Transcriptome Analysis to Understand the Toxicity of Latrodectus tredecimguttatus Eggs
Abstract
:1. Introduction
2. Results
2.1. Illumina Sequencing and Read Assembly
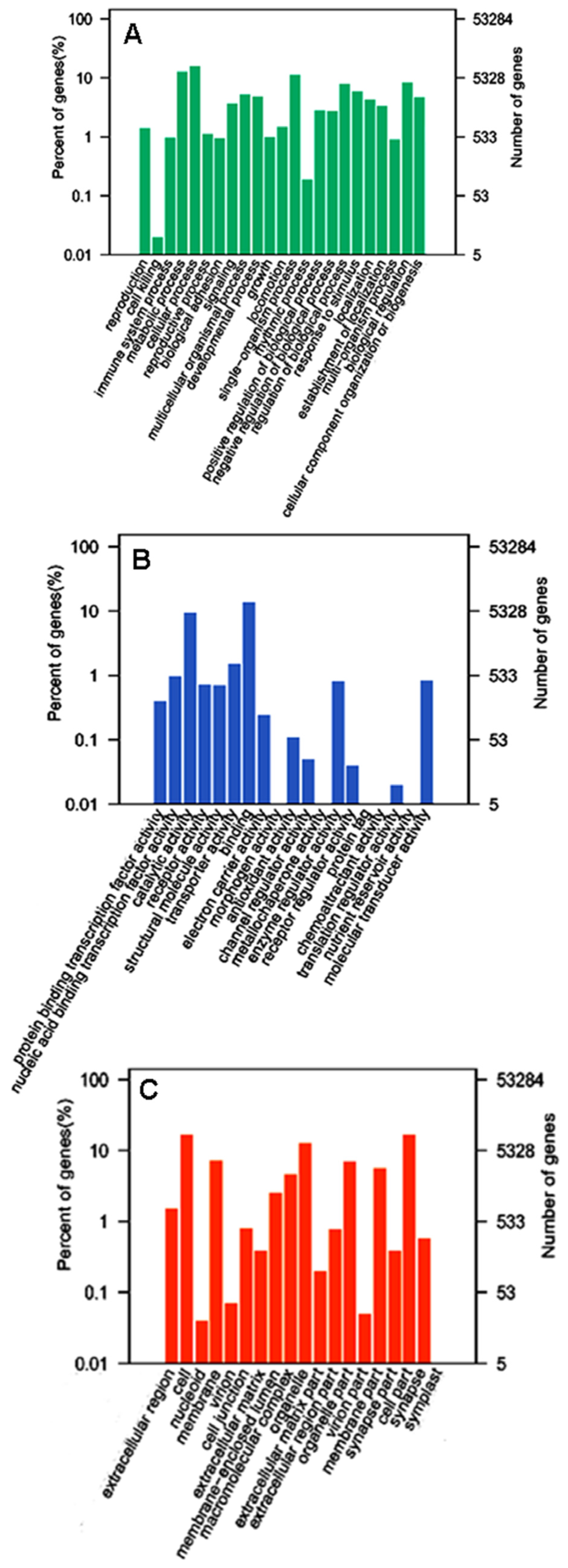
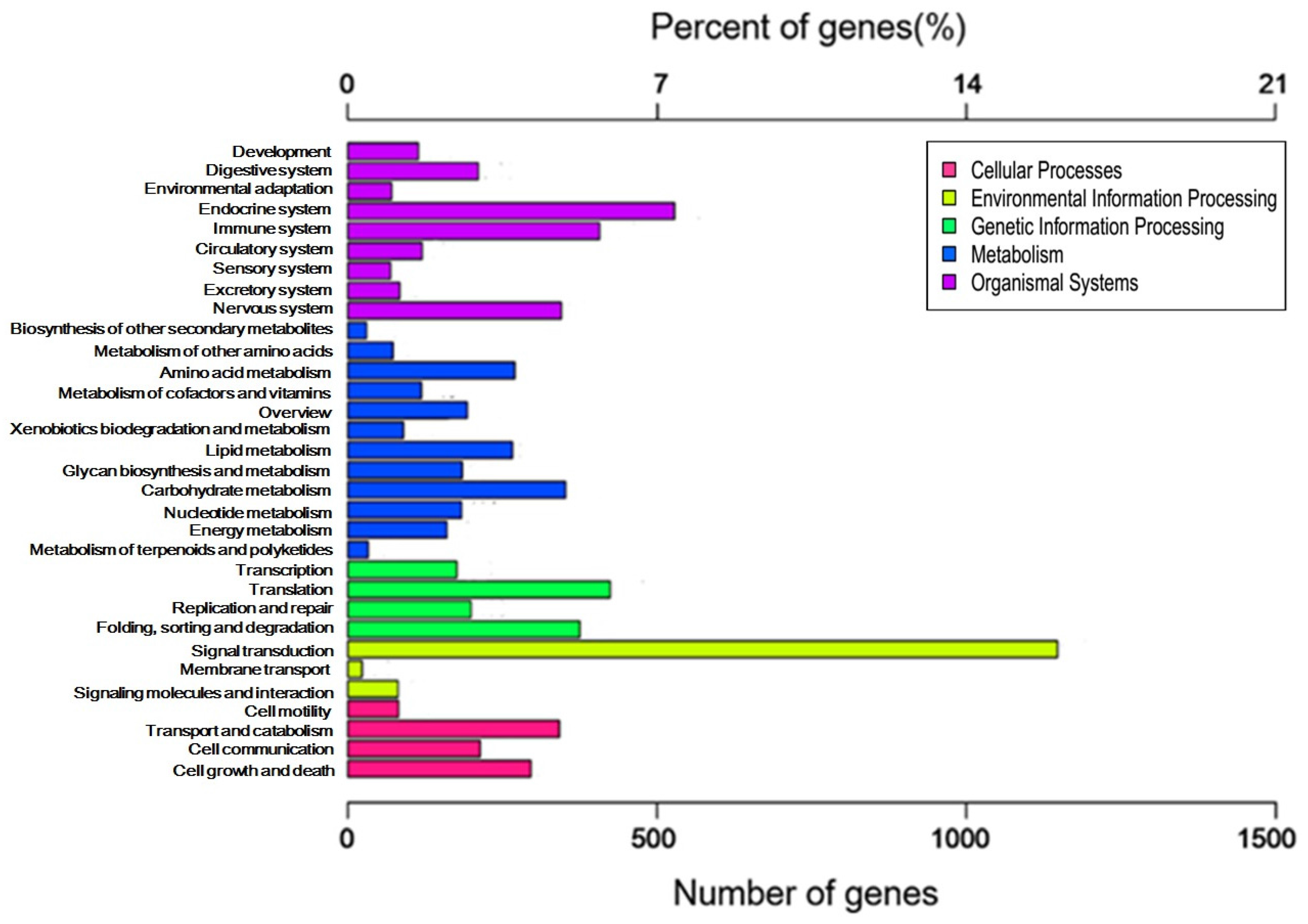
2.2. Categories and Annotations of Unigenes
