Oxidative Stress-Related Transcription Factors in the Regulation of Secondary Metabolism
Abstract
:1. Introduction
2. Stress Activated Signaling Pathways in Yeast and Filamentous Fungi

3. Transcription Factors Related to Oxidative Stress, Which Are Involved in Regulation of Secondary Metabolism
3.1. AP-1
3.2. AtfA
3.3. AtfB
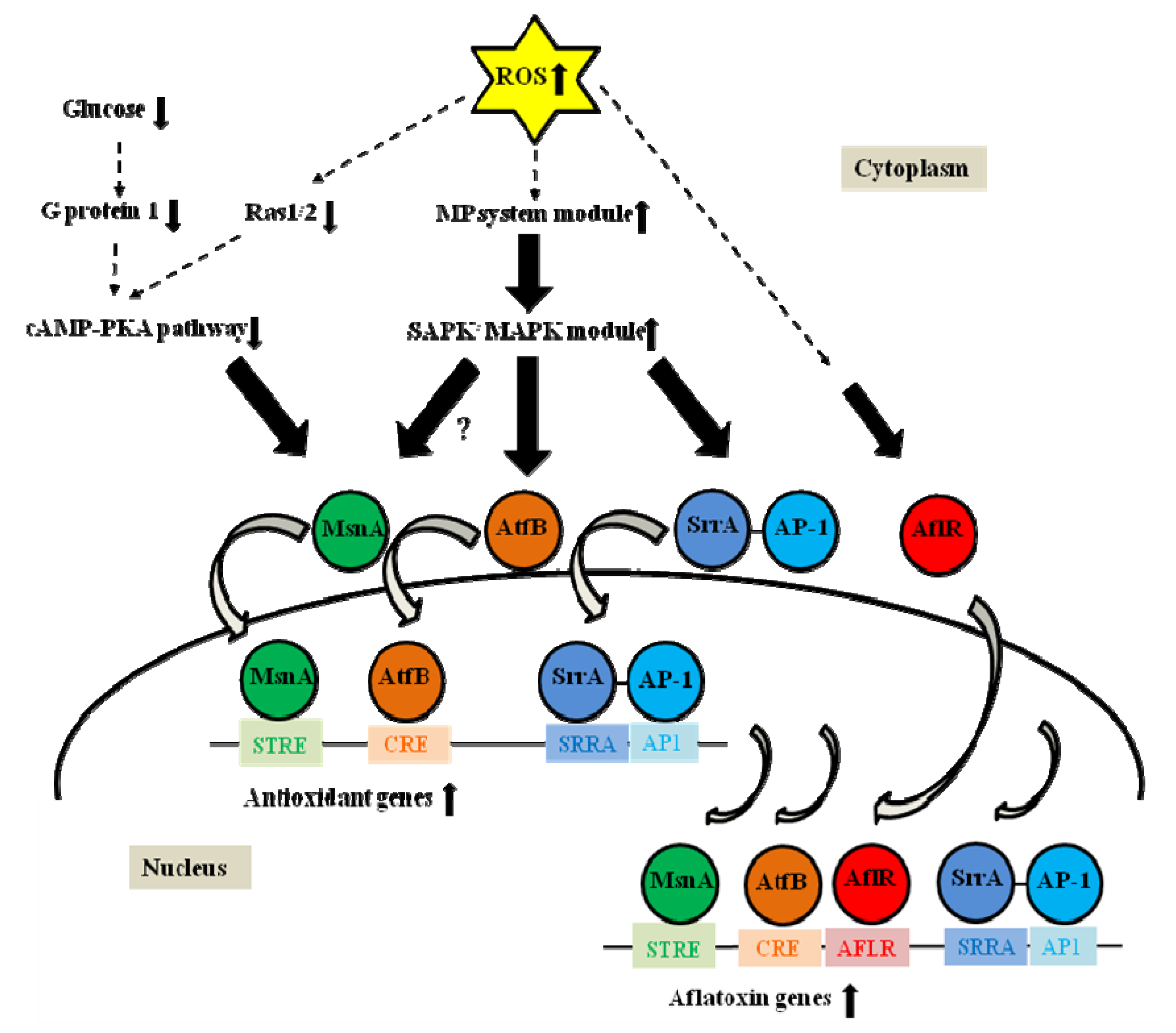
3.4. MsnA
3.5. StuA
3.6. CCAAT-Binding Transcription Factor Complex
4. Transcription Factors with Strong Potential to Coordinate Oxidative Stress and Secondary Metabolism
4.1. Hsf1
4.2. SrrA
4.3. Pcr1
4.4. Myb
5. Conclusions and Future Research
References
- Reverberi, M.; Zjalic, S.; Ricelli, A.; Fabbri, A.A.; Fanelli, C. Oxidant/antioxidant balance in Aspergillus parasiticus affects aflatoxin biosynthesis. Mycotoxin Res. 2006, 22, 39–47. [Google Scholar] [CrossRef]
- Reverberi, M.; Gazzetti, K.; Punelli, F.; Scarpari, M.; Zjalic, S.; Ricelli, A.; Fabbri, A.A.; Fanelli, C. Aoyap1 regulates ota synthesis by controlling cell redox balance in Aspergillus ochraceus. Appl. Microbiol. Biotechnol. 2012, 95, 1293–1304. [Google Scholar] [CrossRef]
- Roze, L.V.; Chanda, A.; Wee, J.; Awad, D.; Linz, J.E. Stress-related transcription factor ATFB integrates secondary metabolism with oxidative stress response in aspergilli. J. Biol. Chem. 2011, 286, 35137–35148. [Google Scholar]
- Yin, W.B.; Amaike, S.; Wohlbach, D.J.; Gasch, A.P.; Chiang, Y.M.; Wang, C.C.; Bok, J.W.; Rohlfs, M.; Keller, N.P. An Aspergillus nidulans bZIP response pathway hardwired for defensive secondary metabolism operates through aflR. Mol. Microbiol. 2012, 83, 1024–1034. [Google Scholar] [CrossRef]
- Hong, S.Y.; Roze, L.V.; Wee, J.; Linz, J.E. Evidence that a transcription factor regulatory network coordinates oxidative stress response and secondary metabolism in aspergilli. Microbiol. Open 2013, 2, 144–160. [Google Scholar]
- Miskei, M.; Karanyi, Z.; Pocsi, I. Annotation of stress-response proteins in the aspergilli. Fungal Genet. Biol. 2009, 46, S105–S120. [Google Scholar] [CrossRef]
- Toone, W.M.; Morgan, B.A.; Jones, N. Redox control of AP-1-like factors in yeast and beyond. Oncogene 2001, 20, 2336–2346. [Google Scholar]
- Ikner, A.; Shiozaki, K. Yeast signaling pathways in the oxidative stress response. Mutat. Res. 2005, 569, 13–27. [Google Scholar] [CrossRef]
- Toone, W.M.; Jones, N. Stress-activated signalling pathways in yeast. Genes Cells 1998, 3, 485–498. [Google Scholar]
- Bahn, Y.S.; Xue, C.; Idnurm, A.; Rutherford, J.C.; Heitman, J.; Cardenas, M.E. Sensing the environment: Lessons from fungi. Nat. Rev. 2007, 5, 57–69. [Google Scholar] [CrossRef]
