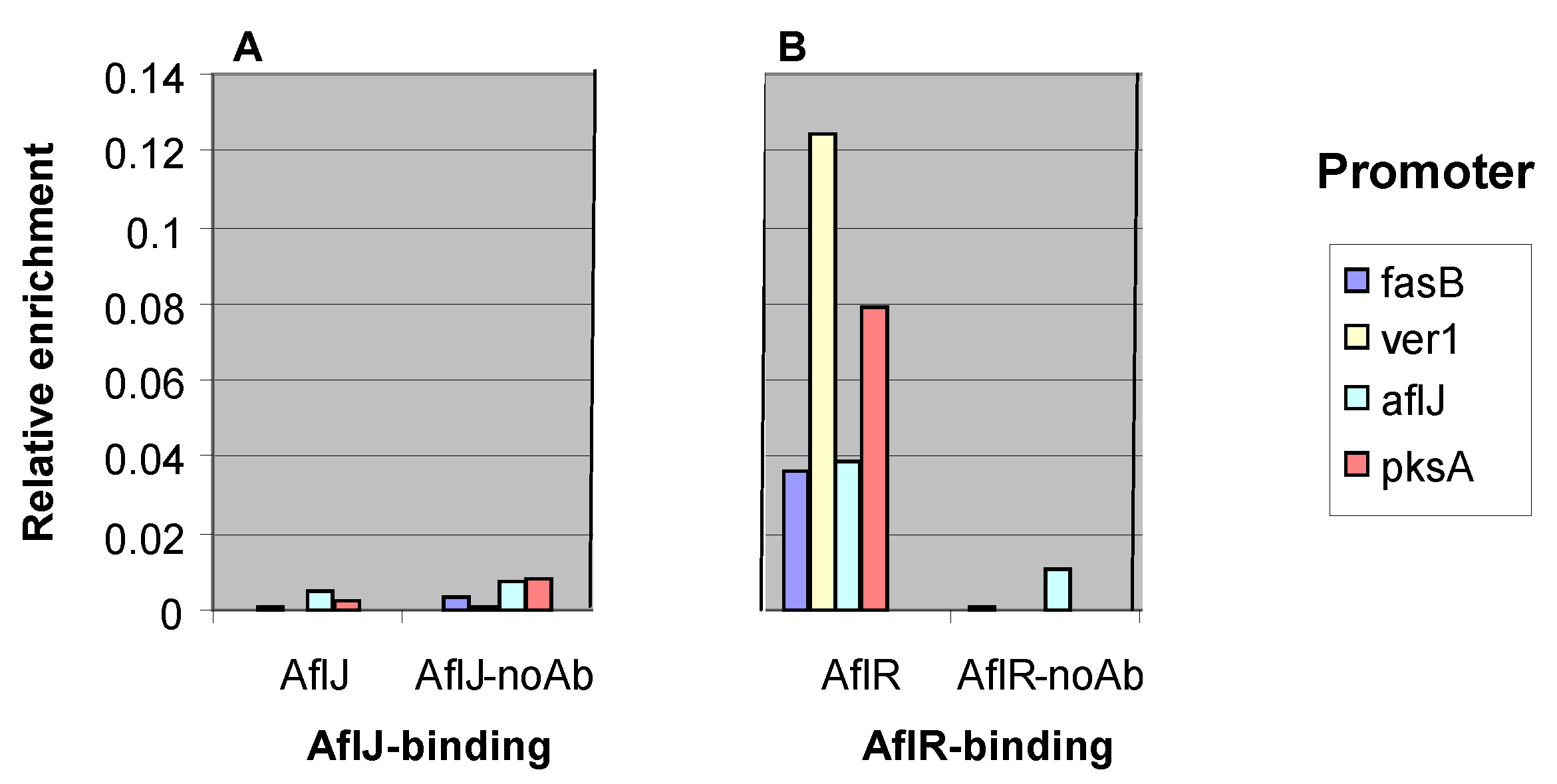
2.3. AflJ Can Not Be Detected at Promoters of Aflatoxin Genes by Chromatin Immunoprecipitation (ChIP)
2.4. Interaction of AflJ with AflR and Cellular Proteins by Yeast Two-Hybrid Assay
| Accession number | Protein |
|---|---|
| XP_002374366 | C6 transcription factor, AmdR |
| XP_002382582 | Endonuclease |
| XP_002379937 | AflL (AvnA) |
| XP_002373959 | PfkA phosphofructokinase |
| XP_002378434 | COP9 signalosome subunit 5 (CsnF) |
| XP_002372709 | Glutamate decaboxylase |
| XP_001822456 | COP9 signalosome subunit 5 (CsnE) |
| XP_002380625 | FAD-dependent oxidoreductase |
| XP_002385336 | FKBP-type peptidyl-prolyl isomerase |
| XP_002377941 | Conidiation-specific protein (Con10) |
| XP_002383331 | Calponin-homology-domain protein |
| XP_002379716 | Nedd8-activating protein (UbaC) |
| XP_001817130 | CFEM domain protein (Pfam03750) |
| XP_001817064 | SteA |
| XP_003190896 | Duf 3752 domain protein (Pfam 12572) |
| XP_001727088 | MFS superfamily (maltose permease) |
| BD | AD | Colonies |
|---|---|---|
| AflJ | AflR | 125 |
| AflJ | AflJ | 0 |
| AflJ | pGADT7 | 0 |
| Lam | AflR | 0 |
| Lam | AflJ | 0 |
2.5. AflJ-GFP Localizes Mainly to Endosome-Like Cellular Structures
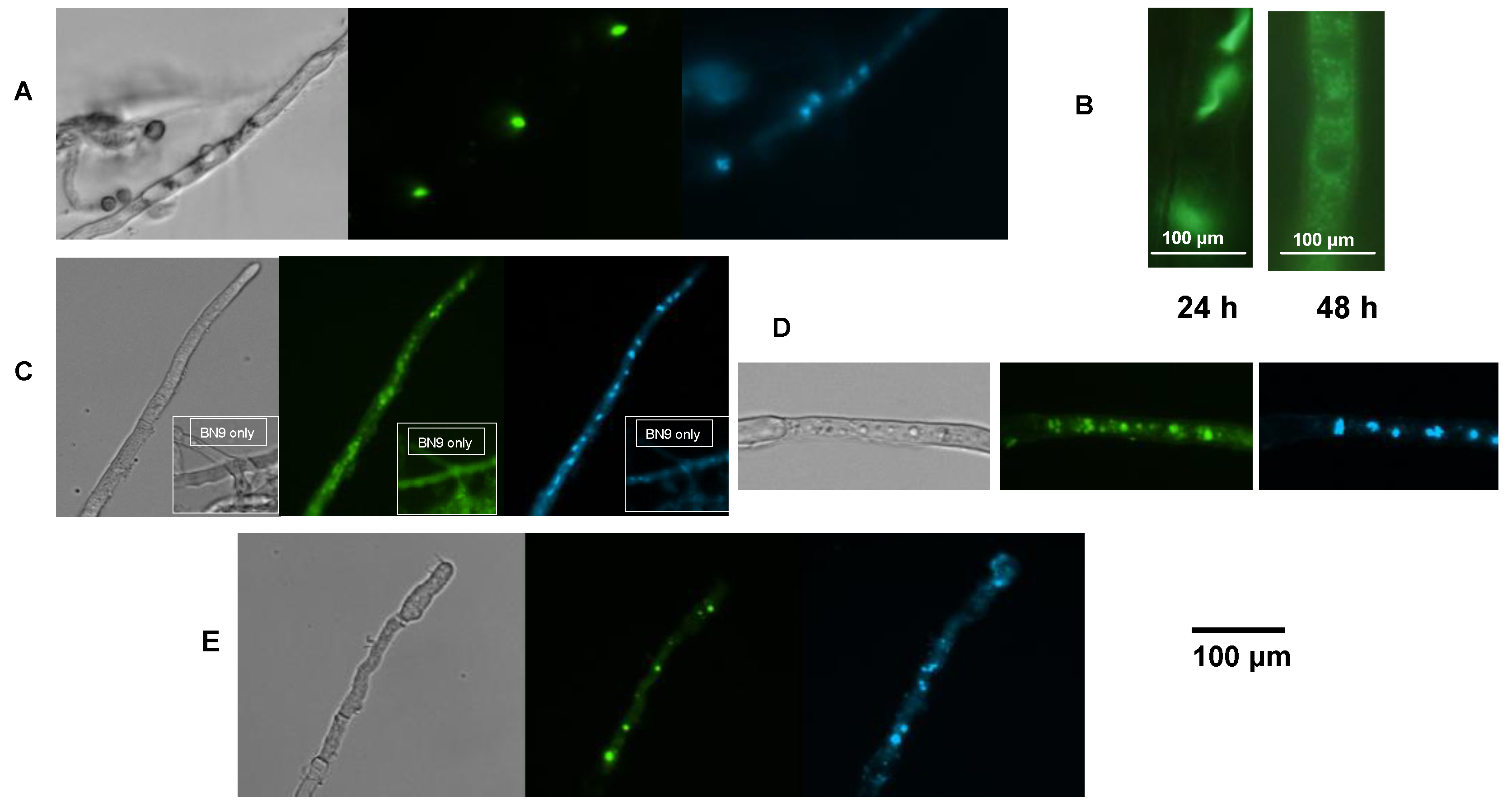
2.6. AflR-GFP Localization
2.7. Bimolecular-Fluorescence Complementation (BiFC, Split eYFP) Assays
2.8. AflJ Is Associated with Endosome Biogenesis and Traffic
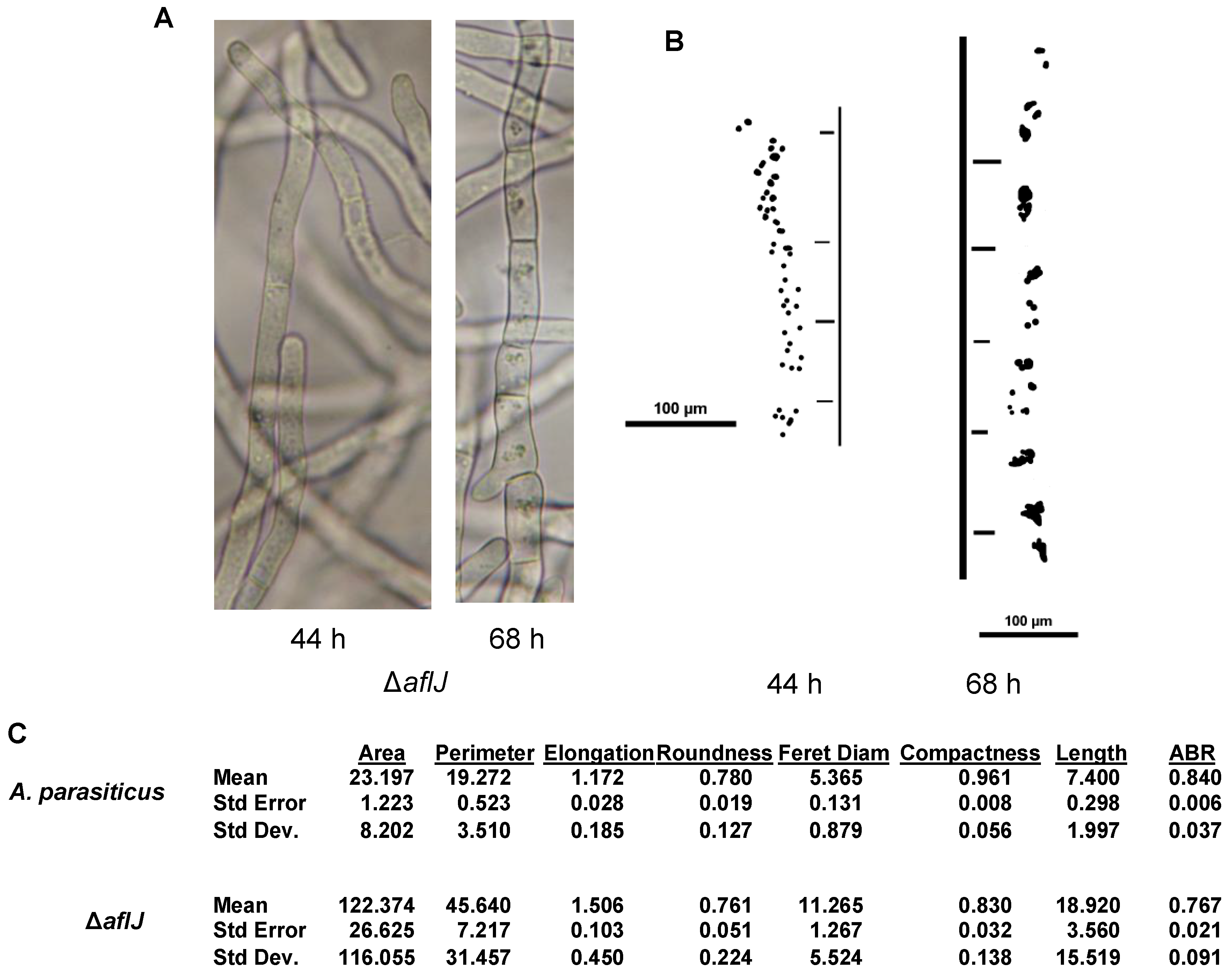
3. Discussion
4. Experimental Section
4.1. Strains, Media, and Growth Conditions
4.2. Chromatin Immunoprecipitation (ChIP)
4.3. Complementation Studies
4.4. Yeast Two-Hybrid Analysis
4.5. Fluorescence Microscopy and Split eYFP Studies (Bimolecular Fluorescence Complementation-BIFC)
