Proteomic Analyses of Agkistrodon contortrix contortrix Venom Using 2D Electrophoresis and MS Techniques
Abstract
:1. Introduction
2. Results
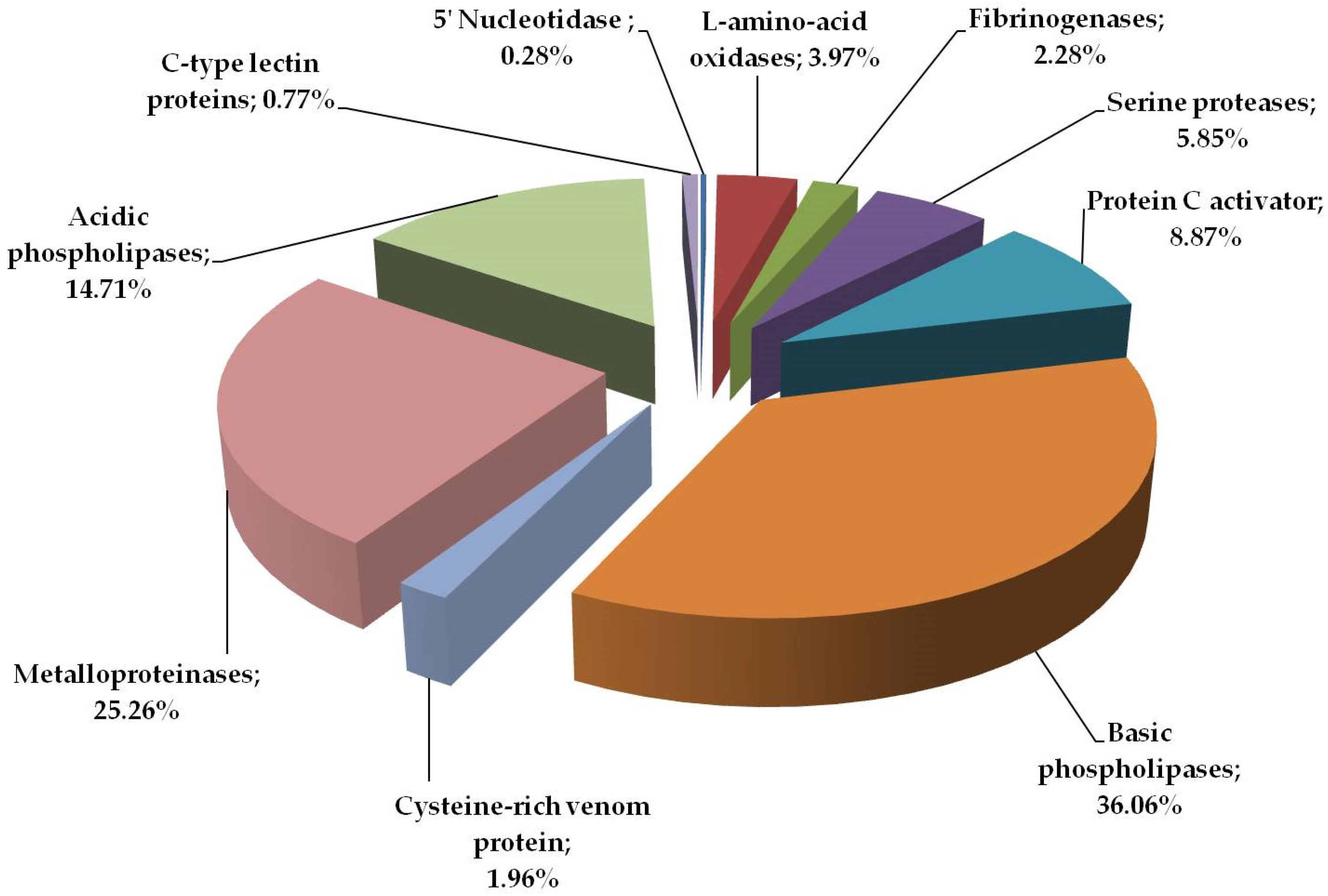
2.1. Proteome Analysis
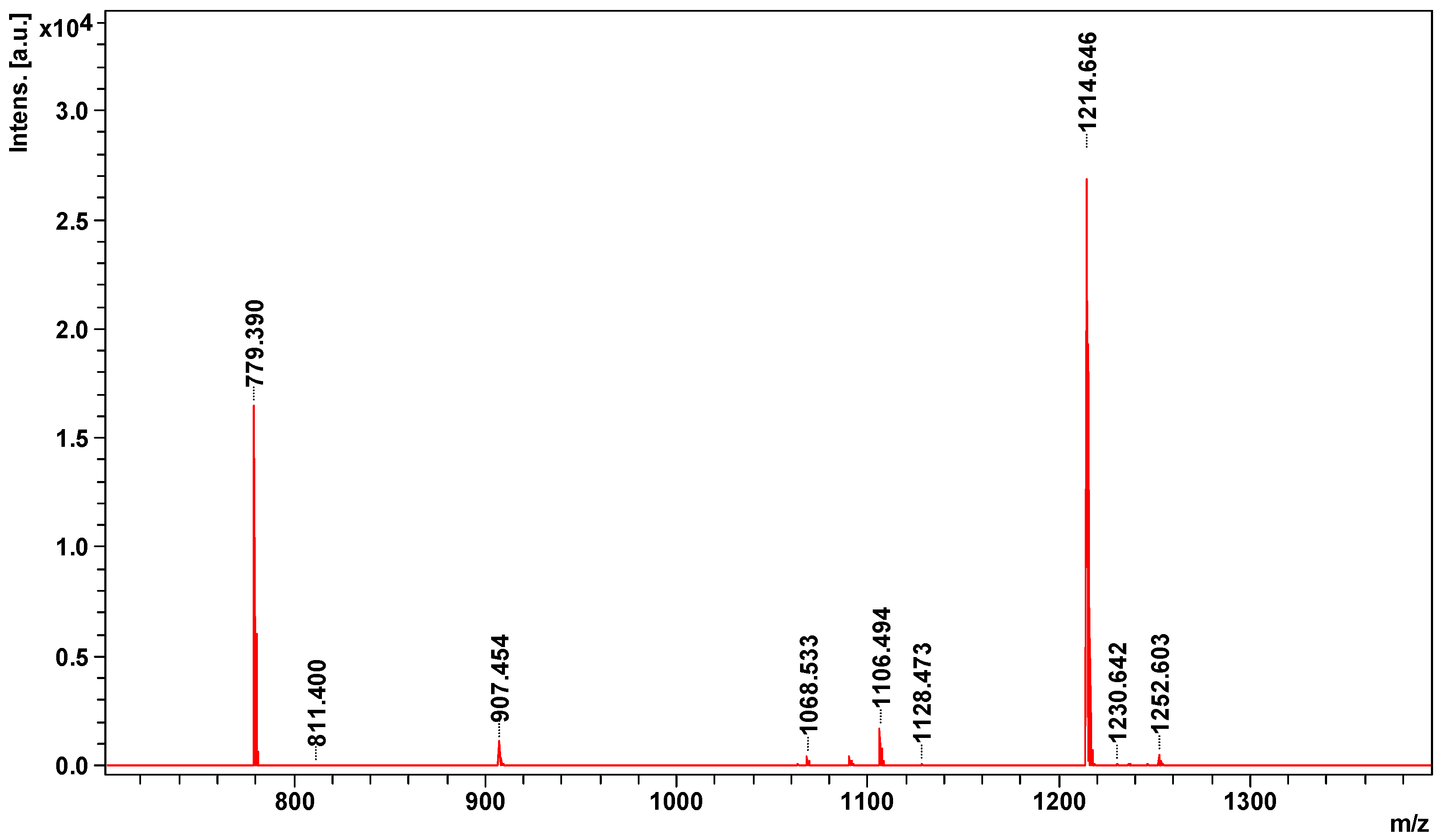
2.2. Peptidome Analysis
3. Discussion
4. Materials and Methods
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Campbell, J.A.; Lamar, W.W. The Venomous Reptiles of the Western Hemisphere; Comstock Publishing Associate: Ithaca, NY, USA, 2004; p. 976. [Google Scholar]
- Gloyd, H.K.; Conant, R. Snakes of the Agkistrodon Complex: A Monographic Review; Society for the Study of Amphibians and Reptiles: St. Louis, MO, USA, 1990; p. 614. [Google Scholar]
- Ernst, C.H.; Ernst, E.M. Copperheads and Cottonmouths. In Snakes of the United States and Canada; Ernst, C.H., Ernst, E.M., Eds.; Smithsonian Books: Washington, DC, USA; London, UK, 2003; pp. 473–486. [Google Scholar]
- Lomonte, B.; Tsai, W.; Ureña-Diaz, J.M.; Sanz, L.; Mora-Obando, D.; Sánchez, E.E.; Fry, B.G.; Gutiérrez, J.M.; Gibbs, H.L.; Sovice, M.G.; et al. Venomics of New World pit vipers: Genus-wide comparisons of venom proteomes across Agkistrodon. J. Proteome 2014, 96, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, N.G.; Cardoso, M.H.; Franco, O.L. Snake venoms: Attractive antimicrobial proteinaceous compounds for therapeutic purposes. Cell. Mol. Life Sci. 2013, 70, 4645–4658. [Google Scholar]
- Kamiguti, A.S.; Zuzel, M.; Theakston, R.D.G. Snake venom metalloproteinases and disintegrins: Interactions with cells. Braz. J. Med. Biol. Res. 1998, 31, 853–862. [Google Scholar] [CrossRef] [PubMed]
- Doley, R.; Zhou, X.; Kini, R.M. Snake venom phospholipase A2 enzymes. In Handbook of Venoms and Toxins of Reptiles; Mackessy, S.P., Ed.; CRC Press: Boca Raton, FL, USA, 2010; pp. 173–206. [Google Scholar]
- Phillips, D.J.; Swenson, S.D.; Markland, F.S., Jr. Thrombin-like snake venom serine proteinases. In Handbook of Venoms and Toxins of Reptiles; Mackessy, S.P., Ed.; CRC Press: Boca Raton, FL, USA, 2010; pp. 139–154. [Google Scholar]
- Heyborne, W.H.; Mackessy, S.P. Cysteine-rich secretory proteins in reptile venoms. In Handbook of Venoms and Toxins of Reptiles; Mackessy, S.P., Ed.; CRC Press: Boca Raton, FL, USA, 2010; pp. 325–336. [Google Scholar]
