Helicobacter pylori vacA Genotypes in Chronic Gastritis and Gastric Carcinoma Patients from Macau, China
Abstract
:1. Introduction
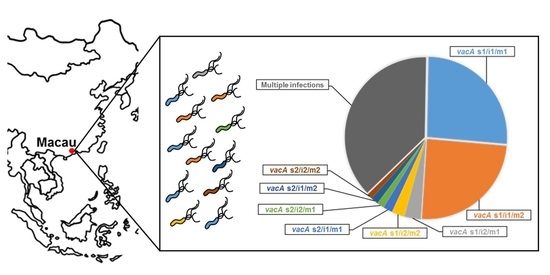
2. Results
2.1. H. pylori vacA Genotypes and cagA Status
2.2. Relationship between vacA i-Region Genotypes, s- and m-Region Genotypes and cagA Status
2.3. Relationship between H. pylori Genotypes and Gastric Carcinoma
3. Discussion
4. Materials and Methods
4.1. Patients and Study Population
4.2. Histopathology
4.3. DNA Isolation
4.4. H. pylori vacA Genotyping and cagA Gene Detection
4.5. Statistical Analyses
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Ervik, M.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Globocan 2012 v1.0, Cancer Incidence and Mortality Worldwide: Iarc Cancerbase no. 11; International Agency for Research on Cancer: Lyon, France, 2013; Available online: http://globocan.iarc.fr (accessed on 14 March 2016).
- Polk, D.B.; Peek, R.M., Jr. Helicobacter pylori: Gastric cancer and beyond. Nat. Rev. Cancer 2010, 10, 403–414. [Google Scholar] [CrossRef] [PubMed]
- Correa, P. Human gastric carcinogenesis: A multistep and multifactorial process—First american cancer society award lecture on cancer epidemiology and prevention. Cancer Res. 1992, 52, 6735–6740. [Google Scholar] [PubMed]
- Figueiredo, C.; Machado, J.C.; Pharoah, P.; Seruca, R.; Sousa, S.; Carvalho, R.; Capelinha, A.F.; Quint, W.; Caldas, C.; van Doorn, L.J.; et al. Helicobacter pylori and interleukin 1 genotyping: An opportunity to identify high-risk individuals for gastric carcinoma. J. Natl. Cancer Inst. 2002, 94, 1680–1687. [Google Scholar] [CrossRef] [PubMed]
- El-Omar, E.M.; Rabkin, C.S.; Gammon, M.D.; Vaughan, T.L.; Risch, H.A.; Schoenberg, J.B.; Stanford, J.L.; Mayne, S.T.; Goedert, J.; Blot, W.J.; et al. Increased risk of noncardia gastric cancer associated with proinflammatory cytokine gene polymorphisms. Gastroenterology 2003, 124, 1193–1201. [Google Scholar] [CrossRef]
- Machado, J.C.; Figueiredo, C.; Canedo, P.; Pharoah, P.; Carvalho, R.; Nabais, S.; Castro Alves, C.; Campos, M.L.; Van Doorn, L.J.; Caldas, C.; et al. A proinflammatory genetic profile increases the risk for chronic atrophic gastritis and gastric carcinoma. Gastroenterology 2003, 125, 364–371. [Google Scholar] [CrossRef]
- Dorer, M.S.; Talarico, S.; Salama, N.R. Helicobacter pylori’s unconventional role in health and disease. PLoS Pathog. 2009, 5, e1000544. [Google Scholar] [CrossRef] [PubMed]
- Odenbreit, S.; Puls, J.; Sedlmaier, B.; Gerland, E.; Fischer, W.; Haas, R. Translocation of helicobacter pylori caga into gastric epithelial cells by type iv secretion. Science 2000, 287, 1497–1500. [Google Scholar] [CrossRef] [PubMed]
- Backert, S.; Tegtmeyer, N.; Fischer, W. Composition, structure and function of the helicobacter pylori cag pathogenicity island encoded type iv secretion system. Fut. Microbiol. 2015, 10, 955–965. [Google Scholar] [CrossRef] [PubMed]
- Peek, R.M., Jr.; Miller, G.G.; Tham, K.T.; Perez-Perez, G.I.; Zhao, X.; Atherton, J.C.; Blaser, M.J. Heightened inflammatory response and cytokine expression in vivo to caga+ helicobacter pylori strains. Lab. Investig. J. Tech. Methods Pathol. 1995, 73, 760–770. [Google Scholar]
- Nogueira, C.; Figueiredo, C.; Carneiro, F.; Gomes, A.T.; Barreira, R.; Figueira, P.; Salgado, C.; Belo, L.; Peixoto, A.; Bravo, J.C.; et al. Helicobacter pylori genotypes may determine gastric histopathology. Am. J. Pathol. 2001, 158, 647–654. [Google Scholar] [CrossRef]