5. Conclusions
Acknowledgments
Conflict of Interest
Supplementary Files
References
- Abnet, C.C. Carcinogenic food contaminants. Cancer Invest. 2007, 25, 189–196. [Google Scholar] [CrossRef]
- Wild, C.P.; Turner, P.C. The toxicology of aflatoxins as a basis for public health decisions. Mutagenesis 2002, 17, 471–481. [Google Scholar] [CrossRef]
- Reddy, K.R.N.; Salleh, B.; Saad, B.; Abbas, H.K.; Abel, C.A.; Shier, W.T. An overview of mycotoxin contamination in foods and its implications for human health. Toxin Rev. 2010, 29, 3–26. [Google Scholar] [CrossRef]
- Chulze, S.N. Strategies to reduce mycotoxin levels in maize during storage: A review. Food Addit. Contam. A 2010, 27, 651–657. [Google Scholar] [CrossRef]
- Reddy, K.R.N.; Abbas, H.K.; Abel, C.A.; Shier, W.T.; Oliveira, C.A.F.; Raghavender, C.R. Mycotoxin contamination of commercially important agricultural commodities. Toxin Rev. 2009, 28, 154–168. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, F. Global burden of aflatoxin-induced hepatocellular carcinoma: A risk assessment. Environ. Health Perspect. 2010, 118, 818–824. [Google Scholar] [CrossRef]
- Jiang, Y.; Jolly, P.E.; Preko, P.; Wang, J.S.; Ellis, W.O.; Phillips, T.D.; Williams, J.H. Aflatoxin-related immune dysfunction in health and in human immunodeficiency virus disease. Clin. Dev. Immunol. 2008, 2008, 790309. [Google Scholar] [PubMed]
- Jolly, P.E.; Shuaib, F.M.; Jiang, Y.; Preko, P.; Baidoo, J.; Stiles, J.K.; Wang, J.S.; Phillips, T.D.; Williams, J.H. Association of high viral load and abnormal liver function with high aflatoxin B1-albumin adduct levels in HIV-positive Ghanaians: Preliminary observations. Food Addit. Contam. A 2011, 28, 1224–1234. [Google Scholar] [CrossRef]