- Yamazaki, Y.; Morita, T. Structure and function of snake venom cysteine-rich secretory proteins. Toxicon 2004, 44, 227–231. [Google Scholar] [CrossRef] [PubMed]
- Komori, Y.; Nikai, T.; Tohkai, T.; Sugihara, H. Primary structure and biological activity of snake venom lectin (APL) from Agkistrodon. p. piscivorus (Eastern cottonmouth). Toxicon 1999, 37, 1053–1064. [Google Scholar] [CrossRef]
- Calvette, J.J. Snake venomics, antivenomics, and venom phenotyping: The ménage à trois of proteomic tools aimed at understanding the biodiversity of venoms. In Toxins and Hemostasis; Kini, R.M., Clemetson, K.J., Markland, F.S., McLane, M.A., Morita, T., Eds.; Springer: Dordrecht, The Netherlands; Heidelberg, Germany; London, UK; New York, NY, USA, 2010; pp. 45–72. [Google Scholar]
- Vejayan, J.; Shin Yee, L.; Ponnudurai, G.; Ambu, S.; Ibrahim, I. Protein profile analysis of Malaysian snake venoms by two-dimensional gel electrophoresis. J. Venom. Anim. Toxins Incl. Trop. Dis. 2010, 16, 623–630. [Google Scholar] [CrossRef]
- Menezes, M.C.; Furtado, M.F.; Travaglia-Cardoso, S.R.; Camargo, A.C.M.; Serrano, S.M.T. Sex-based individual variation of snake venom proteome among eighteen Bothrops jararaca siblings. Toxicon 2006, 47, 304–312. [Google Scholar] [CrossRef] [PubMed]
- Igci, N.; Demiralp, D.O. A preliminary investigation into the venom proteome of Macrovipera lebetina obtuse. (Dwigubsky, 1832) from Southeastern Anatolia by MALDI-TOF mass spectrometry and comparison of venom protein profiles with Macrovipera lebetina lebetina. (Linnaeus, 1758) from Cyprus by 2D-PAGE. Arch. Toxicol. 2012, 86, 441–451. [Google Scholar] [PubMed]
- Serrano, S.M.T.; Shannon, J.D.; Wang, D.; Camargo, A.C.M.; Fox, J.W. A multifaceted analysis of viperid snake venoms by two-dimensional gel electrophoresis: An approach to understanding venom proteomics. Proteomics 2005, 5, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Correa-Netto, C.; Teixeira-Araujo, R.; Aguiar, A.S.; Melgarejo, A.R.; De-Simone, S.G.; Soares, M.R.; Foguel, D.; Zingali, R.B. Immunome and venome of Bothrops jararacussu: A proteomic approach to study the molecular immunology of snake toxins. Toxicon 2010, 55, 1222–1235. [Google Scholar] [CrossRef] [PubMed]