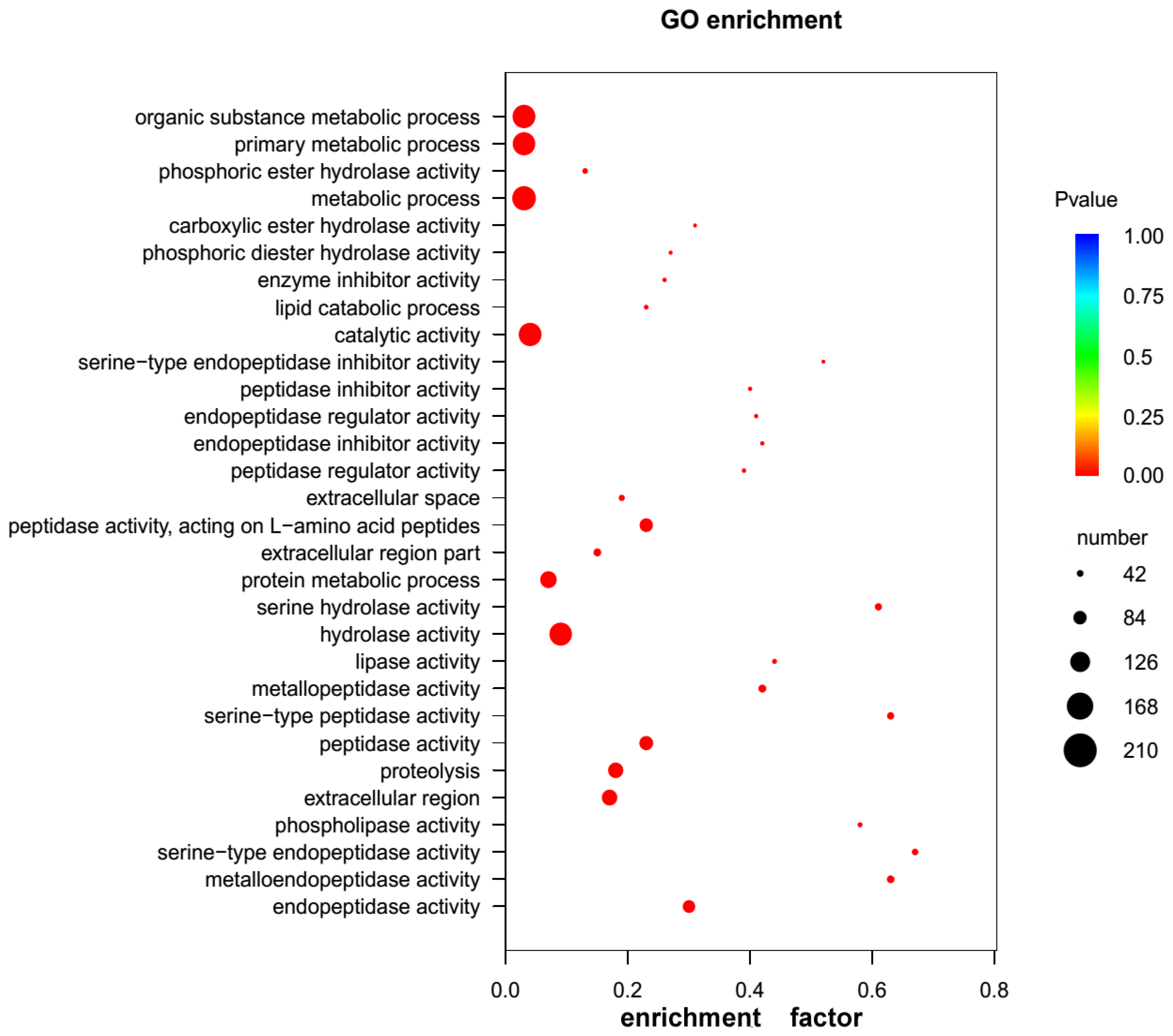
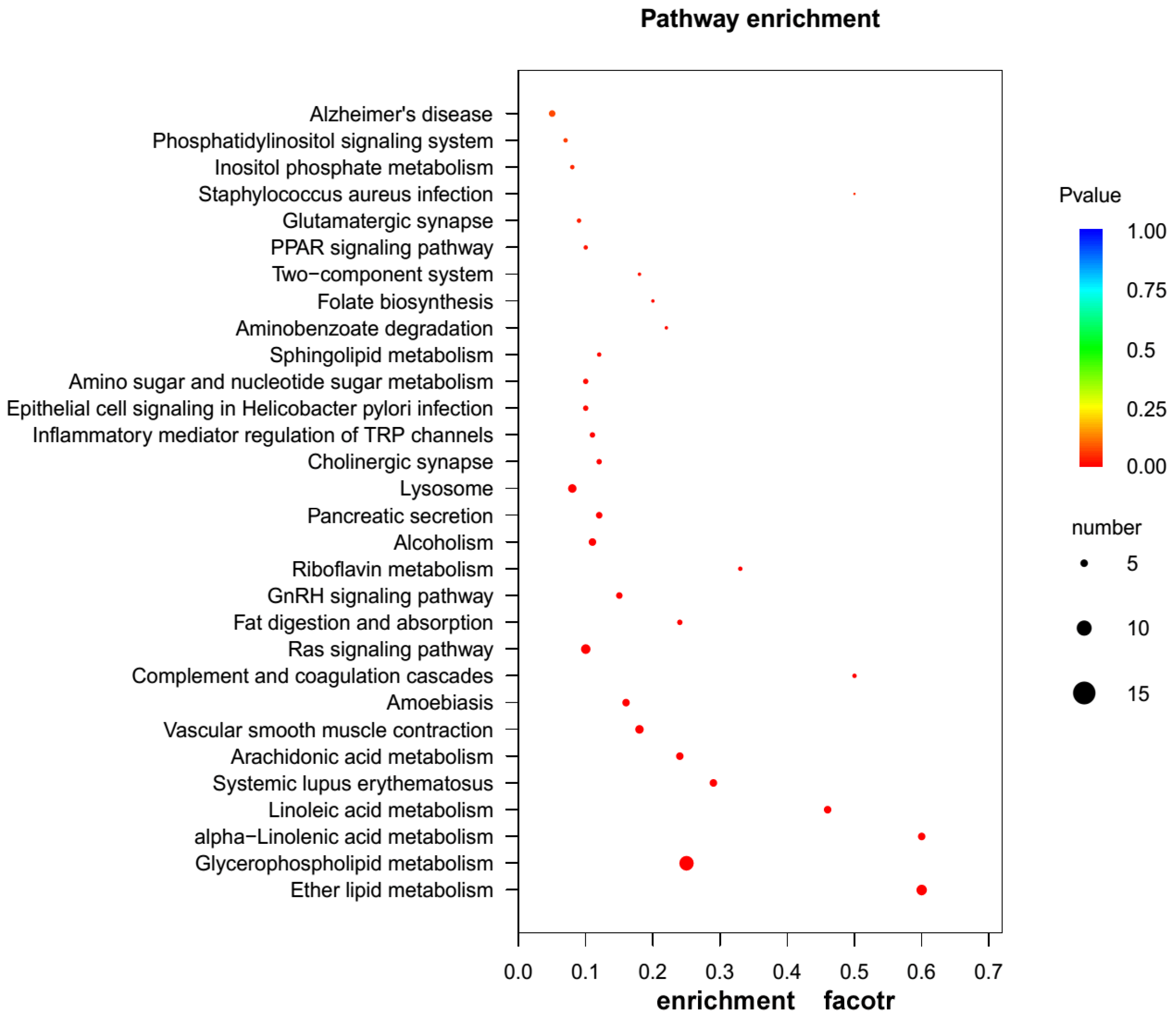
2.3. GO and KEGG Pathway Enrichment Analyses of Unigenes
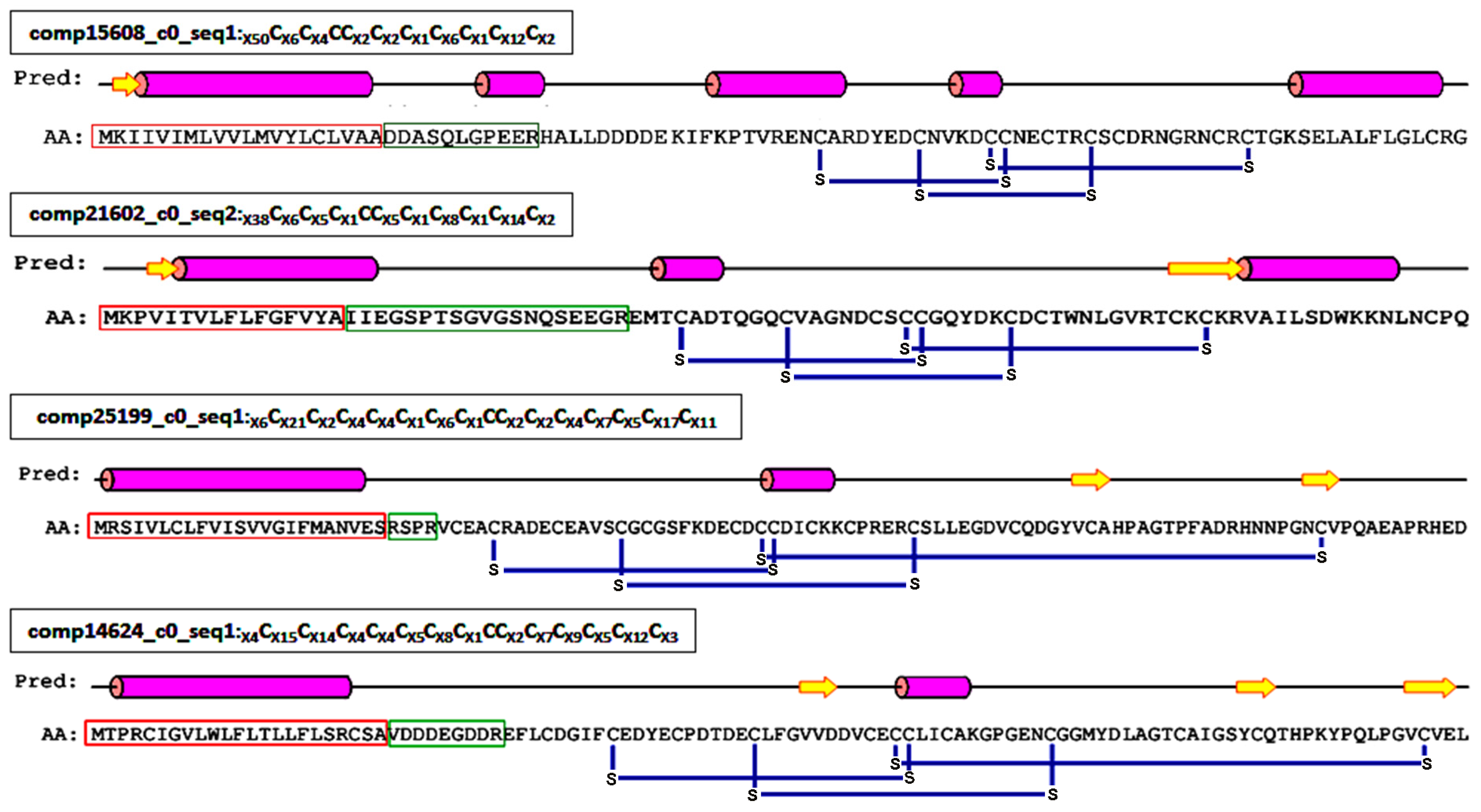
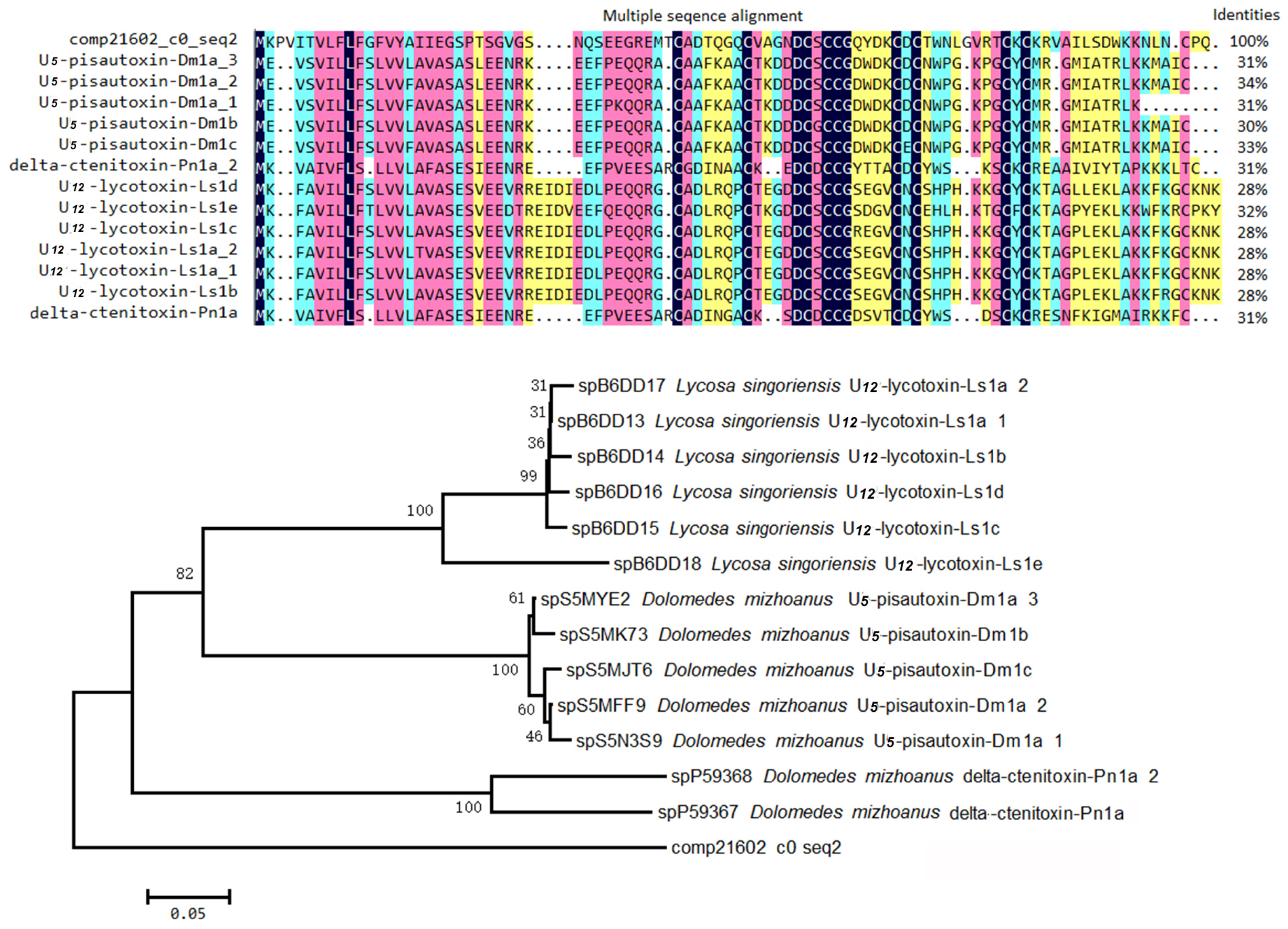
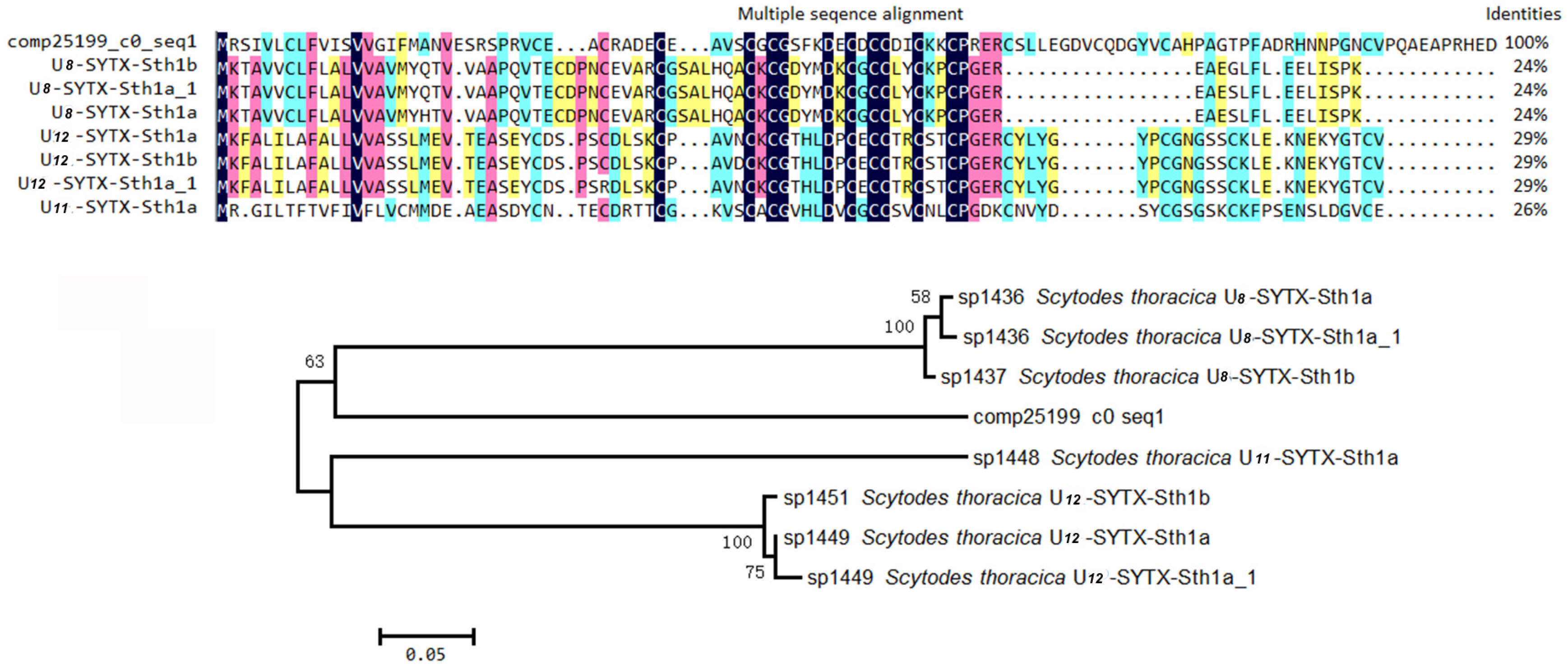
2.4. Proteinaceous Toxins in the Eggs
2.4.1. ICK Motif Peptide Toxins
2.4.2. Non-ICK Motif Peptide Toxins
2.4.3. Antimicrobial Peptides and Precursors
2.4.4. Toxin-Like Proteins or Enzymes
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. cDNA Library Construction, Sequencing and De Novo Assembly
5.2. Annotation and Identification of Proteinaceous Toxins
5.3. Other Bioinformatics Analyses
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- World Spider Catalog (2016). World Spider Catalog. Natural History Museum Bern, Version 17.0. Available online: http://wsc.nmbe.ch (accessed on 19 December 2016).