- Heikens, G.T.; Manary, M. Wasting disease in African children: The challenges ahead. Malawi Med. J. 2009, 21, 101–105. [Google Scholar] [PubMed]
- Shuaib, F.M.; Jolly, P.E.; Ehiri, J.E.; Jiang, Y.; Ellis, W.O.; Stiles, J.K.; Yatich, N.J.; Funkhouser, E.; Person, S.D.; Wilson, C.; Williams, J.H. Association between anemia and aflatoxin B1 biomarker levels among pregnant women in Kumasi, Ghana. Am. J. Trop. Med. Hyg. 2010, 83, 1077–1083. [Google Scholar] [CrossRef]
- Williams, J.H.; Phillips, T.D.; Jolly, P.E.; Stiles, J.K.; Jolly, C.M.; Aggarwal, D. Human aflatoxicosis in developing countries: A review of toxicology, exposure, potential health consequences, and interventions. Am. J. Clin. Nutr. 2004, 80, 1106–1122. [Google Scholar] [PubMed]
- Wild, C.P.; Gong, Y.Y. Mycotoxins and human disease: A largely ignored global health issue. Carcinogenesis 2010, 31, 71–82. [Google Scholar] [CrossRef]
- Ehrlich, K.C.; Chang, P.K.; Yu, J.; Cary, J.W.; Bhatnagar, D. Control of Aflatoxin Biosynthesis in Aspergillus Species. In Aflatoxins—Biochemistry and Molecular Biology; Guevara-Gonzalez, R.G., Ed.; InTech: Rijeka, Croatia, 2011; pp. 21–40. [Google Scholar]
- Ehrlich, K.C.; Montalbano, B.G.; Cary, J.W. Binding of the C6-zinc cluster protein, AFLR, to the promoters of aflatoxin pathway biosynthesis genes in Aspergillus parasiticus. Gene 1999, 230, 249–257. [Google Scholar] [CrossRef]
- Lee, J.W.; Roze, L.V.; Linz, J.E. Evidence that a wortmannin-sensitive signal transduction pathway regulates aflatoxin biosynthesis. Mycologia 2007, 99, 562–568. [Google Scholar] [CrossRef]