- Calvete, J.J.; Juarez, P.; Sanz, L. Snake venomics. Strategy and applications. J. Mass Spectrom. 2007, 42, 1405–1414. [Google Scholar] [CrossRef] [PubMed]
- Vejayan, J.; Tang, M.S.; Halijah, I. The role of conventional two-dimensional electrophoresis (2DE) and its newer applications in the study of snake venoms. In Proteomic Applications in Biology; Heazlewood, J.L., Petzold, C.J., Eds.; IN TECH: Rijeka, Croatia, 2012; pp. 226–252. [Google Scholar]
- Bocian, A.; Urbanik, M.; Hus, K.; Łyskowski, A.; Petrilla, V.; Andrejčáková, Z.; Petrillová, M.; Legath, J. Proteome and peptidome of Vipera berus berus. Venom Mol. 2016, 21, 1398. [Google Scholar] [CrossRef] [PubMed]
- Walsh, G.; Jefferis, R. Post-translational modifications in the context of therapeutic proteins. Nat. Biotechnol. 2006, 24, 1241–1252. [Google Scholar] [CrossRef] [PubMed]
- Calvete, J.J. Antivenomics and venom phenotyping: A marriage of convenience to address the performance and range of clinical use of antivenoms. Toxicon 2010, 56, 1284–1291. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.H.; Tan, K.Y.; Fung, S.Y.; Tan, N.H. Venom-gland transcriptome and venomproteome of the Malaysian king cobra (Ophiophagus hannah). BMC Genom. 2015, 16, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Trummal, K.; Samel, M.; Aaspõllu, A.; Tõnismägi, K.; Titma, T.; Subbi, J.; Siigur, J.; Siigur, E. 5′-Nucleotidase from Vipera lebetina venom. Toxicon 2015, 93, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Dhananjaya, B.L.; Nataraju, A.; Rajesh, R.; Raghavendra Gowda, C.D.; Sharath, B.K.; Vishwanath, B.S.; D’Souza, C.J.M. Anticoagulant effect of Naja naja venom 5′-Nucleotidase: Demonstration through the use of novel specific inhibitor, vanillic acid. Toxicon 2006, 48, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Rael, E.D. Venom phosphatases and 5′-Nucleotidase. In Enzymes from Snake Venom; Bailey, G.S., Ed.; Alaken Inc.: Fort Collins, CO, USA, 1998; pp. 405–423. [Google Scholar]
- Da Silva, N.J., Jr.; Aird, S.D. Prey specificity, comparative lethality and compositional differences of coral snake venoms. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2001, 128, 425–456. [Google Scholar] [CrossRef]
- Aird, S.D. Ophidian envenomation strategies and the role of purines. Toxicon 2002, 40, 335–393. [Google Scholar] [CrossRef]