- Banuett, F. Signalling in the yeasts: An informational cascade with links to the filamentous fungi. Microbiol. Mol. Biol. Rev. 1998, 62, 249–274. [Google Scholar]
- Aguirre, J.; Hansberg, W.; Navarro, R. Fungal responses to reactive oxygen species. Med. Mycol. 2006, 44, S101–S107. [Google Scholar] [CrossRef]
- Bahn, Y.S. Master and commander in fungal pathogens: The two-component system and the hog signaling pathway. Eukaryot. Cell 2008, 7, 2017–2036. [Google Scholar] [CrossRef]
- Eaton, C.J.; Jourdain, I.; Foster, S.J.; Hyams, J.S.; Scott, B. Functional analysis of a fungal endophyte stress-activated MAP kinase. Curr. Genet. 2008, 53, 163–174. [Google Scholar] [CrossRef]
- Segmuller, N.; Ellendorf, U.; Tudzynski, B.; Tudzynski, P. BcSak1, a stress-activated mitogen-activated protein kinase, is involved in vegetative differentiation and pathogenicity in Botrytis cinerea. Eukaryot. Cell 2007, 6, 211–221. [Google Scholar] [CrossRef]
- Heller, J.; Ruhnke, N.; Espino, J.J.; Massaroli, M.; Collado, I.G.; Tudzynski, P. The mitogen-activated protein kinase BcSak1 of Botrytis cinerea is required for pathogenic development and has broad regulatory functions beyond stress response. Mol. Plant Microbe Interact. 2012, 25, 802–816. [Google Scholar] [CrossRef]
- Nguyen, T.V.; Schafer, W.; Bormann, J. The stress-activated protein kinase FgOS-2 is a key regulator in the life cycle of the cereal pathogen Fusarium graminearum. Mol. Plant Microbe Interact. 2012, 25, 1142–1156. [Google Scholar] [CrossRef]
- Moye-Rowley, W.S. Transcription factors regulating the response to oxidative stress in yeast. Antioxid. Redox Signal. 2002, 4, 123–140. [Google Scholar] [CrossRef]
- Zhulin, I.B.; Taylor, B.L.; Dixon, R. PAS domain S-boxes in Archaea, Bacteria and sensors for oxygen and redox. Trends Biochem. Sci. 1997, 22, 331–333. [Google Scholar] [CrossRef]
- Nguyen, A.N.; Lee, A.; Place, W.; Shiozaki, K. Multistep phosphorelay proteins transmit oxidative stress signals to the fission yeast stress-activated protein kinase. Mol. Biol. Cell 2000, 11, 1169–1181. [Google Scholar]
- Chen, D.; Wilkinson, C.R.; Watt, S.; Penkett, C.J.; Toone, W.M.; Jones, N.; Bahler, J. Multiple pathways differentially regulate global oxidative stress responses in fission yeast. Mol. Biol. Cell 2008, 19, 308–317. [Google Scholar] [CrossRef]
- Thon, M.; Al Abdallah, Q.; Hortschansky, P.; Scharf, D.H.; Eisendle, M.; Haas, H.; Brakhage, A.A. The CCAAT-binding complex coordinates the oxidative stress response in eukaryotes. Nucleic Acids Res. 2010, 38, 1098–1113. [Google Scholar] [CrossRef]
- Karimpour, S.; Lou, J.; Lin, L.L.; Rene, L.M.; Lagunas, L.; Ma, X.; Karra, S.; Bradbury, C.M.; Markovina, S.; Goswami, P.C.; et al. Thioredoxin reductase regulates AP-1 activity as well as thioredoxin nuclear localization via active cysteines in response to ionizing radiation. Oncogene 2002, 21, 6317–6327. [Google Scholar] [CrossRef]