- Ushkaryov, Y.A.; Volynski, K.E.; Ashton, A.C. The multiple actions of black widow spider toxins and their selective use in neurosecretion studies. Toxicon 2004, 43, 527–542. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.S. The Spiders of China Arachnida Araneae Theridiidae; Science Press: Beijing, China, 1998; pp. 293–294. (In Chinese) [Google Scholar]
- Camp, N.E. Black widow spider envenomation. J. Emerg. Nurs. 2014, 40, 193–194. [Google Scholar] [CrossRef] [PubMed]
- Vassilevski, A.A.; Kozlov, S.A.; Grishin, E.V. Molecular Diversity of Spider venom. Biochemistry 2009, 74, 1505–1534. [Google Scholar] [CrossRef] [PubMed]
- Windley, M.J.; Herzig, V.; Dziemborowicz, S.A.; Hardy, M.C.; King, G.F.; Nicholson, G.M. Spider-Venom Peptides as Bioinsecticides. Toxins (Basel) 2012, 4, 192–227. [Google Scholar] [CrossRef] [PubMed]
- Klint, J.K.; Senff, S.; Rupasinghem, D.B.; Er, S.Y.; Herzig, V.; Nicholson, G.M.; King, G.F. Spider-venom peptides that target voltage-gated sodium channels: pharmacological tools and potential therapeutic leads. Toxicon 2012, 60, 478–491. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Duan, Z.; Yu, Y.; Liu, Z.; Liu, Z.; Liang, S. The venom gland transcriptome of Latrodectus tredecimguttatus revealed by deep sequencing and cDNA library analysis. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.C.; Duan, Z.G.; Yang, J.; Yan, X.J.; Zhou, H.; He, X.Z.; Liang, S.P. Physiological and biochemical analysis of L. tredecimguttatus venom collected by electrical stimulation. J. Physiol. Biochem. 2007, 63, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Duan, Z.G.; Yan, X.J.; He, X.Z.; Zhou, H.; Chen, P.; Cao, R.; Xiong, J.X.; Hu, W.J.; Wang, X.C.; Liang, S.P. Extraction and protein component analysis of venom from the dissected venom glands of Latrodectus tredecimguttatus. Comp. Biochem. Physiol. Part B 2006, 145, 350–357. [Google Scholar] [CrossRef] [PubMed]
- Duan, Z.G.; Yang, J.; Yan, X.J.; Zeng, X.Z.; Wang, X.C.; Liang, S.P. Venom properties of the spider Latrodectus tredecimguttatus and comparison of two venom-collecting methods. Zool. Res. 2009, 30, 381–388. (In Chinese) [Google Scholar] [CrossRef]
- Buffkin, D.C.; Russell, F.E.; Deshmukh, A. Preliminary study on the toxicity of black widow spider eggs. Toxicon 1971, 9, 393–402. [Google Scholar] [CrossRef]
- Russell, F.E.; Maretić, Z. Effects of Latrodectus egg poison on web building. Toxicon 1979, 17, 649–650. [Google Scholar] [CrossRef]
- Yan, Y.; Li, J.; Zhang, Y.; Peng, X.; Guo, T.; Wang, J.; Hu, W.; Duan, Z.; Wang, X. Physiological and biochemical characterization of egg extract of black widow spiders to uncover molecular basis of egg toxicity. Biol. Res. 2014, 47. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, H.; Duan, Z.; Cao, R.; Wang, X.; Liang, S. Protein compositional analysis of the eggs of black widow spider (Latrodectus tredecimguttatus): Implications for the understanding of egg toxicity. J. Biochem. Mol. Toxicol. 2012, 26, 510–515. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yan, Y.; Wang, J.; Guo, T.; Hu, W.; Duan, Z.; Wang, X.; Liang, S. Purification and partial characterization of a novel neurotoxic protein from eggs of black widow spiders (Latrodectus tredecimguttatus). J. Biochem. Mol. Toxicol. 2013, 27, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yan, Y.; Yu, H.; Peng, X.; Zhang, Y.; Hu, W.; Duan, Z.; Wang, X.; Liang, S. Isolation and identification of a sodium channel-inhibiting protein from eggs of black widow spiders. Int. J. Boil. Macromol. 2014, 65, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Lei, Q.; Yu, H.; Peng, X.; Yan, S.; Wang, J.; Yan, Y.; Wang, X. Isolation and Preliminary Characterization of Proteinaceous Toxins with Insecticidal and Antibacterial Activities from Black Widow Spider (L. tredecimguttatus) Eggs. Toxins (Basel) 2015, 7, 886–899. [Google Scholar] [CrossRef] [PubMed]
- Mutz, K.O.; Heilkenbrinker, A.; Lönne, M.; Walter, J.G.; Stahl, F. Transcriptome analysis using next-generation sequencing. Curr. Opin. Biotechnol. 2013, 24, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Jungo, F.; Bougueleret, L.; Xenarios, I.; Poux, S. The UniProtKB/Swiss-Prot Tox-Prot program: A central hub of integrated venom protein data. Toxicon 2012, 60, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Jungo, F.; Estreicher, A.; Bairoch, A.; Bougueleret, L.; Xenarios, I. Animal Toxins: How is Complexity Represented in Databases? Toxins (Basel) 2010, 2, 262–282. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.A.; Wang, Z. Next-generation transcriptome assembly. Nat. Rev. Genet. 2011, 12, 671–682. [Google Scholar] [CrossRef] [PubMed]
- Pineda, S.S.; Undheim, E.A.; Rupasinghe, D.B.; Ikonomopoulou, M.P.; King, G.F. Spider venomics: implications for drug discovery. Future Med. Chem. 2014, 6, 1699–1714. [Google Scholar] [CrossRef] [PubMed]
- Daly, N.L.; Craik, D.J. Bioactive cystine knot proteins. Curr. Opin. Chem. Biol. 2011, 15, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Haney, R.A.; Ayoub, N.A.; Clarke, T.H.; Hayashi, C.Y.; Garb, J.E. Dramatic expansion of the black widow toxin arsenal uncovered by multi-tissue transcriptomics and venom proteomics. BMC Genom. 2014, 15, 366–383. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, S.; Malyavka, A.; Mccutchen, B.A.; Lu, A.; Schepers, E.; Herrmann, R.; Grishin, E. A novel strategy for the identification of toxinlike structures in spider venom. Proteins 2005, 59, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, S.; Grishin, E. Classification of spider neurotoxins using structural motifs by primary structure features. Single residue distribution analysis and pattern analysis techniques. Toxicon 2005, 46, 672–686. [Google Scholar] [CrossRef] [PubMed]