- Schmidt-Heydt, M.; Abdel-Hadi, A.; Magan, N.; Geisen, R. Complex regulation of the aflatoxin biosynthesis gene cluster of Aspergillus flavus in relation to various combinations of water activity and temperature. Int. J. Food Microbiol. 2009, 135, 231–237. [Google Scholar] [CrossRef] [Green Version]
- Du, W.; Obrian, G.R.; Payne, G.A. Function and regulation of aflJ in the accumulation of aflatoxin early pathway intermediate in Aspergillus flavus. Food Addit. Contam. 2007, 24, 1043–1050. [Google Scholar] [CrossRef]
- Chang, P.K. The Aspergillus parasiticus protein AFLJ interacts with the aflatoxin pathway-specific regulator AFLR. Mol. Genet. Genomics 2003, 268, 711–719. [Google Scholar] [PubMed]
- Chang, P.K. Lack of interaction between AFLR and AFLJ contributes to nonaflatoxigenicity of Aspergillus sojae. J. Biotechnol. 2004, 107, 245–253. [Google Scholar] [CrossRef]
- Kiyota, T.; Hamada, R.; Sakamoto, K.; Iwashita, K.; Yamada, O.; Mikami, S. Aflatoxin non-productivity of Aspergillus oryzae caused by loss of function in the aflJ gene product. J. Biosci. Bioeng. 2011, 111, 512–517. [Google Scholar] [CrossRef]
- Meyers, D.M.; Obrian, G.; Du, W.L.; Bhatnagar, D.; Payne, G.A. Characterization of aflJ, a gene required for conversion of pathway intermediates to aflatoxin. Appl. Environ. Microbiol. 1998, 64, 3713–3717. [Google Scholar] [PubMed]
- Yin, W.B.; Amaike, S.; Wohlbach, D.J.; Gasch, A.P.; Chiang, Y.M.; Wang, C.C.; Bok, J.W.; Rohlfs, M.; Keller, N.P. An Aspergillus nidulans bZIP response pathway hardwired for defensive secondary metabolism operates through aflR. Mol. Microbiol. 2012, 83, 1024–1034. [Google Scholar] [CrossRef]