- Dhananjaya, B.L.; D’Souza, C.J.M. The pharmacological role of nucleotidases in snake venoms. Cell Biochem. Funct. 2010, 28, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Kini, R.M. Excitement ahead: Structure, function and mechanism of snake venom phospholipase A2 enzymes. Toxicon 2003, 42, 827–840. [Google Scholar] [CrossRef] [PubMed]
- Atanasov, V.N.; Danchev, D.; Mitewa, M.; Petrova, S. Hemolytic and anticoagulant study of the neurotoxin vipoxin and its components-basic phospholipase A2 and an acidic inhibitor. Biochemistry 2009, 74, 276–280. [Google Scholar] [CrossRef] [PubMed]
- Teng, C.M.; Chen, Y.H.; Ouyang, C. Biphasic effect on platelet aggregation by phospholipase a purified from Vipera russellii snake venom. Biochim. Biophys. Acta 1984, 772, 393–402. [Google Scholar] [CrossRef]
- Kovalchuk, S.I.; Ziganshin, R.H.; Starkov, V.G.; Tsetlin, V.I.; Utkin, Y.N. Quantitative proteomic analysis of venoms from Russian vipers of Pelias group: Phospholipases A2 are the main venom components. Toxins 2016, 8, 105. [Google Scholar] [CrossRef] [PubMed]
- Moura-da-Silva, A.M.; Butera, D.; Tanjoni, I. Importance of snake venom metalloproteinases in cell biology: Effects on platelets, inflammatory and endothelial cells. Curr. Pharm. Des. 2007, 13, 2893–2905. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.W.; Serrano, S.M. Insights into and speculations about snake venom metalloproteinase (SVMP) synthesis, folding and disulfide bond formation and their contribution to venom complexity. FEBS J. 2008, 275, 3016–3030. [Google Scholar] [CrossRef] [PubMed]
- Sajevic, T.; Leonardi, A.; Križaj, I. Haemostatically active proteins in snake venoms. Toxicon 2011, 57, 627–645. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; di Cera, E. Serine peptidases: Classification, structure and function. Cell. Mol. Life Sci. 2008, 65, 1220–1236. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, V.G.; Redford, D.T.; Boyle, P.K. Effect of iron and carbon monoxide on fibrinogenase-like degradation of plasmatic coagulation by venoms of Six Agkistrodon Species. Basic Clin. Pharmacol. Toxicol. 2016, 118, 390–395. [Google Scholar] [CrossRef] [PubMed]