- King, G.F.; Hardy, M.C. Spider-venom peptides: Structure, pharmacology, and potential for control of insect pests: Annu. Rev. Entomol. 2013, 58, 475–496. [Google Scholar] [CrossRef] [PubMed]
- Zobel-Thropp, P.A.; Correa, S.M.; Garb, J.E.; Binford, G.J. Spit and venom from scytodes spiders: A diverse and distinct cocktail. J. Proteome Res. 2014, 13, 817–835. [Google Scholar] [CrossRef] [PubMed]
- Vray, B.; Hartmann, S.; Hoebeke, J. Immunomodulatory properties of cystatins. Cell. Mol. Life Sci. 2002, 59, 1503–1512. [Google Scholar] [CrossRef] [PubMed]
- Turk, V.; Stoka, V.; Turk, D. Cystatins: Biochemical and structural properties, and medical relevance. Front. Biosci. 2008, 13, 5406–5420. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, A.; Valdés, J.J.; Kotsyfakis, M. The role of cystatins in tick physiology and blood feeding. Ticks Tick Borne Dis. 2012, 3, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Richards, R.; St Pierre, L.; Trabi, M.; Johnson, L.A.; de Jersey, J.; Masci, P.P.; Lavin, M.F. Cloning and characterisation of novel cystatins from elapid snake venom glands. Biochimie 2011, 93, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Zavasnik-Bergant, T. Cystatin protease inhibitors and immune functions. Front. Biosci. 2008, 13, 4625–4637. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Zhang, J.; Feng, W.; Bao, N.; Song, D.; Zhu, B.C. Pharmacological characterization of spider antimicrobial peptides. Protein Pept. Lett. 2005, 12, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Saez, N.J.; Senff, S.; Jensen, J.E.; Er, S.Y.; Herzig, V.; Rash, L.D.; King, G.F. Spider-venom peptides as therapeutics. Toxins (Basel) 2010, 2, 2851–2871. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, X.; Wang, Z. APD2: the updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 2009, 37, D933–D937. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Chen, Z.W. Isolation, characterization and anti-cancer activity of SK84, a novel glycine-rich antimicrobial peptide from Drosophila virilis. Peptides 2010, 31, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.K.; Lee, M.J.; Go, H.J.; Kim, G.D.; Jeong, H.D.; Nam, B.H.; Park, N.G. Purification and antimicrobial function of ubiquitin isolated from the gill of Pacific oyster, Crassostrea gigas. Mol. Immunol. 2013, 53, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.B.; Shan, B.; Bai, H.M.; Tang, J.; Yan, L.Z.; Ma, Y.B. Hydrophilic/hydrophobic characters of antimicrobial peptides derived from animals and their effects on multidrug resistant clinical isolates. Zool. Res. 2015, 36, 41–47. [Google Scholar] [PubMed]
- Fernandes, J.M.; Kemp, G.D.; Molle, M.G.; Smith, V.J. Anti-microbial properties of histone H2A from skin secretions of rainbow trout, Oncorhynchus mykiss. Biochem. J. 2002, 368, 611–620. [Google Scholar] [CrossRef] [PubMed]
- Papareddy, P.; Rydengård, V.; Pasupuleti, M.; Walse, B.; Mörgelin, M.; Chalupka, A.; Malmsten, M.; Schmidtchen, A. Proteolysis of human thrombin generates novel host defense peptides. PLoS Pathog. 2010, 6. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.K.; Lee, M.J.; Go, H.J.; Kim, Y.J.; Park, N.G. Antimicrobial function of the GAPDH-related antimicrobial peptide in the skin of skipjack tuna, Katsuwonus pelamis. Fish Shellfish Immunol. 2014, 36, 571–581. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.K.; Lee, M.J.; Go, H.J.; Park, T.H.; Park, N.G. Purification and characterization of YFGAP, a GAPDH-related novel antimicrobial peptide, from the skin of yellowfin tuna, Thunnus albacares. Fish Shellfish Immunol. 2012, 33, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Trevisan-Silva, D.; Gremski, L.H.; Chaim, O.M.; da Silveira, R.B.; Meissner, G.O.; Mangili, O.C.; Barbaro, K.C.; Gremski, W.; Veiga, S.S.; Senff-Ribeiro, A. Astacin-like metalloproteases are a gene family of toxins present in the venom of different species of the brown spider (genus Loxosceles). Biochimie 2010, 92, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Gremski, L.H.; da Silveira, R.B.; Chaim, O.M.; Probst, C.M.; Ferrer, V.P.; Nowatzki, J.; Weinschutz, H.C.; Madeira, H.M.; Gremski, W.; Nader, H.B.; et al. A novel expression profile of the Loxosceles intermedia spider venomous gland revealed by transcriptome analysis. Mol. Biosyst. 2010, 6, 2403–2416. [Google Scholar] [CrossRef] [PubMed]
- Chaim, O.M.; Trevisan-Silva, D.; Chaves-Moreira, D.; Wille, A.C.; Ferrer, V.P.; Matsubara, F.H.; Mangili, O.C.; da Silveira, R.B.; Gremski, L.H.; Gremski, W.; et al. Brown spider (Loxosceles genus) venom toxins: tools for biological purposes. Toxins (Basel) 2011, 3, 309–344. [Google Scholar] [CrossRef] [PubMed]
- Gremski, L.H.; Trevisan-Silva, D.; Ferrer, V.P.; Matsubara, F.H.; Meissner, G.O.; Wille, A.C.; Vuitika, L.; Dias-Lopes, C.; Ullah, A.; de Moraes, F.R.; et al. Recent advances in the understanding of brown spider venoms: From the biology of spiders to the molecular mechanisms of toxins. Toxicon 2014, 83, 91–120. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, J.M.; Lomonte, B. Phospholipases A2: unveiling the secrets of a functionally versatile group of snake venom toxins. Toxicon 2013, 62, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.H.; He, Q.Y.; Peng, K.; Diao, J.B.; Jiang, L.P.; Tang, X.; Liang, S.P. Discovery of a distinct superfamily of Kunitz-type toxin (KTT) from tarantulas. PLoS ONE 2008, 3, e3414. [Google Scholar] [CrossRef]
- Chen, Z.Y.; Hu, Y.T.; Yang, W.S.; He, Y.W.; Feng, J.; Wang, B.; Zhao, R.M.; Ding, J.P.; Cao, Z.J.; Li, W.X.; et al. Hg1, novel peptide inhibitor specific for Kv1.3 channels from first scorpion Kunitz-type potassium channel toxin family. J. Biol. Chem. 2012, 287, 13813–13821. [Google Scholar] [CrossRef] [PubMed]