- Shaaban, M.I.; Bok, J.W.; Lauer, C.; Keller, N.P. Suppressor mutagenesis identifies a velvet complex remediator of Aspergillus nidulans secondary metabolism. Eukaryot. Cell 2010, 9, 1816–1824. [Google Scholar] [CrossRef]
- Roze, L.V.; Chanda, A.; Linz, J.E. Compartmentalization and molecular traffic in secondary metabolism: A new understanding of established cellular processes. Fungal Genet. Biol. 2011, 48, 35–48. [Google Scholar] [CrossRef]
- Chanda, A.; Roze, L.V.; Kang, S.; Artymovich, K.A.; Hicks, G.R.; Raikhel, N.V.; Calvo, A.M.; Linz, J.E. A key role for vesicles in fungal secondary metabolism. Proc. Natl. Acad. Sci. USA 2009, 106, 19533–19538. [Google Scholar] [CrossRef] [PubMed]
- Chanda, A.; Roze, L.V.; Linz, J.E. A possible role for exocytosis in aflatoxin export in Aspergillus parasiticus. Eukaryot. Cell 2010, 9, 1724–1727. [Google Scholar] [CrossRef]
- Linz, J.E.; Chanda, A.; Hong, S.Y.; Whitten, D.A.; Wilkerson, C.; Roze, L.V. Proteomic and biochemical evidence support a role for transport vesicles and endosomes in stress response and secondary metabolism in Aspergillus parasiticus. J. Proteome Res. 2012, 11, 767–775. [Google Scholar] [CrossRef]
- Chiang, Y.M.; Szewczyk, E.; Davidson, A.D.; Entwistle, R.; Keller, N.P.; Wang, C.C.; Oakley, B.R. Characterization of the Aspergillus nidulans monodictyphenone gene cluster. Appl. Environ. Microbiol. 2010, 76, 2067–2074. [Google Scholar] [CrossRef]
- Schwelm, A.; Bradshaw, R.E. Genetics of Dothistromin Biosynthesis of Dothistroma septosporum: An Update. Toxins 2010, 2, 2680–2698. [Google Scholar] [CrossRef]
- Chen, H.; Lee, M.H.; Daub, M.E.; Chung, K.R. Molecular analysis of the cercosporin biosynthetic gene cluster in Cercospora nicotianae. Mol. Microbiol. 2007, 64, 755–770. [Google Scholar] [CrossRef]