- Thon, M.; Al-Abdallah, Q.; Hortschansky, P.; Brakhage, A.A. The thioredoxin system of the filamentous fungus Aspergillus nidulans: Impact on development and oxidative stress response. J. Biol. Chem. 2007, 282, 27259–27269. [Google Scholar] [CrossRef]
- Reverberi, M.; Zjalic, S.; Ricelli, A.; Punelli, F.; Camera, E.; Fabbri, C.; Picardo, M.; Fanelli, C.; Fabbri, A.A. Modulation of antioxidant defense in Aspergillus parasiticus is involved in aflatoxin biosynthesis: A role for the ApyapA gene. Eukaryot. Cell 2008, 7, 988–1000. [Google Scholar] [CrossRef]
- Reverberi, M.; Zjalic, S.; Punelli, F.; Ricelli, A.; Fabbri, A.A.; Fanelli, C. Apyap1 affects aflatoxin biosynthesis during Aspergillus parasiticus growth in maize seeds. Food Addit. Contam. 2007, 24, 1070–1075. [Google Scholar]
- Yin, W.B.; Reinke, A.W.; Szilagyi, M.; Emri, T.; Chiang, Y.M.; Keating, A.E.; Pocsi, I.; Wang, C.C.; Keller, N.P. bZIP transcription factors affecting secondary metabolism, sexual development and stress responses in Aspergillus nidulans. Microbiology 2013, 159, 77–88. [Google Scholar] [CrossRef]
- Wu, A.L.; Moye-Rowley, W.S. GSH1, which encodes gamma-glutamylcysteine synthetase, is a target gene for yAP-1 transcriptional regulation. Mol. Cell. Biol. 1994, 14, 5832–5839. [Google Scholar] [CrossRef]
- Karin, M.; Liu, Z.; Zandi, E. AP-1 function and regulation. Curr. Opin. Cell biol. 1997, 9, 240–246. [Google Scholar] [CrossRef]
- Fernandes, L.; Rodrigues-Pousada, C.; Struhl, K. Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol. Cell. Biol. 1997, 17, 6982–6993. [Google Scholar]
- Delaunay, A.; Isnard, A.D.; Toledano, M.B. H2O2 sensing through oxidation of the Yap1 transcription factor. Embo J. 2000, 19, 5157–5166. [Google Scholar] [CrossRef]
- Lee, J.; Godon, C.; Lagniel, G.; Spector, D.; Garin, J.; Labarre, J.; Toledano, M.B. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 1999, 274, 16040–16046. [Google Scholar]
- Abate, C.; Patel, L.; Rauscher, F.J., 3rd; Curran, T. Redox regulation of fos and jun DNA-binding activity in vitro. Science 1990, 249, 1157–1161. [Google Scholar]
- He, X.J.; Fassler, J.S. Identification of novel Yap1p and Skn7p binding sites involved in the oxidative stress response of Saccharomyces cerevisiae. Mol. Microbiol. 2005, 58, 1454–1467. [Google Scholar] [CrossRef]
- Toda, T.; Shimanuki, M.; Saka, Y.; Yamano, H.; Adachi, Y.; Shirakawa, M.; Kyogoku, Y.; Yanagida, M. Fission yeast pap1-dependent transcription is negatively regulated by an essential nuclear protein, crm1. Mol. Cell. Biol. 1992, 12, 5474–5484. [Google Scholar]
