1. Introduction
Russell’s viper (
Daboia russelii) is a medically important snake, variants of which are distributed throughout East and Southeast Asia. A Russell’s viper bite has a 60% morbidity rate and the fatality rate is 8.2% in Myanmar [
1]. The cause of death includes shock, massive bleeding and renal failure. Snake venom metalloproteinases (SVMPs) play a major role in the local and systemic clinical manifestations: blistering, necrosis and bleeding from the fang marks and incoagulable blood, thrombocytopenia, spontaneous systemic bleeding, hypotension, increased permeability and reduced urine output [
2]. Although Russell’s viper antivenoms are available, their efficacy in reversal tissue damage, such as acute renal failure, is limited [
3]. Novel treatment modalities are required.
Snake venom metalloproteinases (SVMPs) play major roles in pathogenesis of Russell’s viper bites [
4,
5]. SVMPs are categorised into P-I to P-III classes according to their domain organization with different molecular weights [
6]: Class I (P-I) contains only a prodomain and a metalloproteinase domain (20–30 kDa); Class II (P-II) contains a prodomain, metalloproteinase domain followed by disintegrin domain (30–60 kDa); Class III (P-III) contains a pro, metalloproteinase, disintegrin-like and cysteine-rich domain (60–100 kDa). There are subclasses in P-II and P-III depending on post-translational modifications. The variation in domain composition between SVMP classes contributes to a wide spectrum of substrate specific proteolytic activity. The active site of the metalloproteinase domain has a consensus H
142EXXHXXGXXH
152 sequence. The catalytic zinc-ion is located at the bottom of the active-site cleft, and tetrahedrally coordinated by His
142, His
146, His
152, and a water molecule anchored to Glu
143 [
7]. The degradation of endothelial cell membrane proteins (integrin and cadherin), basement membrane components (fibronectin, laminin and collagen) and blood coagulation proteins (fibrinogen, factor X and prothrombin) leads to haemorrhage. Generally, P-III SVMPs have more potent haemorrhagic activity than P-I and P-II SVMPs [
8].
In order to protect against auto-digestion by SVMPs, snake venom of several species are found to contain natural protease inhibitors: citrate and small peptides. The latter bind selectively to SVMPs in the venom glands to protect glandular tissues and venom factors from self-digestion by SVMPs [
9]. Three endogenous peptides: pyroGlu-Lys-Trp (pEKW), pyroGlu-Asn-Trp (pENW) and pyroGlu-Gln-Trp (pEQW) isolated from venom of Taiwan habu (
Trimeresurus mucrosquamatus) showed an inhibitory action on proteolytic activity of metalloproteinases present in the crude venom [
10]. It is reported that these peptide inhibitors regulate the proteolytic activities of their SVMPs in a reversible manner under physiological conditions [
11]. Other pit vipers, such as
Bothrops asper [
12] and some rattlesnakes [
13], also have venoms containing endogenous tripeptides: pEQW and pENW. African vipers,
Echis ocellatus and
Cerastes cerastes cerastes, have pEKW tripeptides. These tripeptides are encoded by tandemly repeating elements from the transcripts which also contain a CNP (C-type natriuretic peptide) homologous sequence at the C-terminus [
14]. Two peptides: PtA (pENW) and PtB (pEQW) isolated from venom liquor of
Deinagkistrodon acutus (Hundred-pacer viper) showed anti-human platelet aggregation activity in vitro and protection effects on ADP-induced paralysis and formation of pulmonary thrombosis in mice [
15].
We hypothesized that Myanmar Russell’s viper venom might contain endogenous peptides to neutralise its own potent SVMPs. The goal of this research was to purify and identify specific SVMP inhibitors (SVMPIs) from the venom as well as from venom glands and to determine their inhibitory action on purified SVMPs from same source of venom. From the transcriptome of the snake, novel SVMPI transcripts containing tripeptide motifs and ANP (atrial natriuretic peptide) sequences were found. Two tripeptides were purified from the venom and identified as pERW and pEKW. Their effect on biological activities of two SVMPs: RVV-X and Daborhagin from the same venom, purified through newly developed strategy, were examined. Both synthetic peptides showed complete inhibitory action on the gelatinolytic activity of RVV-X and fibrinogenolytic activity of Daborhagin at 5 mM concentration (approximate protease to inhibitor molar ratio of 1:500). The results might contribute to the development of complementary candidates for current antivenom therapy of Russell’s viper bites, as well as for novel therapeutic agents for cardiovascular diseases.