- Yamaoka, Y.; Kodama, T.; Gutierrez, O.; Kim, J.G.; Kashima, K.; Graham, D.Y. Relationship between helicobacter pylori icea, caga, and vaca status and clinical outcome: Studies in four different countries. J. Clin. Microbiol. 1999, 37, 2274–2279. [Google Scholar] [PubMed]
- Wang, J.; van Doorn, L.J.; Robinson, P.A.; Ji, X.; Wang, D.; Wang, Y.; Ge, L.; Telford, J.L.; Crabtree, J.E. Regional variation among vaca alleles of helicobacter pylori in china. J. Clin. Microbiol. 2003, 41, 1942–1945. [Google Scholar] [CrossRef] [PubMed]
- Cover, T.L.; Tummuru, M.K.; Cao, P.; Thompson, S.A.; Blaser, M.J. Divergence of genetic sequences for the vacuolating cytotoxin among helicobacter pylori strains. J. Biol. Chem. 1994, 269, 10566–10573. [Google Scholar] [PubMed]
- Atherton, J.C.; Cao, P.; Peek, R.M., Jr.; Tummuru, M.K.; Blaser, M.J.; Cover, T.L. Mosaicism in vacuolating cytotoxin alleles of helicobacter pylori. Association of specific vaca types with cytotoxin production and peptic ulceration. J. Biol. Chem. 1995, 270, 17771–17777. [Google Scholar] [PubMed]
- Rhead, J.L.; Letley, D.P.; Mohammadi, M.; Hussein, N.; Mohagheghi, M.A.; Eshagh Hosseini, M.; Atherton, J.C. A new helicobacter pylori vacuolating cytotoxin determinant, the intermediate region, is associated with gastric cancer. Gastroenterology 2007, 133, 926–936. [Google Scholar] [CrossRef] [PubMed]
- van Doorn, L.J.; Figueiredo, C.; Sanna, R.; Pena, S.; Midolo, P.; Ng, E.K.; Atherton, J.C.; Blaser, M.J.; Quint, W.G. Expanding allelic diversity of helicobacter pylori vaca. J. Clin. Microbiol. 1998, 36, 2597–2603. [Google Scholar] [PubMed]
- Ferreira, R.M.; Machado, J.C.; Letley, D.; Atherton, J.C.; Pardo, M.L.; Gonzalez, C.A.; Carneiro, F.; Figueiredo, C. A novel method for genotyping the helicobacter pylori vaca intermediate region directly in gastric biopsy specimens. J. Clin. Microbiol. 2012, 50, 3983–3989. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, C.A.; Figueiredo, C.; Lic, C.B.; Ferreira, R.M.; Pardo, M.L.; Ruiz Liso, J.M.; Alonso, P.; Sala, N.; Capella, G.; Sanz-Anquela, J.M. Helicobacter pylori caga and vaca genotypes as predictors of progression of gastric preneoplastic lesions: A long-term follow-up in a high-risk area in spain. Am. J. Gastroenterol. 2011, 106, 867–874. [Google Scholar] [CrossRef] [PubMed]
- Matos, J.I.; de Sousa, H.A.; Marcos-Pinto, R.; Dinis-Ribeiro, M. Helicobacter pylori caga and vaca genotypes and gastric phenotype: A meta-analysis. Eur. J. Gastroenterol. Hepatol. 2013, 25, 1431–1441. [Google Scholar] [CrossRef] [PubMed]
- Basso, D.; Zambon, C.F.; Letley, D.P.; Stranges, A.; Marchet, A.; Rhead, J.L.; Schiavon, S.; Guariso, G.; Ceroti, M.; Nitti, D.; et al. Clinical relevance of helicobacter pylori caga and vaca gene polymorphisms. Gastroenterology 2008, 135, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Memon, A.A.; Hussein, N.R.; Miendje Deyi, V.Y.; Burette, A.; Atherton, J.C. Vacuolating cytotoxin genotypes are strong markers of gastric cancer and duodenal ulcer-associated helicobacter pylori strains: A matched case-control study. J. Clin. Microbiol. 2014, 52, 2984–2989. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, R.M.; Figueiredo, C.; Bonet, C.; Pardo, M.L.; Liso, J.M.; Alonso, P.; Sala, N.; Capella, G.; Sanz-Anquela, J.M.; Gonzalez, C.A. Helicobacter pylori vaca intermediate region genotyping and progression of gastric preneoplastic lesions. Am. J. Gastroenterol. 2012, 107, 145–146. [Google Scholar] [CrossRef] [PubMed]