- Asano, Y.; Hagiwara, D.; Yamashino, T.; Mizuno, T. Characterization of the bZip-type transcription factor NapA with reference to oxidative stress response in Aspergillus nidulans. Biosci. Biotechnol. Biochem. 2007, 71, 1800–1803. [Google Scholar] [CrossRef]
- Mulford, K.E.; Fassler, J.S. Association of the Skn7 and Yap1 transcription factors in the Saccharomyces cerevisiae oxidative stress response. Eukaryot. Cell 2011, 10, 761–769. [Google Scholar] [CrossRef]
- Balazs, A.; Pocsi, I.; Hamari, Z.; Leiter, E.; Emri, T.; Miskei, M.; Olah, J.; Toth, V.; Hegedus, N.; Prade, R.A.; et al. AtfA bZIP-type transcription factor regulates oxidative and osmotic stress responses in Aspergillus nidulans. Mol. Genet. Genomics 2010, 283, 289–303. [Google Scholar] [CrossRef]
- Lara-Rojas, F.; Sanchez, O.; Kawasaki, L.; Aguirre, J. Aspergillus nidulans transcription factor AtfA interacts with the MAPK SakA to regulate general stress responses, development and spore functions. Mol. Microbiol. 2011, 80, 436–454. [Google Scholar] [CrossRef]
- Hagiwara, D.; Asano, Y.; Yamashino, T.; Mizuno, T. Characterization of bZip-type transcription factor AtfA with reference to stress responses of conidia of Aspergillus nidulans. Biosci. Biotechnol. Biochem. 2008, 72, 2756–2760. [Google Scholar] [CrossRef]
- Temme, N.; Oeser, B.; Massaroli, M.; Heller, J.; Simon, A.; Collado, I.G.; Viaud, M.; Tudzynski, P. Bcatf1, a global regulator, controls various differentiation processes and phytotoxin production in Botrytis cinerea. Mol. Plant Pathol. 2012, 13, 704–718. [Google Scholar] [CrossRef]
- Watanabe, Y.; Yamamoto, M. Schizosaccharomyces pombe pcr1+ encodes a CREB/ATF protein involved in regulation of gene expression for sexual development. Mol. Cell. Biol. 1996, 16, 704–711. [Google Scholar]
- Sanso, M.; Gogol, M.; Ayte, J.; Seidel, C.; Hidalgo, E. Transcription factors Pcr1 and Atf1 have distinct roles in stress- and sty1-dependent gene regulation. Eukaryot. Cell 2008, 7, 826–835. [Google Scholar] [CrossRef]
- Proft, M.; Struhl, K. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol. Cell 2002, 9, 1307–1317. [Google Scholar] [CrossRef]
- Sakamoto, K.; Iwashita, K.; Yamada, O.; Kobayashi, K.; Mizuno, A.; Akita, O.; Mikami, S.; Shimoi, H.; Gomi, K. Aspergillus oryzae atfA controls conidial germination and stress tolerance. Fungal Genet. Biol. 2009, 46, 887–897. [Google Scholar] [CrossRef]
- Roze, L.V.; Miller, M.J.; Rarick, M.; Mahanti, N.; Linz, J.E. A novel cAMP-response element, CRE1, modulates expression of nor-1 in Aspergillus parasiticus. J. Biol. Chem. 2004, 279, 27428–27439. [Google Scholar]