3. Discussion
In the current study, we have developed a new method which can be used to simultaneously isolate the two SVMPs, RVV-X and Daborhagin, from Myanmar Russell’s viper venom. The relative amounts of these enzymes in the venom were determined. In addition, four novel RNA sequences of SVMP inhibitor (MRV1-4) were derived from the venom gland transcriptome. These sequences are different from those of previously reports in other snakes. For the first time in studies of Russell’s viper venom, two tripeptide SVMP inhibitors, pERW and pEKW have been isolated. Evidence for the complete inhibition of RVV-X and Daborhagin activities by these tripeptides is presented to support our hypothesis.
Russell’s viper is a venomous species of the South-East Asian region. The clinical manifestations of its bites reflect the high content of proteases such as snake venom serine proteases and snake venom metalloproteinases (SVMPs). It has been shown that the SVMPs comprise approximately 11% to 65% of the total protein in the Viperidae venoms [
17]. In Myanmar Russell’s viper, SVMPs contribute to 20% of the crude venom (data not shown) and Class III SVMPs are found to be the major component. In comparison with other species, the Myanmar species have 6–7 times more Daborhagin than Indian species [
16] and SVMPs, mainly RVV-X, in Sri Lankan species comprise just 6.9% of the crude venom [
18]. Thus, it can be noted that the Myanmar venom contains greater amounts of SVMPs than that of the Indian and Sri Lankan species. The variations in types and amounts of SVMPs in venom among different subspecies of Russell’s viper might be due to diversity in their prey at different locations and this could lead to the dissimilar severity or clinical presentations of snakebite patients.
Russell’s viper venom factor X activator (RVV-X) is a well-characterised Class III metalloproteinase (formally known as Class IV) which specifically activates coagulation factor X by hydrolysis of an Arg-Ile bond in factor X. It is a glycoprotein consisting of a heavy chain (α-chain, 57.6 kDa) and two light chains (β- and γ-chains, 19.4 kDa and 16.4 kDa) linked by disulfide bonds [
19]. In addition to proteolytic activity on factor X and IX, RVV-X also inhibits collagen- and ADP-stimulated platelet aggregation [
20] and has a strong affinity for protein S [
21]. Factor X activators are also found in
Vipera lebetina (blunt-nosed viper) in which it exhibits specific proteolytic activity towards human factor X and also factor IX, but it is not active against prothrombin nor fibrinogen [