- Castro, H.C.; Zingali, R.B.; Albuquerque, M.G.; Pujol-Luz, M.; Rodrigues, C.R. Snake venom thrombin-like enzymes: From reptilase to now. Cell. Mol. Life Sci. 2004, 61, 843–856. [Google Scholar] [CrossRef] [PubMed]
- You, W.K.; Choi, W.S.; Koh, Y.S.; Shin, H.C.; Jang, Y.; Chung, K.H. Functional characterization of recombinant batroxobin, a snake venom thrombin-like enzyme, expressed from Pichia pastoris. FEBS Lett. 2004, 571, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Kini, R.M. Anticoagulant proteins from snake venoms: Structure, function and mechanism. Biochem. J. 2006, 397, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Serrano, S.M.T.; Maroun, R.C. Snake venom serine proteinases: Sequence homology vs substrate specificity, a paradox to be solved. Toxicon 2005, 45, 1115–1132. [Google Scholar] [CrossRef] [PubMed]
- Du, X.Y.; Clemetson, K.J. Snake venom L-amino acid oxidases. Toxicon 2002, 40, 659–665. [Google Scholar] [CrossRef]
- Torii, S.; Naito, M.; Tsuruo, T. Apoxin I, a novel apoptosis-inducing factor with L-amino acid oxidase activity purified from Western diamondback rattlesnake venom. J. Biol. Chem. 1997, 272, 9539–9542. [Google Scholar] [PubMed]
- Ramazanova, A.S.; Starkov, V.G.; Osipov, A.V.; Ziganshin, R.H.; Filkin, S.Y.; Tsetlin, V.I.; Utkin, Y.N. Cysteine-rich venom proteins from the snakes of Viperinae subfamily—Molecular cloning and phylogenetic relationship. Toxicon 2009, 53, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Ozeki, Y.; Matsui, T.; Hamako, J.; Suzuki, M.; Fujimura, Y.; Yoshida, E.; Nishida, S.; Titani, K. C-type galactoside-binding lectin from Bothrops jararaca venom: Comparison of its structure and function with those of botrocetin. Arch. Biochem. Biophys. 1994, 308, 306–310. [Google Scholar] [CrossRef] [PubMed]
- Ogilvie, M.L.; Byl, J.W.; Gartner, T.K. Platelet-aggregation is stimulated by lactose-inhibitable snake venom lectins. Thromb. Haemost. 1989, 62, 704–707. [Google Scholar] [PubMed]
- Munawar, A.; Zahid, A.; Negm, A.; Akrem, A.; Spencer, P.; Betzel, C. Isolation and characterization of Bradykinin potentiating peptides from Agkistrodon bilineatus venom. Proteome Sci. 2016, 14, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Munawar, A.; Trusch, M.; Georgieva, D.; Spencer, P.; Frochaux, V.; Harder, S.; Arni, R.K.; Duhalov, D.; Genov, N.; Schlüter, H.; et al. Venom peptide analysis of Vipera ammodytes meridionalis (Viperinae) and Bothrops jararacussu (Crotalinae) demonstrates subfamily-specificity of the peptidome in the family Viperidae. Mol. Biosyst. 2011, 7, 3298–3307. [Google Scholar] [CrossRef] [PubMed]