- Martinez-Pastor, M.T.; Marchler, G.; Schuller, C.; Marchler-Bauer, A.; Ruis, H.; Estruch, F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 1996, 15, 2227–2235. [Google Scholar]
- Chang, P.K.; Scharfenstein, L.L.; Luo, M.; Mahoney, N.; Molyneux, R.J.; Yu, J.; Brown, R.L.; Campbell, B.C. Loss of msnA, a putative stress regulatory gene, in Aspergillus parasiticus and Aspergillus flavus increased production of conidia, aflatoxins and kojic acid. Toxins 2011, 3, 82–104. [Google Scholar] [CrossRef]
- Scherer, M.; Wei, H.; Liese, R.; Fischer, R. Aspergillus nidulans catalase-peroxidase gene (cpea) is transcriptionally induced during sexual development through the transcription factor StuA. Eukaryot. Cell 2002, 1, 725–735. [Google Scholar] [CrossRef]
- Twumasi-Boateng, K.; Yu, Y.; Chen, D.; Gravelat, F.N.; Nierman, W.C.; Sheppard, D.C. Transcriptional profiling identifies a role for BrlA in the response to nitrogen depletion and for StuA in the regulation of secondary metabolite clusters in Aspergillus fumigatus. Eukaryot. Cell 2009, 8, 104–115. [Google Scholar] [CrossRef]
- Brakhage, A.A.; Andrianopoulos, A.; Kato, M.; Steidl, S.; Davis, M.A.; Tsukagoshi, N.; Hynes, M.J. HAP-like CCAAT-binding complexes in filamentous fungi: Implications for biotechnology. Fungal Genet. Biol. 1999, 27, 243–252. [Google Scholar] [CrossRef]
- Romier, C.; Cocchiarella, F.; Mantovani, R.; Moras, D. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J. Biol. Chem. 2003, 278, 1336–1345. [Google Scholar]
- Laloum, T.; de Mita, S.; Gamas, P.; Baudin, M.; Niebel, A. CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. 2013, 18, 157–166. [Google Scholar] [CrossRef]
- Dolfini, D.; Gatta, R.; Mantovani, R. Nf-y and the transcriptional activation of CCAAT promoters. Crit. Rev. Biochem. Mol. Biol. 2012, 47, 29–49. [Google Scholar] [CrossRef]
- Petroni, K.; Kumimoto, R.W.; Gnesutta, N.; Calvenzani, V.; Fornari, M.; Tonelli, C.; Holt, B.F., 3rd; Mantovani, R. The promiscuous life of plant nuclear factor y transcription factors. Plant Cell 2012, 24, 4777–4792. [Google Scholar] [CrossRef]
- Gatta, R.; Mantovani, R. NF-Y affects histone acetylation and H2A.Z deposition in cell cycle promoters. Epigenetics 2011, 6, 526–534. [Google Scholar] [CrossRef]
- Hahn, S.; Guarente, L. Yeast HAP2 and HAP3: Transcriptional activators in a heteromeric complex. Science 1988, 240, 317–321. [Google Scholar]
- Forsburg, S.L.; Guarente, L. Identification and characterization of HAP4: A third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 1989, 3, 1166–1178. [Google Scholar] [CrossRef]
- Huber, E.M.; Scharf, D.H.; Hortschansky, P.; Groll, M.; Brakhage, A.A. DNA minor groove sensing and widening by the CCAAT-binding complex. Structure 2012, 20, 1757–1768. [Google Scholar] [CrossRef]