22].
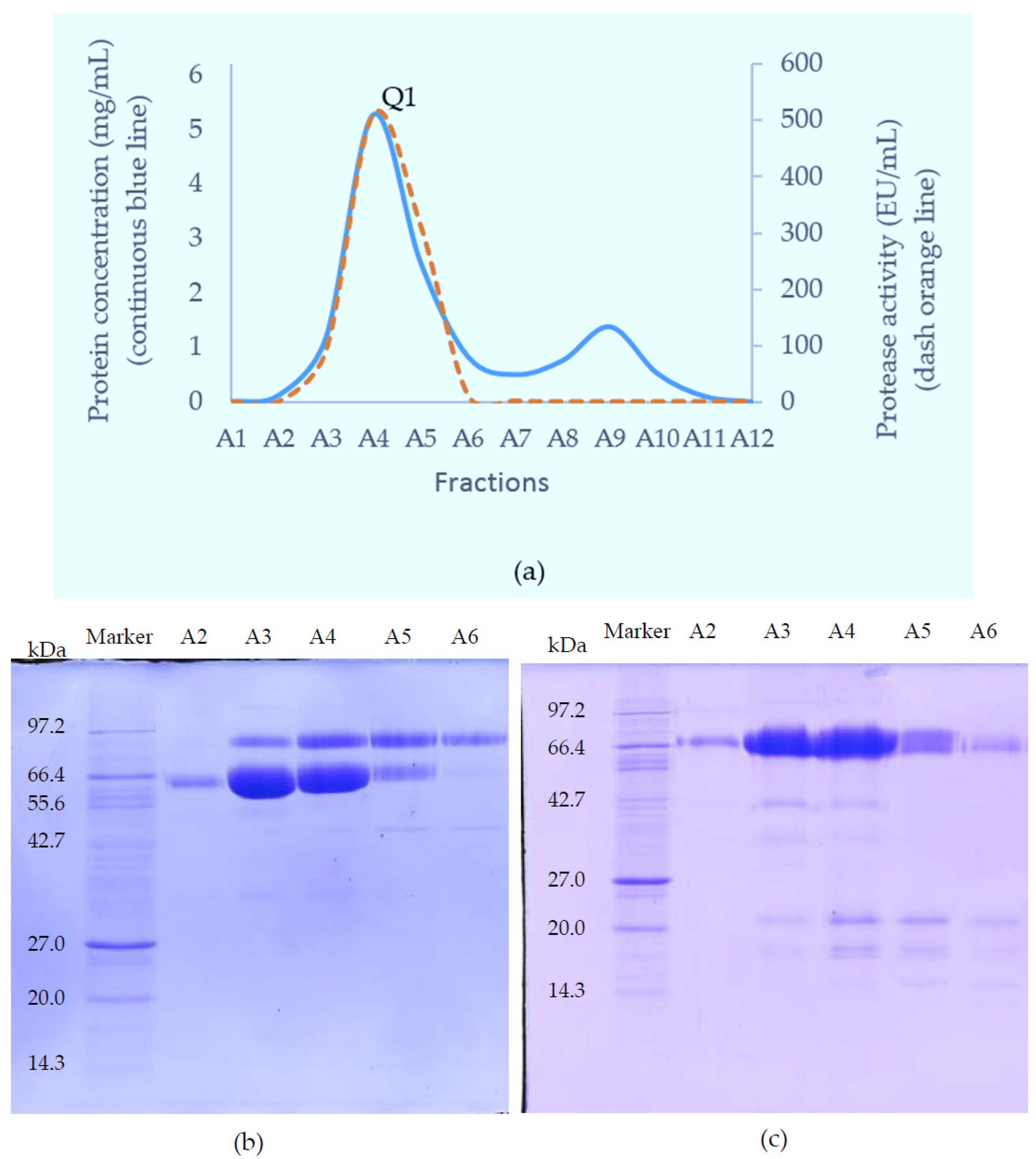
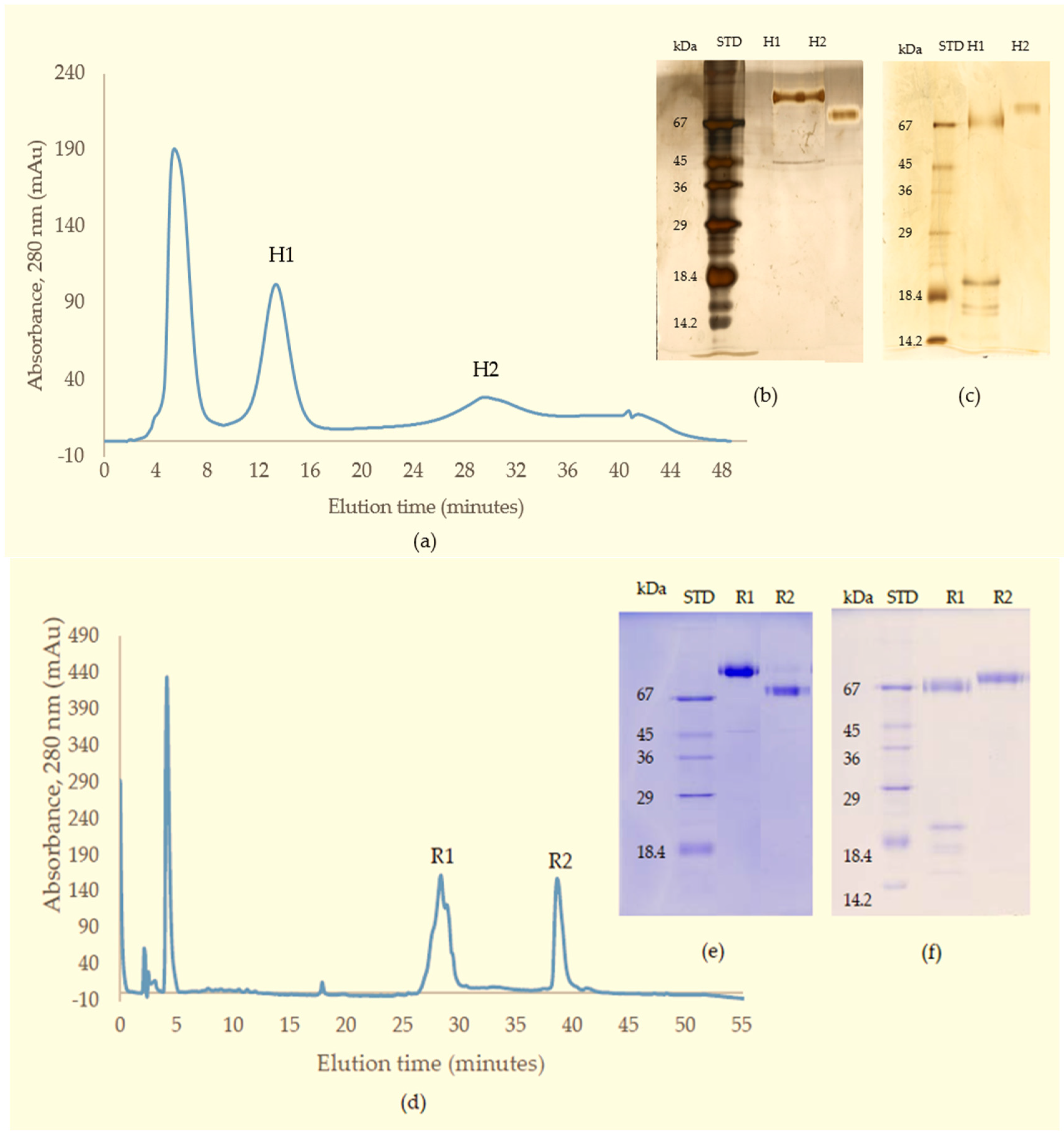
In the present study, the purified RVV-X was shown to be composed of a heavy chain (67 kDa) and two light chains (20 kDa and 15 kDa). The two thin bands on SDS-PAGE at around 15 kDa level suggested that the γ-light chain in Myanmar species might exist as 2 forms, likely due to either amino acid variation or differences in N-glycosylation. Our experiments showed that MRV RVV-X possesses hydrolytic activity to gelatin (Type I collagen, bovine), which had not been characterised before for RVV-X.
Another potent Class III SVMP, Daborhagin, composed of metalloproteinase, disintegrin and cysteine-rich domains, was also purified from MRV venom. The Daborhagin-M from Myanmar Russell’s viper venom specifically digested the α-chain of fibrinogen, fibronectin and type IV collagen in vitro and exhibited haemorrhagic [
16], edema inducing and myonecrotic activity in mice [
23]. In our studies, a 67 kDa metalloproteinase was isolated and matched to Daborhagin-K from Indian species using mass spectrometric analysis of tryptic peptides. This MRV Daborhagin exhibited potent α-fibrinogenolytic activity, but did not digest gelatin.