- Yamaoka, Y. Mechanisms of disease: Helicobacter pylori virulence factors. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Ogiwara, H.; Sugimoto, M.; Ohno, T.; Vilaichone, R.K.; Mahachai, V.; Graham, D.Y.; Yamaoka, Y. Role of deletion located between the intermediate and middle regions of the helicobacter pylori vaca gene in cases of gastroduodenal diseases. J. Clin. Microbiol. 2009, 47, 3493–3500. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.; Jones, K.R.; Olsen, C.H.; Joo, Y.M.; Yoo, Y.J.; Chung, I.S.; Cha, J.H.; Merrell, D.S. Epidemiological link between gastric disease and polymorphisms in vaca and caga. J. Clin. Microbiol. 2010, 48, 559–567. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, N.F.; Guimaraes, N.; Figueiredo, C.; Keevil, C.W.; Vieira, M.J. A new model for the transmission of helicobacter pylori: Role of environmental reservoirs as gene pools to increase strain diversity. Crit. Rev. Microbiol. 2007, 33, 157–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uemura, N.; Okamoto, S.; Yamamoto, S.; Matsumura, N.; Yamaguchi, S.; Yamakido, M.; Taniyama, K.; Sasaki, N.; Schlemper, R.J. Helicobacter pylori infection and the development of gastric cancer. N. Engl. J. Med. 2001, 345, 784–789. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, R.M.; Machado, J.C.; Figueiredo, C. Clinical relevance of helicobacter pylori vaca and caga genotypes in gastric carcinoma. Best Pract. Res. Clin. Gastroenterol. 2014, 28, 1003–1015. [Google Scholar] [CrossRef] [PubMed]
- Winter, J.A.; Letley, D.P.; Cook, K.W.; Rhead, J.L.; Zaitoun, A.A.; Ingram, R.J.; Amilon, K.R.; Croxall, N.J.; Kaye, P.V.; Robinson, K.; et al. A role for the vacuolating cytotoxin, vaca, in colonisation and helicobacter pylori-induced metaplasia in the stomach. J. Infect. Dis. 2014, 210, 954–963. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.; Olivares, A.; Torres, E.; Yilmaz, O.; Cohen, H.; Perez-Perez, G. Diversity of vaca intermediate region among helicobacter pylori strains from several regions of the world. J. Clin. Microbiol. 2010, 48, 690–696. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Kim, N.; Nam, R.H.; Suh, J.H.; Chang, H.; Lee, J.W.; Kim, Y.S.; Kim, J.M.; Choi, J.W.; Park, J.G.; et al. Association of polymorphisms in virulence factor of helicobacter pylori and gastroduodenal diseases in south korea. J. Gastroenterol. Hepatol. 2014, 29, 984–991. [Google Scholar] [CrossRef] [PubMed]
- Yamaoka, Y.; Orito, E.; Mizokami, M.; Gutierrez, O.; Saitou, N.; Kodama, T.; Osato, M.S.; Kim, J.G.; Ramirez, F.C.; Mahachai, V.; et al. Helicobacter pylori in north and south america before columbus. FEBS Lett. 2002, 517, 180–184. [Google Scholar] [CrossRef]
- Wong, B.C.; Yin, Y.; Berg, D.E.; Xia, H.H.; Zhang, J.Z.; Wang, W.H.; Wong, W.M.; Huang, X.R.; Tang, V.S.; Lam, S.K. Distribution of distinct vaca, caga and icea alleles in helicobacter pylori in hong kong. Helicobacter 2001, 6, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.J.; van der Hulst, R.W.; Feller, M.; Xiao, S.D.; Tytgat, G.N.; Dankert, J.; van der Ende, A. Equally high prevalences of infection with caga-positive helicobacter pylori in chinese patients with peptic ulcer disease and those with chronic gastritis-associated dyspepsia. J. Clin. Microbiol. 1997, 35, 1344–1347. [Google Scholar] [PubMed]