- Kato, M. An overview of the CCAAT-box binding factor in filamentous fungi: Assembly, nuclear translocation, and transcriptional enhancement. Biosci. Biotechnol. Biochem. 2005, 69, 663–672. [Google Scholar] [CrossRef]
- Steidl, S.; Tuncher, A.; Goda, H.; Guder, C.; Papadopoulou, N.; Kobayashi, T.; Tsukagoshi, N.; Kato, M.; Brakhage, A.A. A single subunit of a heterotrimeric CCAAT-binding complex carries a nuclear localization signal: Piggy back transport of the pre-assembled complex to the nucleus. J. Mol. Biol. 2004, 342, 515–524. [Google Scholar] [CrossRef]
- Goda, H.; Nagase, T.; Tanoue, S.; Sugiyama, J.; Steidl, S.; Tuncher, A.; Kobayashi, T.; Tsukagoshi, N.; Brakhage, A.A.; Kato, M. Nuclear translocation of the heterotrimeric CCAAT binding factor of Aspergillus oryzae is dependent on two redundant localising signals in a single subunit. Arch. Microbiol. 2005, 184, 93–100. [Google Scholar] [CrossRef]
- Litzka, O.; Then Bergh, K.; Brakhage, A.A. The Aspergillus nidulans penicillin-biosynthesis gene aat (penDE) is controlled by a CCAAT-containing DNA element. Eur. J. Biochem. 1996, 238, 675–682. [Google Scholar]
- Litzka, O.; Papagiannopolous, P.; Davis, M.A.; Hynes, M.J.; Brakhage, A.A. The penicillin regulator PENR1 of Aspergillus nidulans is a HAP-like transcriptional complex. Eur. J. Biochem. 1998, 251, 758–767. [Google Scholar]
- Sprote, P.; Hynes, M.J.; Hortschansky, P.; Shelest, E.; Scharf, D.H.; Wolke, S.M.; Brakhage, A.A. Identification of the novel penicillin biosynthesis gene aatb of Aspergillus nidulans and its putative evolutionary relationship to this fungal secondary metabolism gene cluster. Mol. Microbiol. 2008, 70, 445–461. [Google Scholar]
- Caruso, M.L.; Litzka, O.; Martic, G.; Lottspeich, F.; Brakhage, A.A. Novel basic-region helix-loop-helix transcription factor (AnBH1) of Aspergillus nidulans counteracts the CCAAT-binding complex AnCF in the promoter of a penicillin biosynthesis gene. J. Mol. Biol. 2002, 323, 425–439. [Google Scholar]
- Li, W.X.; Oono, Y.; Zhu, J.; He, X.J.; Wu, J.M.; Iida, K.; Lu, X.Y.; Cui, X.; Jin, H.; Zhu, J.K. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 2008, 20, 2238–2251. [Google Scholar] [CrossRef]
- Zeilinger, S.; Schmoll, M.; Pail, M.; Mach, R.L.; Kubicek, C.P. Nucleosome transactions on the Hypocrea jecorina (Trichoderma reesei) cellulase promoter cbh2 associated with cellulase induction. Mol. Genet. Genomics 2003, 270, 46–55. [Google Scholar] [CrossRef]
- Rauscher, R.; Wurleitner, E.; Wacenovsky, C.; Aro, N.; Stricker, A.R.; Zeilinger, S.; Kubicek, C.P.; Penttila, M.; Mach, R.L. Transcriptional regulation of xyn1, encoding xylanase I, in Hypocrea jecorina. Eukaryot. Cell 2006, 5, 447–456. [Google Scholar]