- Calvete, J.J.; Fasoli, E.; Sanz, L.; Boschetti, E.; Righetti, P.G. Exploring the venom proteome of the western diamondback rattlesnake, Crotalus atrox, via snake venomics and combinatorial peptide ligand library approaches. J. Proteome Res. 2009, 8, 3055–3067. [Google Scholar] [CrossRef] [PubMed]
- Graham, R.L.; Graham, C.; McClean, S.; Chen, T.; O’Rourke, M.; Hirst, D.; Theakston, D.; Shaw, C. Identification and functional analysis of a novel bradykinin inhibitory peptide in the venoms of New World Crotalinae pit vipers. Biochem. Biophys. Res. Commun. 2005, 338, 1587–1592. [Google Scholar] [CrossRef] [PubMed]
- Całkosiński, I.; Seweryn, E.; Zasadowski, A.; Małolepsza-Jarmołowska, K.; Dzierzba, K.; Bronowicka-Szydełko, A.; Mierzchała, M.; Ceremuga, I.; Rosińczuk-Tonderys, J.; Dobrzyński, M.; et al. The composition, biochemical properties and toxicity of snake venoms. PHMD 2010, 64, 262–272. [Google Scholar] [PubMed]
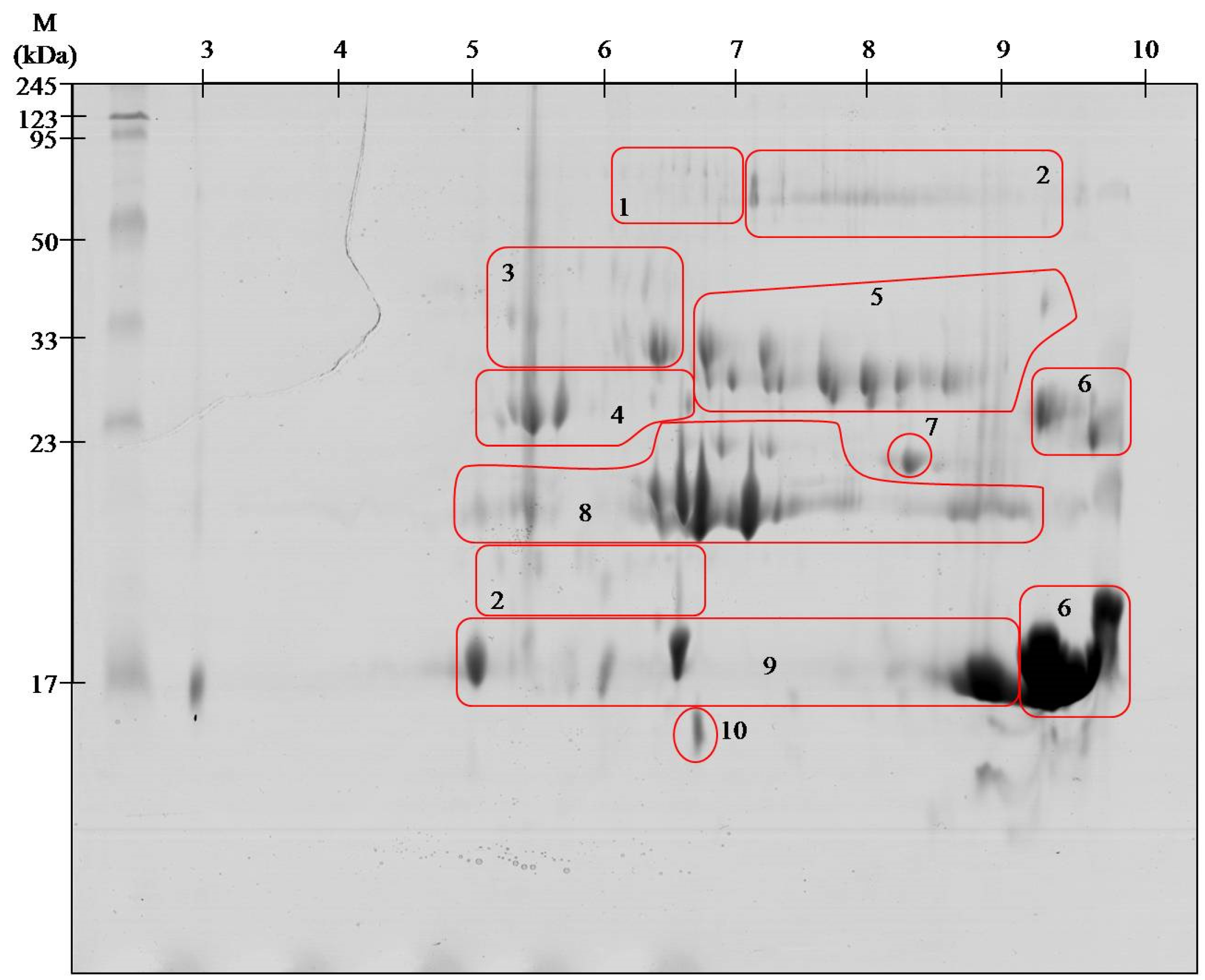
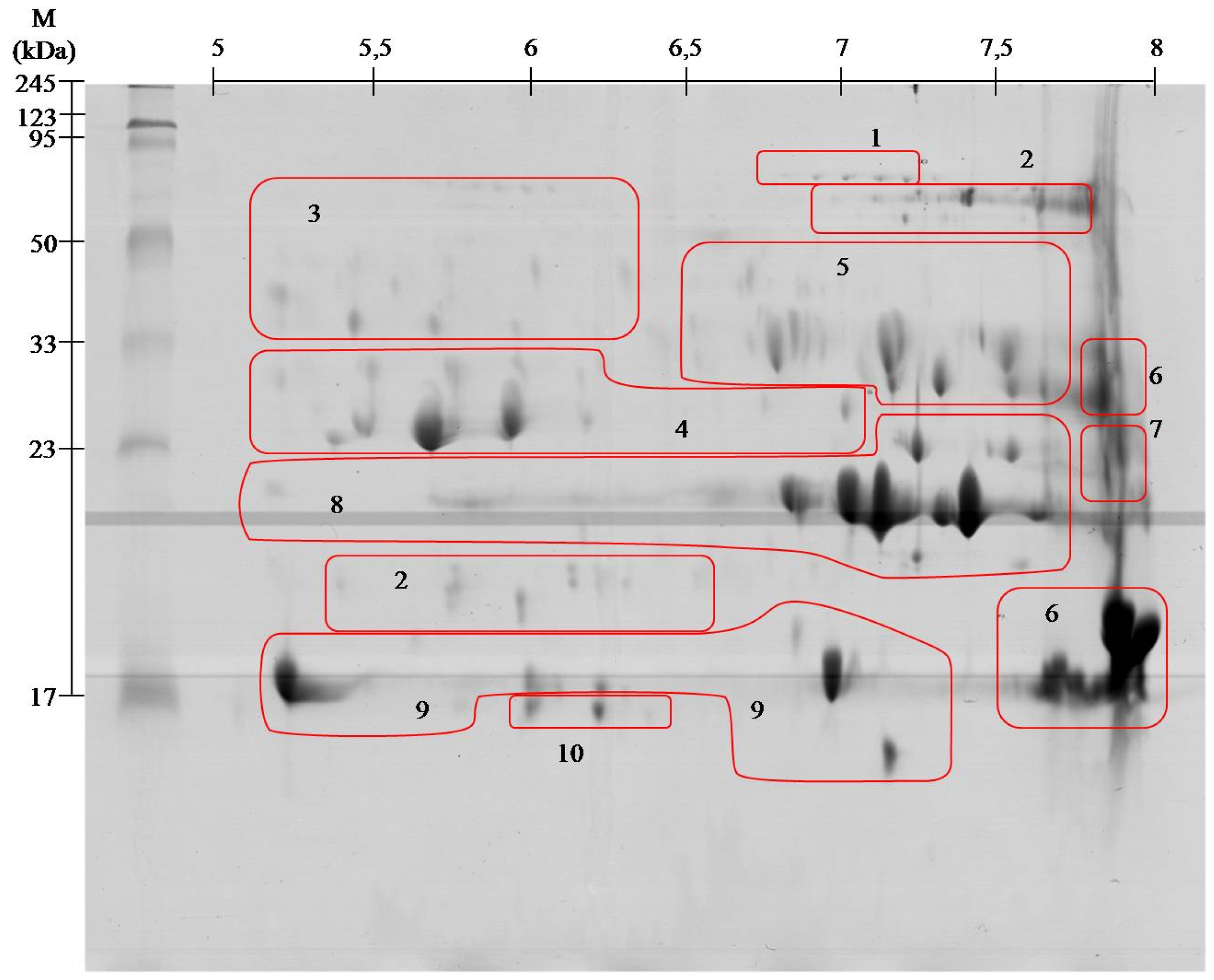
| Spot no. # | Identified Protein & | Accession * | Organism ¥ | Mass [kDa] £ | S ± | Peptide Sequence ¤ |
|---|---|---|---|---|---|---|
| 1 | Snake venom 5′-nucleotidase | V5NTD_CROAD | Crotalus adamanteus | 65.2 | 97 | ETPVLSNPEGPYLEFR (1719.099) |
| V5NTD_CROAD | Crotalus adamanteus | 65.2 | 39 | QAFEHSVHR (1110.651) | ||
| V5NTD_GLOBB | Gloydius blomhoffii blomhoffii | 6 | 115 | SFELTILHTNDVMAR (1753.091) | ||
| V5NTD_GLOBR | Gloydius brevicaudus | 65 | 65 | PMF SC 13.3% | ||
| 2 | l-amino-acid oxidase | OXLA_GLOHA | Gloydius halys | 57.4 | 116 | ETDYEEFLEIAR (1514.762) |
| OXLA_PSEAU | Pseudechis austarlis | 59 | 54 | PMS SC 10.4% | ||
| 3 | Beta-fibrinogenase brevinase | VSPB_GLOBL | Gloydius blomhoffii | 26.3 | 65 | VIGGDECNINEHR (1512.767) |
| Beta-fibrinogenase | VSPBF_MACLB | Macrovipera lebetina | 28.2 | 26 | FFCLSSK (888.452) | |
| 4 | Thrombin-like enzyme bilineobin | VSP2_AGKBI | Agkistrodon bilineatus | 27.1 | 64 | IIGGDECNINEHR (1526.785) |