- Carbone, I.; Ramirez-Prado, J.H.; Jakobek, J.L.; Horn, B.W. Gene duplication, modularity and adaptation in the evolution of the aflatoxin gene cluster. BMC Evol. Biol. 2007, 7, 111. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Dazzo, F.B.; Glagoleva, O.; Yu, B.; Jain, A. CMEIAS: A computer-aided system for the image analysis of bacterial morphotypes in microbial communities. Microb. Ecol. 2001, 41, 173–194. [Google Scholar] [PubMed]
- Girao, H.; Geli, M.I.; Idrissi, F.Z. Actin in the endocytic pathway: From yeast to mammals. FEBS Lett. 2008, 582, 2112–2119. [Google Scholar] [CrossRef]
- Chen, Y.N.; Slabaugh, E.; Brandizzi, F. Membrane-tethered transcription factors in Arabidopsis thaliana: Novel regulators in stress response and development. Curr. Opin. Plant Biol. 2008, 11, 695–701. [Google Scholar] [CrossRef]
- Gao, H.; Brandizzi, F.; Benning, C.; Larkin, R.M. A membrane-tethered transcription factor defines a branch of the heat stress response in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2008, 105, 16398–16403. [Google Scholar] [CrossRef]
- Seo, P.J.; Lee, A.K.; Xiang, F.; Park, C.M. Molecular and functional profiling of Arabidopsis pathogenesis-related genes: Insights into their roles in salt response of seed germination. Plant Cell Physiol. 2008, 49, 334–344. [Google Scholar] [CrossRef]
- Bayram, O.; Krappmann, S.; Ni, M.; Bok, J.W.; Helmstaedt, K.; Valerius, O.; Braus-Stromeyer, S.; Kwon, N.J.; Keller, N.P.; Yu, J.H.; Braus, G.H. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 2008, 320, 1504–1506. [Google Scholar] [CrossRef] [PubMed]
- Prediction of “hot spots” of aggregation in polypeptides. Available online: http://bioinf.uab.es/aggrescan (accessed on 10 November 2012).