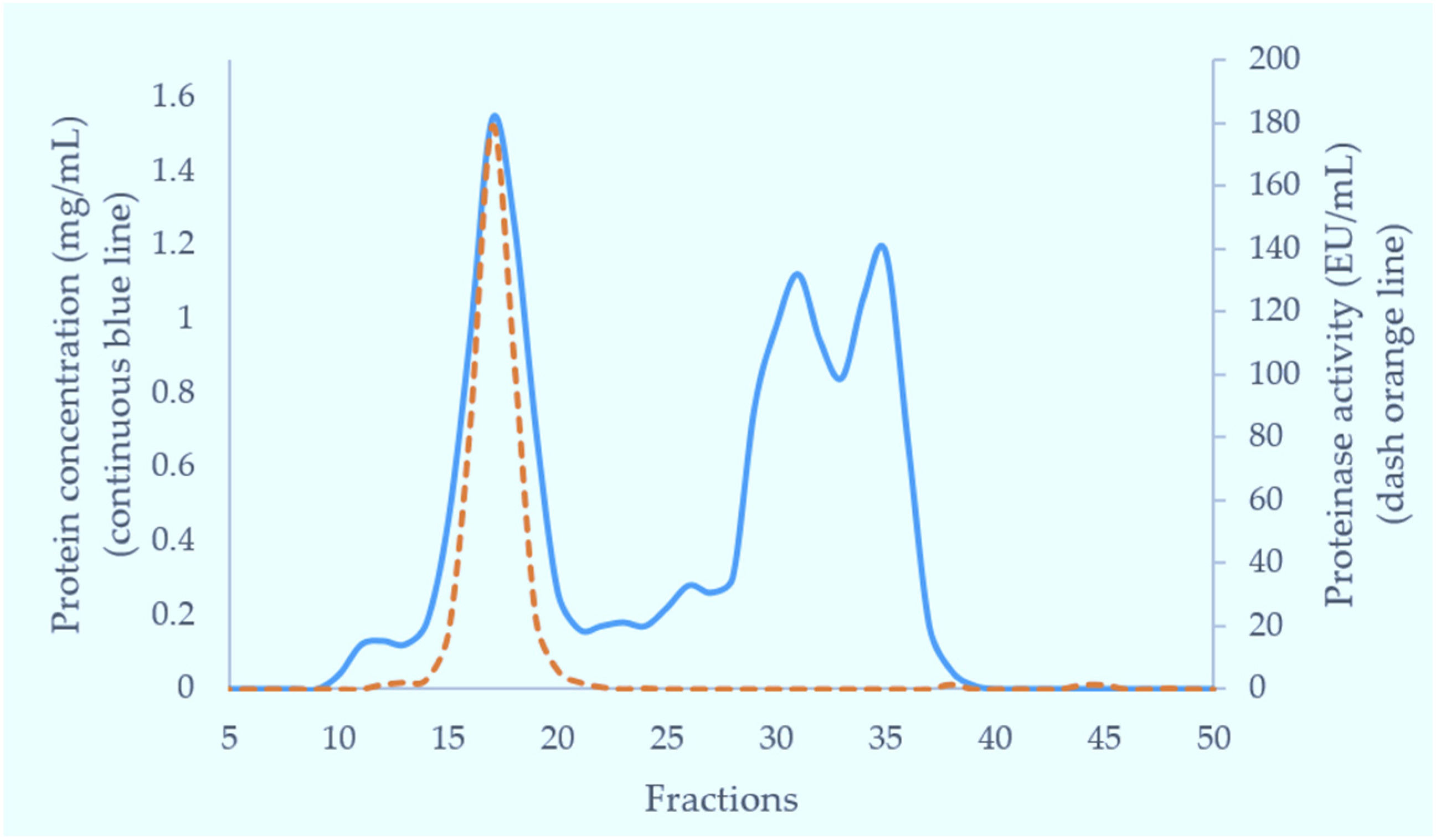
In the current purification strategy, the two SVMPs were co-purified initially, but then could be separated from each other using either hydrophobic interaction chromatography or RP-HPLC. Better resolution was evident on RP-HPLC, and the presence of multiple forms of RVV-X was indicated by the irregularity of the RVV-X RP-HPLC peak, suggesting heterogeneity of the protein (R1,
Figure 3d). Two isoforms of the heavy chain and 6 isoforms of the light chain from RVV-X have been revealed on 2-D electrophoresis in the proteomic study of Risch, M et al. in the same species [
24].
