Detection of Enterotoxigenic Potential and Determination of Clonal Profile in Staphylococcus aureus and Coagulase-Negative Staphylococci Isolated from Bovine Subclinical Mastitis in Different Brazilian States
Abstract
:1. Introduction
2. Results
2.1. Identification of the Strains
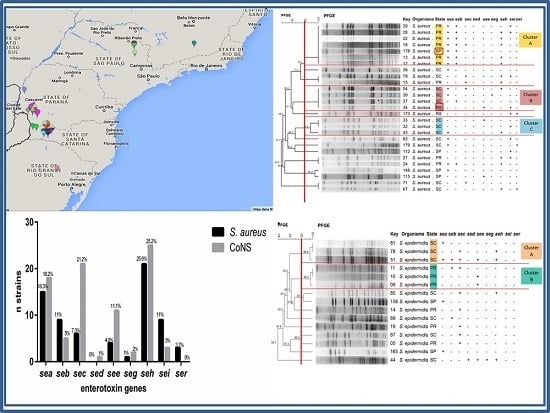
2.2. Detection of Enterotoxin and tst-1 Genes
2.3. Distribution of Staphylococcal Enterotoxin Genes According to the Region Studied
2.4. Determination of Clonal Profile
3. Discussion
4. Experimental Section
4.1. Origin and Identification of the Strains
4.2. Detection of Enterotoxin and tst-1 Genes
4.3. Analysis by Pulsed-Field Gel Electrophoresis
4.4. Spatial Distribution of Staphylococcal Enterotoxin Genes in Different Regions of Brazil
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Radostitis, O.M.; Gay, C.C.; Hinchcliff, K.W.; Constable, P.D. Veterinary medicine. In A Textbook of the Disease of Cattle, Horses, Sheep, Pigs and Goats, 10th ed.; Saunders Elsevier: Philadelphia, PA, USA, 2007; p. 2156. [Google Scholar]
- Rall, V.L.; Miranda, E.S.; Castilho, I.G.; Camargo, C.H.; Langoni, H.; Guimarães, F.F.; Júnior, J.P.A.; Júnior, A.F. Diversity of Staphylococcus species and prevalence of enterotoxin genes isolated from milk of healthy cows and cows with subclinical mastitis. J. Dairy Sci. 2014, 97, 829–837. [Google Scholar] [CrossRef] [PubMed]
- Karahan, M.N.; Açik, B. Çetinkaya investigation of toxin genes by polymerase chain reaction in Staphylococcus aureus strains isolated from bovine mastitis in Turkey. Foodborne Pathog. Dis. 2009, 6, 1029–1035. [Google Scholar] [CrossRef] [PubMed]
- Calsolari, R.A.O.; Pereira, V.C.P.; Júnior, J.P.A.; Cunha, M.L.R.S. Determination of toxigenic capacity by RT-PCR in coagulase-negative staphylococci and Staphylococcus aureus isolated from newborns in Brazil. Microbiol. Immunol. 2011, 55, 394–407. [Google Scholar] [CrossRef] [PubMed]
- Cunha, M.L.R.S.; Peresi, E.; Calsolari, R.A.O.; Júnior, J.P.A. Detection of enterotoxins genes on coagulase-negative staphylococci isolated from foods. Braz. J. Microbiol. 2006, 37, 70–74. [Google Scholar] [CrossRef]
- Omoe, K.; Imanishi, K.; Hu, D.L.; Kato, H.; Takahashi-Omoe, H.; Nakane, A.; Uchiyama, T.; Shinagawa, K. Biological properties of staphylococcal enterotoxin-like toxin type R. Infect. Immun. 2004, 72, 3664–3667. [Google Scholar] [CrossRef] [PubMed]
- List of Prokaryotic Names with Standing in Nomenclature—Genus Staphylococcus. Available online: http://www.bacterio.net/ (accessed on 6 September 2015).