- Dhananjaya, B.L.; D’Souza, C.J. The pharmacological role of phosphatases (acid and alkaline phosphomonoesterases) in snake venoms related to release of purines—A multitoxin. Basic Clin. Pharmacol. Toxicol. 2011, 108, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Schepens, T.; Cammu, G. Neuromuscular blockade: What was, is and will be. Acta Anaesthesiol. Belg. 2014, 65, 151–159. [Google Scholar] [PubMed]
- Fernandes-Pedrosa Mde, F.; Junqueira-de-Azevedo Ide, L.; Gonçalves-de-Andrade, R.M.; Kobashi, L.S.; Almeida, D.D.; Ho, P.L.; Tambourgi, D.V. Transcriptome analysis of Loxosceles laeta (Araneae, Sicariidae) spider venomous gland using expressed sequence tags. BMC Genom. 2008, 9, 279–291. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, L.D.; Dias, N.B.; Roberto, J.; Pinto, A.S.; Palma, M.S. Brown recluse spider venom: proteomic analysis and proposal of a putative mechanism of action. Protein Pept. Lett. 2009, 16, 933–943. [Google Scholar] [CrossRef] [PubMed]
- Duan, Z.; Yan, X.; Cao, R.; Liu, Z.; Wang, X.; Liang, S. Proteomic analysis of Latrodectus Tredecimguttatus venom for uncovering potential latrodectism-related proteins. J. Biochem. Mol. Toxicol. 2008, 22, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Roelants, K.; Champagne, D.E.; Scheib, H.; Tyndall, J.D.; King, G.F.; Nevalainen, T.J.; Norman, J.A.; Lewis, R.J.; Norton, R.S.; et al. The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Annu. Rev. Genom. Hum. Genet. 2009, 10, 483–511. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, G.M.; Roelants, K.; O’Bryan, M.K. The CAP superfamily: cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins-roles in reproduction, cancer, and immune defense. Endocr Rev. 2008, 29, 865–897. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.C.; Hsieh, H.J.; Hsieh, C.H.; Hwang, D.F. Plancitoxin I from the venom of crown-of-thorns starfish (Acanthaster planci) induces oxidative and endoplasmic reticulum stress associated cytotoxicity in A375.S2 cells. Exp. Mol. Pathol. 2015, 99, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.C.; Hsieh, H.J.; Hwang, D.F. Cytotoxic and apoptotic activities of the plancitoxin I from the venom of crown-of-thorns starfish (Acanthaster planci) on A375.S2 cells. J. Appl. Toxicol. 2015, 35, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Negri, L.; Lattanzi, R.; Giannini, E.; Melchiorri, P. Bv8/prokineticin proteins and their receptors. Life Sci. 2007, 81, 1103–1116. [Google Scholar] [CrossRef] [PubMed]
- Di Cera, E. Serine proteases. IUBMB Life 2009, 61, 510–515. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y. Why do we study animal toxins? Zool. Res. 2015, 36, 183–222. [Google Scholar] [PubMed]
- Brahma, R.K.; McCleary, R.J.; Kini, R.M.; Doley, R. Venom gland transcriptomics for identifying, cataloging, and characterizing venom proteins in snakes. Toxicon 2015, 93, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Moreno, M.; Giralt, E. Three valuable peptides from bee and wasp venoms for therapeutic and biotechnological use: melittin, apamin and mastoparan. Toxins (Basel) 2015, 7, 1126–1150. [Google Scholar] [CrossRef] [PubMed]
- Kang, T.S.; Georgieva, D.; Genov, N.; Murakami, M.T.; Sinha, M.; Kumar, R.P.; Kaur, P.; Kumar, S.; Dey, S.; Sharma, S.; et al. Enzymatic toxins from snake venom: Structural characterization and mechanism of catalysis. FEBS J. 2011, 278, 4544–4576. [Google Scholar] [CrossRef] [PubMed]
- Chernetskaia, I.I.; Akhunov, A.; Abduvakhabov, A.A.; Sadykov, A.S. Cholinesterase isolated from the venom of the Latrodectus tredecimguttatus spider using substrate inhibitor analysis. Dokl. Akad. Nauk SSSR 1984, 274, 225–229. [Google Scholar] [PubMed]
- Chernetskaia, I.I.; Akhunov, A.; Sadykov, A.S. Isolation of cholinesterase from the venom of the spider Latrodectus tredecimguttatus and the study of its properties. Dokl. Akad. Nauk SSSR 1983, 269, 1510–1513. [Google Scholar] [PubMed]
- Whelan, N.V.; Kocot, K.M.; Santos, S.R.; Halanych, K.M. Nemertean toxin genes revealed through transcriptome sequencing. Genome Biol. Evol. 2014, 6, 3314–3325. [Google Scholar] [CrossRef] [PubMed]
- Shiomi, K.; Midorikawa, S.; Ishida, M.; Nagashima, Y.; Nagai, H. Plancitoxins, lethal factors from the crown-of-thorns starfish Acanthaster planci, are deoxyribonucleases II. Toxicon 2004, 44, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Ai, W.; Nagai, H.; Nagashima, Y.; Shiomi, K. Structural characterization of plancitoxin I, a deoxyribonuclease II-like lethal factor from the crown-of-thorns starfish Acanthaster planci, by expression in Chinese hamster ovary cells. Jpn. Soc. Fish Sci. 2009, 75, 225–231. [Google Scholar]
- Casewell, N.R.; Wuster, W.; Vonk, F.J.; Harrison, R.A.; Fry, B.G. Complex cocktails: the evolutionary novelty of venoms. Trends Ecol. Evol. 2013, 28, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Stevens, L. Egg proteins: What are their functions? Sci. Prog. 1996, 79, 65–87. [Google Scholar] [PubMed]
- Morgenstern, D.; King, G.F. The venom optimization hypothesis revisited. Toxicon 2013, 63, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Pertea, G.; Huang, X.; Liang, F.; Antonescu, V.; Sultana, R.; Karamycheva, S.; Lee, Y.; White, J.; Cheung, F.; Parvizi, B.; et al. TIGR Gene Indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 2003, 19, 651–652. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L.; et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- The Cluster of Orthologous Groups Database. Available online: http://www.ncbi.nlm.nih.gov/COG (accessed on 4 January 2015).