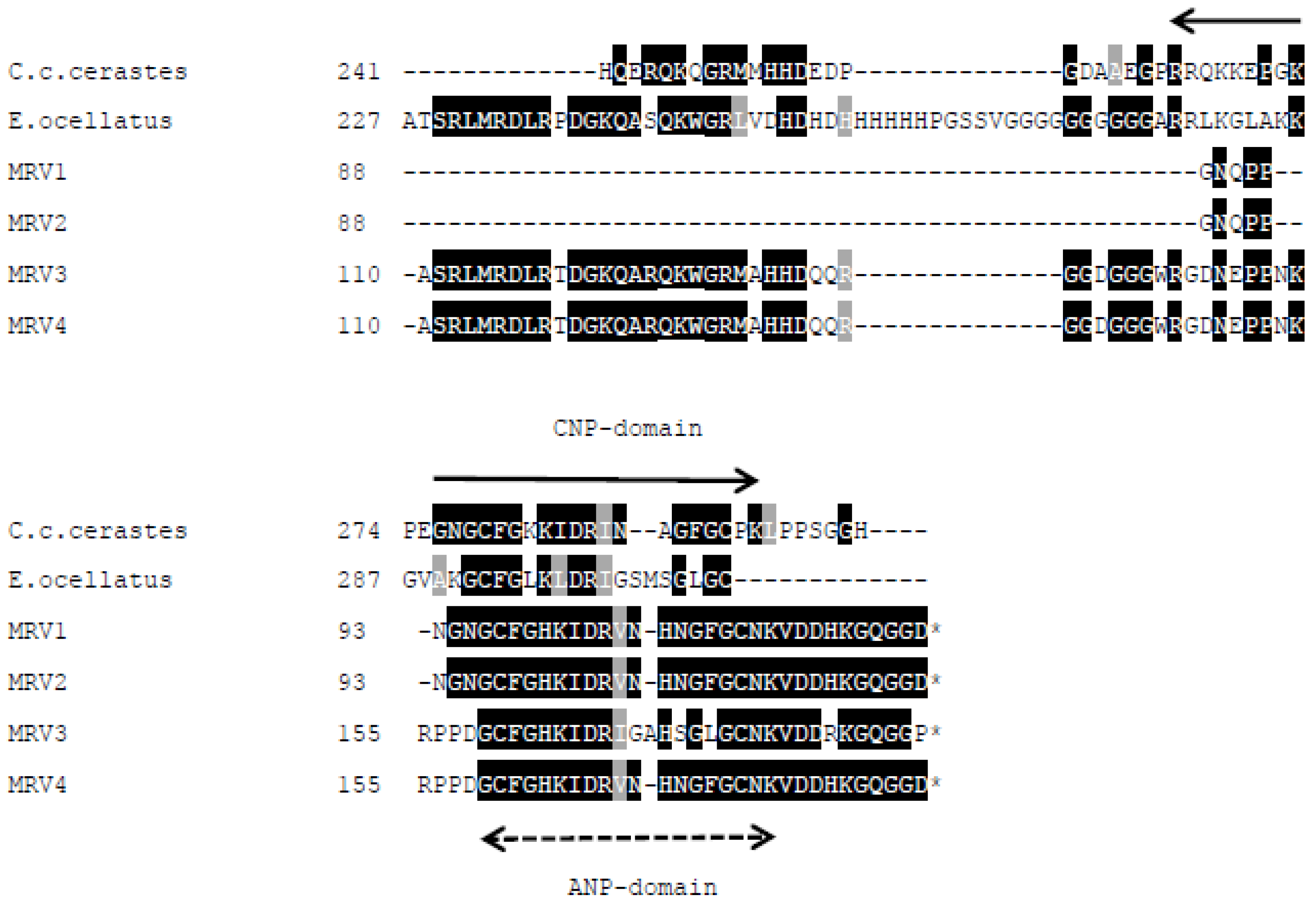
New SVMPI transcripts from Myanmar Russell’s viper were discovered containing novel two inhibitory tripeptides, QKW and QRW. The tripeptide sequences are found in the same transcript as natriuretic peptide sequences, as is the case in African vipers. This assortment of different peptide sequences in the same transcript could be related to independent evolution of toxin genes in snakes. The conserved proline residues in the consensus sequence PXXQ(K/R)WXXP might be a signal point for cleavage of tripeptides from transcripts. The mechanism for release of tripeptides from their transcripts is still unknown. These tripeptides and natriuretic peptides are observed separately in venom, although they are encoded from the same transcript. Since ANP is homologous to hormone, it might be processed near the effective cells. The release and modification of tripeptides [
25] might probably occur during the exocytosis process at an earlier stage than the natriuretic peptides [
26].