- Chen, H.; Crabb, J.W.; Kinsey, J.A. The Neurospora aab-1 gene encodes a CCAAT binding protein homologous to yeast HAP5. Genetics 1998, 148, 123–130. [Google Scholar]
- Camps, S.M.; Dutilh, B.E.; Arendrup, M.C.; Rijs, A.J.; Snelders, E.; Huynen, M.A.; Verweij, P.E.; Melchers, W.J. Discovery of a hapE mutation that causes azole resistance in Aspergillus fumigatus through whole genome sequencing and sexual crossing. PLoS One 2012, 7, e50034. [Google Scholar]
- Fernandes, M.; Xiao, H.; Lis, J.T. Fine structure analyses of the Drosophila and Saccharomyces heat shock factor—Heat shock element interactions. Nucleic Acids Res. 1994, 22, 167–173. [Google Scholar] [CrossRef]
- Liu, X.D.; Thiele, D.J. Oxidative stress induced heat shock factor phosphorylation and HSF-dependent activation of yeast metallothionein gene transcription. Genes Dev. 1996, 10, 592–603. [Google Scholar] [CrossRef]
- Raitt, D.C.; Johnson, A.L.; Erkine, A.M.; Makino, K.; Morgan, B.; Gross, D.S.; Johnston, L.H. The Skn7 response regulator of Saccharomyces cerevisiae interacts with Hsf1 in vivo and is required for the induction of heat shock genes by oxidative stress. Mol. Biol. Cell 2000, 11, 2335–2347. [Google Scholar]
- Yamamoto, A.; Ueda, J.; Yamamoto, N.; Hashikawa, N.; Sakurai, H. Role of heat shock transcription factor in Saccharomyces cerevisiae oxidative stress response. Eukaryot. Cell 2007, 6, 1373–1379. [Google Scholar] [CrossRef]
- Hagiwara, D.; Asano, Y.; Marui, J.; Furukawa, K.; Kanamaru, K.; Kato, M.; Abe, K.; Kobayashi, T.; Yamashino, T.; Mizuno, T. The SskA and SrrA response regulators are implicated in oxidative stress responses of hyphae and asexual spores in the phosphorelay signaling network of Aspergillus nidulans. Biosci. Biotechnol. Biochem. 2007, 71, 1003–1014. [Google Scholar] [CrossRef]
- Vargas-Perez, I.; Sanchez, O.; Kawasaki, L.; Georgellis, D.; Aguirre, J. Response regulators SrrA and SskA are central components of a phosphorelay system involved in stress signal transduction and asexual sporulation in Aspergillus nidulans. Eukaryot. Cell 2007, 6, 1570–1583. [Google Scholar] [CrossRef]
- Fassler, J.S.; West, A.H. Fungal Skn7 stress responses and their relationship to virulence. Eukaryot. Cell 2011, 10, 156–167. [Google Scholar] [CrossRef]
- Morgan, B.A.; Banks, G.R.; Toone, W.M.; Raitt, D.; Kuge, S.; Johnston, L.H. The Skn7 response regulator controls gene expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J. 1997, 16, 1035–1044. [Google Scholar] [CrossRef]
- Ohmiya, R.; Kato, C.; Yamada, H.; Aiba, H.; Mizuno, T. A fission yeast gene (prr1+) that encodes a response regulator implicated in oxidative stress response. J. Biochem. 1999, 125, 1061–1066. [Google Scholar] [CrossRef]