- The Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Database. Available online: http://www.genome.jp/keg (accessed on 14 January 2015).
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Dong, C.; Yu, J.; Liu, W.; Jiang, C.; Liu, J.; Hu, Q.; Fang, X.; Wei, W. Transcriptome profile analysis of young floral buds of fertile and sterile plants from the self-pollinated offspring of the hybrid between novel restorer line NR1 and Nsa CMS line in Brassica napus. BMC Genom. 2013, 14, 26–41. [Google Scholar] [CrossRef] [PubMed]
- Signal4.1 Server. Available online: www.cbs.dtu.dk/services/SignalP-4.1/ (accessed on 5 February 2015).
- Konttin Database. Available online: www.knottin.cbs.cnrs.fr/ (accessed on 5 February 2015).
- PORTER. Available online: http://distill.ucd.ie/porter/ (accessed on 5 February 2015).



| Analysis of Read Assembly | Amount |
|---|---|
| Total number of reads | 47,970,296 |
| Total base pairs (bp) | 5,836,590,233 |
| Average read length (bp) | 121 |
| Total number of transcripts | 69,684 |
| Total number of unigenes | 53,284 |
| Average length of unigenes (bp) | 738 |
| Total number of unigenes >2000 bp in length | 4376 |
| Total number of unigenes annotated in at least one database | 14,185 |
| Sequence ID | Signal Peptide | Score (Bits) | E-Value | Identity | BLAST Annotation |
|---|---|---|---|---|---|
| comp24914_c0_seq1 | N | 64.7 (156) | 8 × 10−11 | 34% | gb|AGA82764.1|toxin-like protein 14 precursor (Urodacus yaschenkoi) |
| comp6833_c0_seq1 | N | 92.0 (227) | 4 × 10−22 | 56% | gb|ABR21046.1|venom toxin-like peptide-6 (Mesobuthus eupeus) |
| comp22890_c0_seq2 | N | 90.9 (224) | 9× 10−22 | 56% | gb|ABR21046.1|venom toxin-like peptide-6 (Mesobuthus eupeus) |
| comp17051_c0_seq1 | N | 53.5 (127) | 3× 10−10 | 32% | as: U15-SYTX-Sth1a||1459 Translation of a toxin from the spider Scytodes thoracica with unknown molecular target and function |
| comp25199_c0_seq1 | Y | 55.8 (133) | 7× 10−11 | 32% | as: U12-SYTX-Sth1a||1449 Translation of a toxin from the spider Scytodes thoracica with unknown molecular target and function |
| comp21232_c0_seq1 | Y | 277 (709) | 5× 10−86 | 98% | gb|ADV40303.1|cystatin-like protein (Latrodectus Hesperus) |
| comp20935_c0_seq1 | Y | 102 (255) | 7× 10−25 | 40% | as: U24-ctenitoxin-Pn1a|sp:P84032|Toxin from venom of the spider Phoneutria nigriventer with unknown molecular target |
| comp213809_c0_seq1 | Y | 65.5 (158) | 1× 10−13 | 29% | as: U24-ctenitoxin-Pn1a|sp:P84032|Toxin from venom of the spider Phoneutria nigriventer with unknown molecular target |
| comp27859_c0_seq1 | Y | 105 (263) | 8× 10−26 | 44% | as: U24-ctenitoxin-Pn1a|sp:P84032|Toxin from venom of the spider Phoneutria nigriventer with unknown molecular target |
| comp96908_c0_seq1 | Y | 48.5 (114) | 1× 10−8 | 35% | as: U16-aranetoxin-Av1a_1||2248 Toxin from venom of the spider Araneus ventricosus with unknown molecular target and function |
| comp5553_c0_seq1 | Y | 53.1 (126) | 9× 10−7 | 32% | sp|Q8MTX1|TXCA_CAEEX U3-aranetoxin-Ce1a OS = Caerostris extrusa PE = 2 SV = 1 |
| Name | Number of Unigenes | Percent of Unigenes (%) | Putative Activities |
|---|---|---|---|
| Metalloprotease | 62 | 25 | Activating proteinogen or zymogen [46,47] |
| Degradating tissue to facilitate the spreading of toxins [46,47] | |||
| Serine protease | 55 | 22.2 | Activating proteinogen or zymogen [46,47,48] |
| Degradating tissue to facilitate the spreading of toxins [46,47,48] | |||
| Phospholipase | 41 | 16.5 | Neurotoxicity, myotoxicity, etc. [46,47,48,49] |
| Serpin | 30 | 12.1 | Inhibiting degradation of proteinaceous toxins by protease [46,47,48] |
| Acting on ion channels, e.g., K+ channel [50,51] | |||
| Phosphatase | 18 | 7.3 | Assisting the liberation of purines [52] |
| Cholinesterase | 12 | 4.8 | Blocking the neuromuscular transmission [53] |
| Allergen/Lipocalin | 11 | 4.4 | Provoking anaphylaxis [29,46,54,55,56] |
| Disrupting hemostasis [57] | |||
| Chitinase | 10 | 4 | Degrading the chitin [57] |
| CAP superfamily | 5 | 2 | Modulating ion channels [57,58] |
| Plancitoxin-1 | 2 | 0.9 | Inducing apoptosis [59,60] |
| Hyaluronidase | 1 | 0.4 | Enhancing tissue permeability to allow the spreading of toxins [46,47,48,57] |
| Prokineticin/AVIT | 1 | 0.4 | Inhibiting the feeding or inducing hyperalgesia [57,61] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, D.; Wang, X. Transcriptome Analysis to Understand the Toxicity of Latrodectus tredecimguttatus Eggs. Toxins 2016, 8, 378. https://doi.org/10.3390/toxins8120378
Xu D, Wang X. Transcriptome Analysis to Understand the Toxicity of Latrodectus tredecimguttatus Eggs. Toxins. 2016; 8(12):378. https://doi.org/10.3390/toxins8120378
Chicago/Turabian StyleXu, Dehong, and Xianchun Wang. 2016. "Transcriptome Analysis to Understand the Toxicity of Latrodectus tredecimguttatus Eggs" Toxins 8, no. 12: 378. https://doi.org/10.3390/toxins8120378