- Ono, H.K.; Omoe, K.; Imanishi, K.; Iwakabe, Y.; Hu, D.L.; Kato, H.; Saito, N.; Nakane, A.; Uchiyama, T.; Shinagawa, K. Identification and characterization of two novel staphylococcal enterotoxins, types S and T. Infect. Immun. 2008, 76, 4999–5005. [Google Scholar] [CrossRef] [PubMed]
- Borst, D.W.; Betley, M.J. Phage-associated differences in staphylococcal enterotoxin A gene (sea) expression correlate with sea allele class. Infect. Immun. 1994, 62, 113–118. [Google Scholar] [PubMed]
- Evenson, M.L.; Hinds, M.W.; Bernstein, R.S.; Bergdoll, M.S. Estimation of human dose of staphylococcal enterotoxin A from a large outbreak of staphylococcal food poisoning involving chocolate milk. Int. J. Food Microbiol. 1988, 7, 311–316. [Google Scholar] [PubMed]
- Balaban, N.; Rasooly, A. Staphylococcal enterotoxins. Int. J. Food Microbiol. 2000, 61, 1–10. [Google Scholar] [CrossRef]
- Tremaine, M.T.; Brockman, D.K.; Betley, M.J. Staphylococcal enterotoxin A gene (sea) expression is not affected by the accessory gene regulator (agr). Infect. Immun. 1993, 61, 56–359. [Google Scholar]
- Marr, J.C.; Lyon, J.D.; Roberson, J.R.; Lupher, M.; Davis, W.C.; Bohach, G.A. Characterization of novel type C staphylococcal enterotoxins: Biological and evolutionary implications. Infect. Immun. 1993, 61, 4254–4262. [Google Scholar] [PubMed]
- Bayles, K.W.; Iandolo, J.J. Genetic and molecular analyses of the gene encoding staphylococcal enterotoxin D. J. Bacteriol. 1989, 171, 4799–4806. [Google Scholar] [PubMed]
- Kokan, N.P.; Bergdoll, M.S. Detection of low-enterotoxin-producing Staphylococcus aureus strains. Appl. Environ. Microbiol. 1987, 53, 2675–2676. [Google Scholar] [PubMed]
- Veras, J.F.; Simeão, L.C.; Lawrence, C.T.; Jefrey, W.S.; Christiano, C.; Santos, D.A.; Cerqueira, M.M.O.P.; Cantini, A.; Nicoli, J.R.; Jett, M. A study of the enterotoxigenicity of coagulase-negative and coagulase-positive staphylococcal isolates from food poisoning outbreaks in Minas Gerais, Brazil. Int. J. Infect. Dis. 2008, 12, 410–415. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, V.; Sawant, A.A.; Gillespie, B.E.; Headrick, S.J.; Ceasaris, L.; Oliver, S.P. Prevalence of enterotoxin and toxic shock syndrome toxin genes in Staphylococcus aureus isolated from milk of cows with mastitis. Foodborne Pathog. Dis. 2006, 3, 274–283. [Google Scholar] [CrossRef] [PubMed]
- Van den Bussche, R.A.; Lyon, J.D.; Bohach, G.A. Molecular evolution of the staphylococcal and streptococcal pyrogenic toxin gene family. Mol. Phylogenet. 1993, 2, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Lawrynowicz-Paciorek, M.; Kochman, M.; Grochowska, A.; Windyga, B. The distribution of enterotoxin and enterotoxin-like genes in Staphylococcus aureus strains isolated from nasal carriers and food samples. Int. J. Food Microbiol. 2007, 117, 319–332. [Google Scholar] [CrossRef] [PubMed]
- Akineden, Ö.; Annemüller, C.; Hassan, A.A.; Lämmler, C.; Wolter, W.; Zschöck, M. Toxin genes and other characteristics of Staphylococcus aureus isolates from milk of cows with mastitis. Clin. Diagn. Lab. Immun. 2001, 8, 959–964. [Google Scholar] [CrossRef] [PubMed]
- Buzzola, F.R.; Quelle, L.; Gomez, M.I.; Catalano, M.; Steele-Moore, L.; Berg, D.; Gentilini, E.; Denamiel, G.; Sordelli, D.O. Genotypic analysis of Staphylococcus aureus from milk of dairy cows with mastitis in Argentina. Epidemiol. Infect. 2001, 126, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Almeida, L.M.D.; Mamizuka, E.M.; Cunha, M.L.R.S.C.; Zafalon, L.F. Fatores de Virulência e Genes Regulatórios agr de Staphylococcus aureus e Outras Espécies Coagulase Positivas Isoladas de Mastites Bovina e Ovina; Universidade de São Paulo: São Paulo, Brazil, 2009. (In Portuguese) [Google Scholar]