| 28 | NSEHIAPLSLPSSPPIVGSVCR (2317.179) | |||||
| Thrombin-like enzyme asperase | VSPL_BOTAS | Bothrops asper | 28.6 | 118 | ETYPDVPHCANINILDHAVCR (2494.238) | |
| Snake venom serine protease PA | VSPP_TRIST | Trimeresurus stajnegri | 28.6 | 40 | VVLNEDEQIR (1115.567) | |
| Snake venom serine proteinase pallabin | VSP1_GLOHA | Gloydius halys | 29.3 | 27 | LDSPVKNSAHIAPLSLPSSPPVGSDCR (2888.600) | |
| Snake venom serine proteinase 9 | VSP9_CROAD | Crotalus adamanteus | 11.7 | 164 | ETYPDVPHCANINILDYEVCR (2578.230) | |
| Snake venom serine proteinase 12 | VSPC_CROAD | Crotalus adamanteus | 29.3 | 113 | DIMLIRLDSPVSNSEHIAPLSLPSSPPSVGSVCR (1823.985) | |
| Thrombin-like enzyme crotalase | VSPCR_CROAD | Crotalus adamanteus | 30.1 | 36 | WDKDIMLIR(1189.656) | |
| 5 | Protein C activator | VSPCA_AGKCO | Agkistrodon contortrix contortrix | 25.7 | 82 | PMF SC 34,2% |
| 91 | NSAHIAPLSLPSNPPSVGSVCR (2260.075) | |||||
| VSPCA_AGKBI | Agkistrodon bilineatus | 2.2 | 68 | VVGGDECNINEHR (1498.711) | ||
| 6 | Basic phospholipase A2 homolog | PA2HB_AGKPI | Agkistrodon piscivorus piscivorus | 14.7 | 80 | PMF SC 49% |
| Basic phospholipase A2 homolog MT1 | PA2H1_AGKCL | Agkistrodon contrortrix laticinctus | 16.5 | 80 | PMF SC 43% | |
| 7 | Cysteine-rich venom protein piscivorin | CRVP_AGKPI | Agkistrodon piscivorus piscivorus | 27.5 | 102 | MEHYPEAAANAER (1537.689) |
| 76 | MEHYPEAAANAER (1553.669) | |||||
| 8 | Snake venom metalloproteinase ACLF | VM1A_AGKCL | Agkistrodon piscivorus loucostroma | 47.1 | 67 | YVELVIIADHR (1327.857) |
| 29 | SHDNAQLLATAIVFDGIIGR (2169.126) | |||||
| VM1A_AGKCL | Agkistrodon contortrix latiematus | 26.7 | 77 | YVELVIVADHR (1313.725 ) | ||
| Zinc metalloproteinase disintegrin | VM2AB_AGKCO | Agkistordon contortrix contortrix | 55.1 | 185 | ISHDNAQLLTAIELDGETIGLANR (2564.272 ) | |
| 41 | YIELVVVADHR (1313, 713) | |||||
| Snake venom metalloproteinase VMP1 | VM1V1_AGKPL | Agkistrodon piscivorus leucostoma | 47.1 | 136 | SHDNAQLLTAIVFDEGIIGR (2169.058) | |
| 37 | APLAGMCDPNR (1201.591) | |||||
| 43 | YVELVIVADHR (1327.784) | |||||
| Zinc metalloproteinase disintegrin-like HR1a | VM3HA_PROFL | Protobothrops flavoviridis | 70.9 | 59 | TWVYEIVNTLNEIYR (1912.993) | |
| Snake venom metalloproteinase fibrolase | VM1F_AGKCO | Akgistrodon contortrix contortrix | 23.2 | 42 | YVQLVIVADHR (1312.613) | |