- Sahara, S.; Sugimoto, M.; Vilaichone, R.K.; Mahachai, V.; Miyajima, H.; Furuta, T.; Yamaoka, Y. Role of helicobacter pylori caga epiya motif and vaca genotypes for the development of gastrointestinal diseases in southeast asian countries: A meta-analysis. BMC Infect. Dis. 2012, 12, 223. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, M.; Yamaoka, Y. The association of vaca genotype and helicobacter pylori-related disease in latin american and african populations. Clin. Microbiol. Infect. 2009, 15, 835–842. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, M.; Zali, M.R.; Yamaoka, Y. The association of vaca genotypes and helicobacter pylori-related gastroduodenal diseases in the middle east. Eur. J. Clin. Microbiol. Infect. Dis. 2009, 28, 1227–1236. [Google Scholar] [CrossRef] [PubMed]
- Plummer, M.; van Doorn, L.J.; Franceschi, S.; Kleter, B.; Canzian, F.; Vivas, J.; Lopez, G.; Colin, D.; Munoz, N.; Kato, I. Helicobacter pylori cytotoxin-associated genotype and gastric precancerous lesions. J. Natl. Cancer Inst. 2007, 99, 1328–1334. [Google Scholar] [CrossRef] [PubMed]
- Alm, R.A.; Ling, L.S.; Moir, D.T.; King, B.L.; Brown, E.D.; Doig, P.C.; Smith, D.R.; Noonan, B.; Guild, B.C.; deJonge, B.L.; et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen helicobacter pylori. Nature 1999, 397, 176–180. [Google Scholar] [CrossRef] [PubMed]
- Salama, N.; Guillemin, K.; McDaniel, T.K.; Sherlock, G.; Tompkins, L.; Falkow, S. A whole-genome microarray reveals genetic diversity among helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 2000, 97, 14668–14673. [Google Scholar] [CrossRef] [PubMed]
- Argent, R.H.; Thomas, R.J.; Letley, D.P.; Rittig, M.G.; Hardie, K.R.; Atherton, J.C. Functional association between the helicobacter pylori virulence factors vaca and caga. J. Med. Microbiol. 2008, 57, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, C.; Van Doorn, L.J.; Nogueira, C.; Soares, J.M.; Pinho, C.; Figueira, P.; Quint, W.G.; Carneiro, F. Helicobacter pylori genotypes are associated with clinical outcome in portuguese patients and show a high prevalence of infections with multiple strains. Scand. J. Gastroenterol. 2001, 36, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Morales-Espinosa, R.; Castillo-Rojas, G.; Gonzalez-Valencia, G.; Ponce de Leon, S.; Cravioto, A.; Atherton, J.C.; Lopez-Vidal, Y. Colonization of mexican patients by multiple helicobacter pylori strains with different vaca and caga genotypes. J. Clin. Microbiol. 1999, 37, 3001–3004. [Google Scholar] [PubMed]
- Wong, B.C.; Wang, W.H.; Berg, D.E.; Fung, F.M.; Wong, K.W.; Wong, W.M.; Lai, K.C.; Cho, C.H.; Hui, W.M.; Lam, S.K. High prevalence of mixed infections by helicobacter pylori in hong kong: Metronidazole sensitivity and overall genotype. Aliment. Pharmacol. Ther. 2001, 15, 493–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben Mansour, K.; Fendri, C.; Battikh, H.; Garnier, M.; Zribi, M.; Jlizi, A.; Burucoa, C. Multiple and mixed helicobacter pylori infections: Comparison of two epidemiological situations in tunisia and france. Infect. Genet. Evolut. 2016, 37, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Krebes, J.; Didelot, X.; Kennemann, L.; Suerbaum, S. Bidirectional genomic exchange between helicobacter pylori strains from a family in coventry, united kingdom. Int. J. Med. Microbiol. IJMM 2014, 304, 1135–1146. [Google Scholar] [CrossRef] [PubMed]
- Loh, J.T.; Shaffer, C.L.; Piazuelo, M.B.; Bravo, L.E.; McClain, M.S.; Correa, P.; Cover, T.L. Analysis of caga in helicobacter pylori strains from colombian populations with contrasting gastric cancer risk reveals a biomarker for disease severity. Cancer Epidemiol. Biomark. Prev. 2011, 20, 2237–2249. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, R.M.; Pinto-Ribeiro, I.; Wen, X.; Marcos-Pinto, R.; Dinis-Ribeiro, M.; Carneiro, F.; Figueiredo, C. Helicobacter pylori caga promoter region sequences influence caga expression and interleukin 8 secretion. J. Infect. Dis. 2016, 213, 669–673. [Google Scholar] [CrossRef] [PubMed]