- Koneman, E.W.; Allen, S.D.; Janda, W.M.; Schreckenberger, P.C.; Winn, W.C., Jr. Color Atlas and Textbook of Diagnostic Microbiology, 5th ed.; Lippincott: Philadelphia, PA, USA, 1997. [Google Scholar]
- Cunha, M.L.R.S.; Sinzato, Y.K.; Silveira, L.V.A. Comparision of methods for identification of Coagulase-negative Staphylococci. Mem. Inst. Oswaldo Cruz 2004, 99, 855–860. [Google Scholar] [CrossRef]
- Barry, T.; Colleran, G.; Glennon, M.; Dunican, L.K.; Gannon, F. The 16S/23S ribosomal spacer region as a target for DNA probes to identify eubacteria. PCR Methods Appl. 1991, 1, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Couto, I.; Pereira, S.; Miragaia, M.; Sanches, I.S.; Lencastre, H. Identification of clinical staphylococcal isolates from humans by Internal Transcribed Spacer PCR. J. Clin. Microbiol. 2001, 39, 3099–3103. [Google Scholar] [CrossRef]
- Straub, J.A.; HerteL, C.; Hammes, W.P. A 23S rDNA-targeted polymerase chain reaction-based system for detection of Staphylococcus aureus in meat starter cultures and dairy products. J. Food Prot. 1999, 62, 1150–1156. [Google Scholar] [PubMed]
- Johnson, W.M.; Tyler, S.D.; Ewan, E.P.; Ashton, F.E.; Pollard, D.R.; Rozee, K.R. Detection of genes for enterotoxins, exfoliative toxins, and toxic shock syndrome toxin 1 in Staphylococcus aureus by the polymerase chain reaction. J. Clin. Microbiol. 1991, 29, 426–430. [Google Scholar] [PubMed]
- Jarraud, S.; Cozon, G.; Vandenesch, F.; Bes, M.; Etienne, J.; Lina, G. Involvement of enterotoxins G and I in staphylococcal toxic shock syndrome and staphylococcal scarlet fever. J. Clin. Microbiol. 1999, 37, 2446–2449. [Google Scholar] [PubMed]
- Jarraud, S.; Mougel, C.; Thioulouse, J.; Lina, G.; Meugnier, H.; Forey, F. Relationships between Staphylococcus genetic background, virulence factors, agr groups (alleles), and human disease. Infect. Imunn. 2002, 70, 631–641. [Google Scholar] [CrossRef]
- Monday, S.R.; Bohach, G.A. Use of multiplex PCR to detect classical and newly described pyrogenic toxin genes in staphylococcal isolates. J. Clin. Microbiol. 1999, 37, 3411–3414. [Google Scholar] [PubMed]
- Chiang, Y.C.; Liao, W.W.; Fan, C.M.; Pai, W.Y.; Chiou, C.S.; Tsen, H.Y. PCR detection of staphylococcal enterotoxins (SEs) N, O, P, Q, R, U, and survey of SE types in Staphylococcus aureus isolates from food-poisoning cases in Taiwan. Int. J. Food Microbiol. 2008, 121, 66–73. [Google Scholar] [CrossRef] [PubMed]
- McDougal, L.K.; Steward, C.D.; Killgore, G.E.; Chaitram, J.M.; McAllister, S.K.; Tenover, F.C. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 2003, 41, 5113–5120. [Google Scholar] [CrossRef] [PubMed]




| Species | (n) | sea | seb | sec | sed | see | seg | seh | sei | sej | ser | ses | set | tst-1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S. aureus | (82) | 15 | 9 | 6 | 0 | 4 | 1 | 21 | 9 | 0 | 3 | 0 | 0 | 0 |
| S. chromogenes | (27) | 5 | 0 | 5 | 0 | 1 | 0 | 10 | 1 | 0 | 0 | 0 | 0 | 0 |
| S. epidermidis | (26) | 3 | 2 | 8 | 1 | 4 | 1 | 6 | 0 | 0 | 0 | 0 | 0 | 0 |
| S. saprophyticus | (17) | 5 | 1 | 1 | 0 | 2 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| S. haemolyticus | (6) | 1 | 1 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| S. simulans | (6) | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| S. warneri | (6) | 0 | 0 | 2 | 0 | 1 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 |