- Lippold, F.; Sanchez, D.H.; Musialak, M.; Schlereth, A.; Scheible, W.R.; Hincha, D.K.; Udvardi, M.K. AtMyb41 regulates transcriptional and metabolic responses to osmotic stress in arabidopsis. Plant Physiol. 2009, 149, 1761–1772. [Google Scholar]
- Borevitz, J.O.; Xia, Y.; Blount, J.; Dixon, R.A.; Lamb, C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 2000, 12, 2383–2394. [Google Scholar]
- Scheible, W.R.; Morcuende, R.; Czechowski, T.; Fritz, C.; Osuna, D.; Palacios-Rojas, N.; Schindelasch, D.; Thimm, O.; Udvardi, M.K.; Stitt, M. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 2004, 136, 2483–2499. [Google Scholar] [CrossRef]
- Peel, G.J.; Pang, Y.; Modolo, L.V.; Dixon, R.A. The LAP1 MYB transcription factor orchestrates anthocyanidin biosynthesis and glycosylation in medicago. Plant J. 2009, 59, 136–149. [Google Scholar] [CrossRef]
- Wieser, J.; Adams, T.H. FlbD encodes a Myb-like DNA-binding protein that coordinates initiation of Aspergillus nidulans conidiophore development. Genes Dev. 1995, 9, 491–502. [Google Scholar] [CrossRef]
- Arratia-Quijada, J.; Sanchez, O.; Scazzocchio, C.; Aguirre, J. FlbD, a Myb transcription factor of Aspergillus nidulans, is uniquely involved in both asexual and sexual differentiation. Eukaryot. Cell 2012, 11, 1132–1142. [Google Scholar] [CrossRef]
- Zhang, M.; Lu, J.; Tao, K.; Ye, W.; Li, A.; Liu, X.; Kong, L.; Dong, S.; Zheng, X.; Wang, Y. A Myb transcription factor of Phytophthora sojae, regulated by MAP kinase PsSAK1, is required for zoospore development. PloS One 2012, 7, e40246. [Google Scholar]
- Cary, J.W.; Harris-Coward, P.Y.; Ehrlich, K.C.; Mack, B.M.; Kale, S.P.; Larey, C.; Calvo, A.M. NsdC and NsdD affect Aspergillus flavus morphogenesis and aflatoxin production. Eukaryot. Cell 2012, 11, 1104–1111. [Google Scholar] [CrossRef]
- Shaaban, M.I.; Bok, J.W.; Lauer, C.; Keller, N.P. Suppressor mutagenesis identifies a velvet complex remediator of Aspergillus nidulans secondary metabolism. Eukaryot. Cell 2010, 9, 1816–1824. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Hong, S.-Y.; Roze, L.V.; Linz, J.E. Oxidative Stress-Related Transcription Factors in the Regulation of Secondary Metabolism. Toxins 2013, 5, 683-702. https://doi.org/10.3390/toxins5040683
Hong S-Y, Roze LV, Linz JE. Oxidative Stress-Related Transcription Factors in the Regulation of Secondary Metabolism. Toxins. 2013; 5(4):683-702. https://doi.org/10.3390/toxins5040683
Chicago/Turabian StyleHong, Sung-Yong, Ludmila V. Roze, and John E. Linz. 2013. "Oxidative Stress-Related Transcription Factors in the Regulation of Secondary Metabolism" Toxins 5, no. 4: 683-702. https://doi.org/10.3390/toxins5040683