- Mueller, D.; Tegtmeyer, N.; Brandt, S.; Yamaoka, Y.; De Poire, E.; Sgouras, D.; Wessler, S.; Torres, J.; Smolka, A.; Backert, S. C-src and c-abl kinases control hierarchic phosphorylation and function of the caga effector protein in western and east asian helicobacter pylori strains. J. Clin. Investig. 2012, 122, 1553–1566. [Google Scholar] [CrossRef] [PubMed]
- Dixon, M.F.; Genta, R.M.; Yardley, J.H.; Correa, P. Classification and grading of gastritis. The updated sydney system. International workshop on the histopathology of gastritis, houston 1994. Am. J. Surg. Pathol. 1996, 20, 1161–1181. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, F.; Seixas, M.; Sobrinho-Simoes, M. New elements for an updated classification of the carcinomas of the stomach. Pathol. Res. Pract. 1995, 191, 571–584. [Google Scholar] [CrossRef]
- Lauren, P. The two histological main types of gastric carcinoma: Diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol. Microbiol. Scand. 1965, 64, 31–49. [Google Scholar] [PubMed]
- Van Doorn, L.J.; Figueiredo, C.; Rossau, R.; Jannes, G.; van Asbroek, M.; Sousa, J.C.; Carneiro, F.; Quint, W.G. Typing of helicobacter pylori vaca gene and detection of caga gene by pcr and reverse hybridization. J. Clin. Microbiol. 1998, 36, 1271–1276. [Google Scholar] [PubMed]
| Chronic Gastritis (n = 234) | Gastric Carcinoma (n = 47) | p-Value | Total | |
|---|---|---|---|---|
| vacA i-region 1 | ||||
| i1 | 172 (85.1%) | 29 (85.3%) | >0.999 | 201 (85.2%) |
| i2 | 30 (14.9%) | 5 (14.7%) | 35 (14.8%) | |
| vacA m-region 2 | ||||
| m1 | 94 (53.7%) | 17 (47.2%) | 0.583 | 111 (52.6%) |
| m2 | 81 (46.3%) | 19 (52.8%) | 100 (47.4%) | |
| vacA s-region 3 | ||||
| s1 | 189 (91.7%) | 38 (88.4%) | 0.553 | 227 (91.2%) |
| s2 | 17 (8.3%) | 5 (11.6%) | 22 (8.8%) | |
| cagA status | ||||
| positive | 208 (88.9%) | 38 (80.9%) | 0.146 | 246 (87.5%) |
| negative | 26 (11.1%) | 9 (19.1%) | 35 (12.5%) | |
| vacA i-Region | vacA s-Region | vacA m-Region | cagA Status | |||
|---|---|---|---|---|---|---|
| s1 | s2 | m1 | m2 | Positive | Negative | |
| vacA i1 | 138 (89.6%) | 9 (56.3%) | 76 (85.4%) | 71 (87.7%) | 140 (90.9%) | 7 (43.8%) |
| vacA i2 | 16 (10.4%) | 7 (43.8%) | 13 (14.6%) | 10 (12.3%) | 14 (9.1%) | 9 (56.2%) |
| p-value | 0.002 | 0.823 | <0.001 | |||
| H. pylori | Chronic Gastritis | Gastric Carcinoma | p-Value | Total |
|---|---|---|---|---|
| Single infections 1 | 142 (62.0%) | 28 (65.1%) | 0.735 | 170 (62.5%) |
| Multiple infections 2 | 87 (38.0%) | 15 (34.9%) | 102 (37.5%) |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinto-Ribeiro, I.; Ferreira, R.M.; Batalha, S.; Hlaing, T.; Wong, S.I.; Carneiro, F.; Figueiredo, C. Helicobacter pylori vacA Genotypes in Chronic Gastritis and Gastric Carcinoma Patients from Macau, China. Toxins 2016, 8, 142. https://doi.org/10.3390/toxins8050142
Pinto-Ribeiro I, Ferreira RM, Batalha S, Hlaing T, Wong SI, Carneiro F, Figueiredo C. Helicobacter pylori vacA Genotypes in Chronic Gastritis and Gastric Carcinoma Patients from Macau, China. Toxins. 2016; 8(5):142. https://doi.org/10.3390/toxins8050142
Chicago/Turabian StylePinto-Ribeiro, Ines, Rui M. Ferreira, Sellma Batalha, Thazin Hlaing, Sio In Wong, Fatima Carneiro, and Ceu Figueiredo. 2016. "Helicobacter pylori vacA Genotypes in Chronic Gastritis and Gastric Carcinoma Patients from Macau, China" Toxins 8, no. 5: 142. https://doi.org/10.3390/toxins8050142