| 9 | Acidic phospholipase A2 BpirPLA2-I | PA2A1_BOTPI | Bothrops pirajai | 14.4 | 48 | CCFVMDCCYGK (1505.585) |
| 49 | QICECDR (980.433) | |||||
| Acidic phospholipase A2 S1E6-b | PA2AB_CALRH | Calloselasma rhodostoma | 14.3 | 56 | PMF SC 30.2% | |
| Acidic phospholipase A2 | PA2A_GLOHA | Gloydius halys | 14.7 | 57 | PMF SC 25.8 % | |
| Acidic phospholipase A2 1 | PA2A1_PROFL | Protobothrops flavoviridis | 15.5 | 30 | AAAICFR (808.334) | |
| 10 | C-type lectin APL | LECG_AGKPI | Agkistrodon piscivorus piscivorus | 16.7 | 103 | PMF SC 51.1% |
| 54 | DFSWEWTDR (1241.611) | |||||
| 105 | EFCVELVSLTGYR (1572.785) | |||||
| 101 | GQAEVWIGLWDK (1401.639) | |||||
| C- type lectin PAL | LECG_BITAR | Bitis arietans | 16.6 | 56 | PMF SC 58.5% |
| Parent ion m/z | Identified Protein & | Accession * | Organism ¥ | Peptide Sequence ≠ | Mass [Da] £ | S ± |
|---|---|---|---|---|---|---|
| 1063.5343 | Bradykinin inhibitor peptide | BKIP_AGKBI | Agkistrodon bilineatus | TPPAGPDVGPR | 1063 | 9 |
| 1214.6465 | Bradykinin-potentiating peptide POL-236 | BPP36_CROAT | Crotalus atrox | QLWPRPQIPP- + Gln- > pyro-Glu (N-term Q) | 1231 | 59 |
| 1230.6420 | Bradykinin potentiating peptide E | gi|229310 | Gloydius blomhoffii | EKWDPPPVSPP- + Glu- > pyro-Glu (N-term E) | 1248 | 13 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bocian, A.; Urbanik, M.; Hus, K.; Łyskowski, A.; Petrilla, V.; Andrejčáková, Z.; Petrillová, M.; Legáth, J. Proteomic Analyses of Agkistrodon contortrix contortrix Venom Using 2D Electrophoresis and MS Techniques. Toxins 2016, 8, 372. https://doi.org/10.3390/toxins8120372
Bocian A, Urbanik M, Hus K, Łyskowski A, Petrilla V, Andrejčáková Z, Petrillová M, Legáth J. Proteomic Analyses of Agkistrodon contortrix contortrix Venom Using 2D Electrophoresis and MS Techniques. Toxins. 2016; 8(12):372. https://doi.org/10.3390/toxins8120372
Chicago/Turabian StyleBocian, Aleksandra, Małgorzata Urbanik, Konrad Hus, Andrzej Łyskowski, Vladimír Petrilla, Zuzana Andrejčáková, Monika Petrillová, and Jaroslav Legáth. 2016. "Proteomic Analyses of Agkistrodon contortrix contortrix Venom Using 2D Electrophoresis and MS Techniques" Toxins 8, no. 12: 372. https://doi.org/10.3390/toxins8120372