- Conchillo-Sole, O.; de Groot, N.S.; Aviles, F.X.; Vendrell, J.; Daura, X.; Ventura, S. AGGRESCAN: A server for the prediction and evaluation of “hot spots” of aggregation in polypeptides. BMC Bioinform. 2007, 8, 65. [Google Scholar] [CrossRef]
- Yu, J.; Fedorova, N.D.; Montalbano, B.G.; Bhatnagar, D.; Cleveland, T.E.; Bennett, J.W.; Nierman, W.C. Tight control of mycotoxin biosynthesis gene expression in Aspergillus flavus by temperature as revealed by RNA-Seq. FEMS Microbiol. Lett. 2011, 322, 145–149. [Google Scholar] [CrossRef]
- O’Brian, G.R.; Georgianna, D.R.; Wilkinson, J.R.; Yu, J.; Abbas, H.K.; Bhatnagar, D.; Cleveland, T.E.; Nierman, W.; Payne, G.A. The effect of elevated temperature on gene transcription and aflatoxin biosynthesis. Mycologia 2007, 99, 232–239. [Google Scholar] [CrossRef]
- Chang, P.K.; Yu, J.; Bhatnagar, D.; Cleveland, T.E. Repressor-AFLR interaction modulates aflatoxin biosynthesis in Aspergillus parasiticus. Mycopathologia 1999, 147, 105–112. [Google Scholar] [CrossRef]
- Tsitsigiannis, D.I.; Zarnowski, R.; Keller, N.P. The lipid body protein, PpoA, coordinates sexual and asexual sporulation in Aspergillus nidulan. J. Biol. Chem. 2003, 29, 29. [Google Scholar]
- Chanda, A.; Roze, L.V.; Pastor, A.; Frame, M.K.; Linz, J.E. Purification of a vesicle-vacuole fraction functionally linked to aflatoxin synthesis in Aspergillus parasiticus. J. Microbiol. Methods 2009, 78, 28–33. [Google Scholar] [CrossRef]
- Ehrlich, K.C.; Li, P.; Scharfenstein, L.; Chang, P.-K. HypC is the anthrone oxidase involved in aflatoxin production. Appl. Environ. Microbiol. 2010, 76, 3374–3377. [Google Scholar] [CrossRef]
- Chang, P.-K.; Scharfenstein, L.L.; Wei, Q.; Bhatnagar, D. Development and refinement of a high-efficiency gene-targeting system for Aspergillus flavus. J. Microbiol. Methods. 2010, 81, 240–246. [Google Scholar] [CrossRef]
- Bayman, P.; Cotty, P.J. Improved media for selecting nitrate-nonutilizing mutants in Aspergillus flavus. Mycologia 1991, 83, 311–316. [Google Scholar] [CrossRef]
- Cary, J.W.; Harris-Coward, P.Y.; Ehrlich, K.C.; Mack, B.M.; Kale, S.P.; Larey, C.; Calvo, A.M. NsdC and NsdD affect Aspergillus flavus morphogenesis and aflatoxin production. Eukaryot. Cell 2012, 11, 1104–1111. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; Vandesompele, J.; Wittwer, C.T. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Hoshida, H.; Fujita, T.; Murata, K.; Kubo, K.; Akada, R. Copper-dependent production of a Pycnoporus coccineus extracellular laccase in Aspergillus oryzae and Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 2005, 69, 1090–1097. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ehrlich, K.C.; Mack, B.M.; Wei, Q.; Li, P.; Roze, L.V.; Dazzo, F.; Cary, J.W.; Bhatnagar, D.; Linz, J.E. Association with AflR in Endosomes Reveals New Functions for AflJ in Aflatoxin Biosynthesis. Toxins 2012, 4, 1582-1600. https://doi.org/10.3390/toxins4121582
Ehrlich KC, Mack BM, Wei Q, Li P, Roze LV, Dazzo F, Cary JW, Bhatnagar D, Linz JE. Association with AflR in Endosomes Reveals New Functions for AflJ in Aflatoxin Biosynthesis. Toxins. 2012; 4(12):1582-1600. https://doi.org/10.3390/toxins4121582
Chicago/Turabian StyleEhrlich, Kenneth C., Brian M. Mack, Qijian Wei, Ping Li, Ludmila V. Roze, Frank Dazzo, Jeffrey W. Cary, Deepak Bhatnagar, and John E. Linz. 2012. "Association with AflR in Endosomes Reveals New Functions for AflJ in Aflatoxin Biosynthesis" Toxins 4, no. 12: 1582-1600. https://doi.org/10.3390/toxins4121582