| S. hyicus | (5) | 2 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| S. hominis | (4) | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| S. xylosus | (2) | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 181 | 33 | 14 | 27 | 1 | 15 | 3 | 46 | 12 | 0 | 3 | 0 | 0 | 0 |
| Species | Cluster | No. of Isolates | Enterotoxin Genes | Origin of Isolates |
|---|---|---|---|---|
| S. aureus | A | 7 | sea (7); seb (7); sec (1); seh (5); sei (2) | PR; SP |
| B | 4 | sea (2); seb (1); sec (1); see (1); seg (1); seh (1); sei (1) | SC; RS | |
| C | 3 | sea (1); see (1); seh (1); sei (1) | SC | |
| S. epidermidis | A | 3 | sea (2); seb (1); sec (2); see (1); seh (2) | SC |
| B | 3 | seb (1); sec (1); see (2) | PR |
| Name | Product | 5′ to 3′ Nucleotide Sequence | Reference | Amplicon Size (bp) |
|---|---|---|---|---|
| sea1 | Enterotoxin A | TTGGAAACGGTTAAAACGAA | [28] | 120 |
| sea2 | GAACCTTCCCATCAAAAACA | |||
| seb1 | Enterotoxin B | TCGCATCAAACTGACAAACG | [28] | 478 |
| seb2 | GCAGGTACTCTATAAGTGCC | |||
| sec1 | Enterotoxin C | GACATAAAAGCTAGGAATTT | [28] | 257 |
| sec2 | AAATCGGATTAACATTATCC | |||
| sed1 | Enterotoxin D | CTAGTTTGGTAATATCTCCT | [28] | 317 |
| sed2 | TAATGCTATATCTTATAGGG | |||
| see1 | Enterotoxin E | CAAAGAAATGCTTTAAGCAATCTTAGGCCAC | [29] | 170 |
| see2 | CTTACCGCCAAAGCTG | |||
| seg1 | Enterotoxin G | AATTATGTGAATGCTCAACCCGATC | [29] | 642 |
| seg2 | AAACTTATATGGAACAAAAGGTACTAGTTC | |||
| seh1 | Enterotoxin H | CAATCACATCATATGCGAAAGCAG | [30] | 375 |
| seh2 | CATCTACCCAAACATTAGCACC | |||
| sei1 | Enterotoxin I | CTCAAGGTGATATTGGTGTAGG | [29] | 576 |
| sei2 | AAAAAACTTACAGGCAGTCCATCTC | |||
| selj1 | Enterotoxin J | CATCAGAACTGTTGTTCCGCTAG | [31] | 146 |
| selj2 | CTGAATTTTACCATCAAAGGTAC | |||
| ser1 | Enterotoxin R | AGATGTGTTTGGAATACCCTAT | [32] | 123 |
| ser2 | CTATCAGCTGTGGAGTGCAT | |||
| ses1 | Enterotoxin S | TTCAGAAATAGCCAATCATTTCAA | [8] | 195 |
| ses2 | CCTTTTTGTTGAGAGCCGTC | |||
| set1 | Enterotoxin T | GGTGATTATGTAGATGCTTGGG | [8] | 170 |
| set2 | TCGGGTGTTACTTCTGTTTGC | |||
| tst1 | Toxic shock syndrome toxin | ATGGCAGCATCAGCTTGATA | [28] | 350 |
| tst2 | TTTCCAATAACCACCCGTTT |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mello, P.L.; Moraes Riboli, D.F.; Pinheiro, L.; De Almeida Martins, L.; Vasconcelos Paiva Brito, M.A.; Ribeiro de Souza da Cunha, M.D.L. Detection of Enterotoxigenic Potential and Determination of Clonal Profile in Staphylococcus aureus and Coagulase-Negative Staphylococci Isolated from Bovine Subclinical Mastitis in Different Brazilian States. Toxins 2016, 8, 104. https://doi.org/10.3390/toxins8040104
Mello PL, Moraes Riboli DF, Pinheiro L, De Almeida Martins L, Vasconcelos Paiva Brito MA, Ribeiro de Souza da Cunha MDL. Detection of Enterotoxigenic Potential and Determination of Clonal Profile in Staphylococcus aureus and Coagulase-Negative Staphylococci Isolated from Bovine Subclinical Mastitis in Different Brazilian States. Toxins. 2016; 8(4):104. https://doi.org/10.3390/toxins8040104
Chicago/Turabian StyleMello, Priscila Luiza, Danilo Flávio Moraes Riboli, Luiza Pinheiro, Lisiane De Almeida Martins, Maria Aparecida Vasconcelos Paiva Brito, and Maria De Lourdes Ribeiro de Souza da Cunha. 2016. "Detection of Enterotoxigenic Potential and Determination of Clonal Profile in Staphylococcus aureus and Coagulase-Negative Staphylococci Isolated from Bovine Subclinical Mastitis in Different Brazilian States" Toxins 8, no. 4: 104. https://doi.org/10.3390/toxins8040104