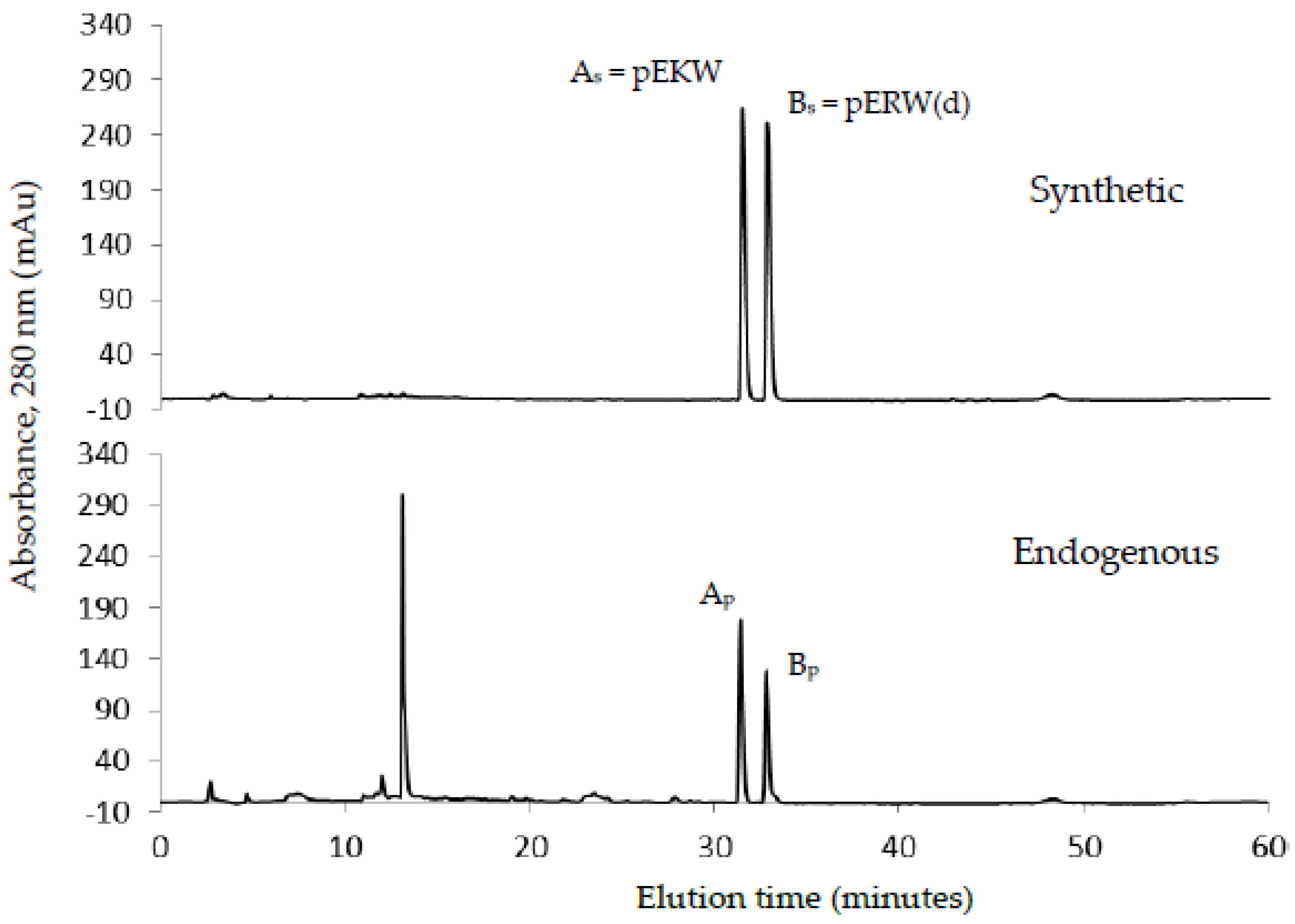
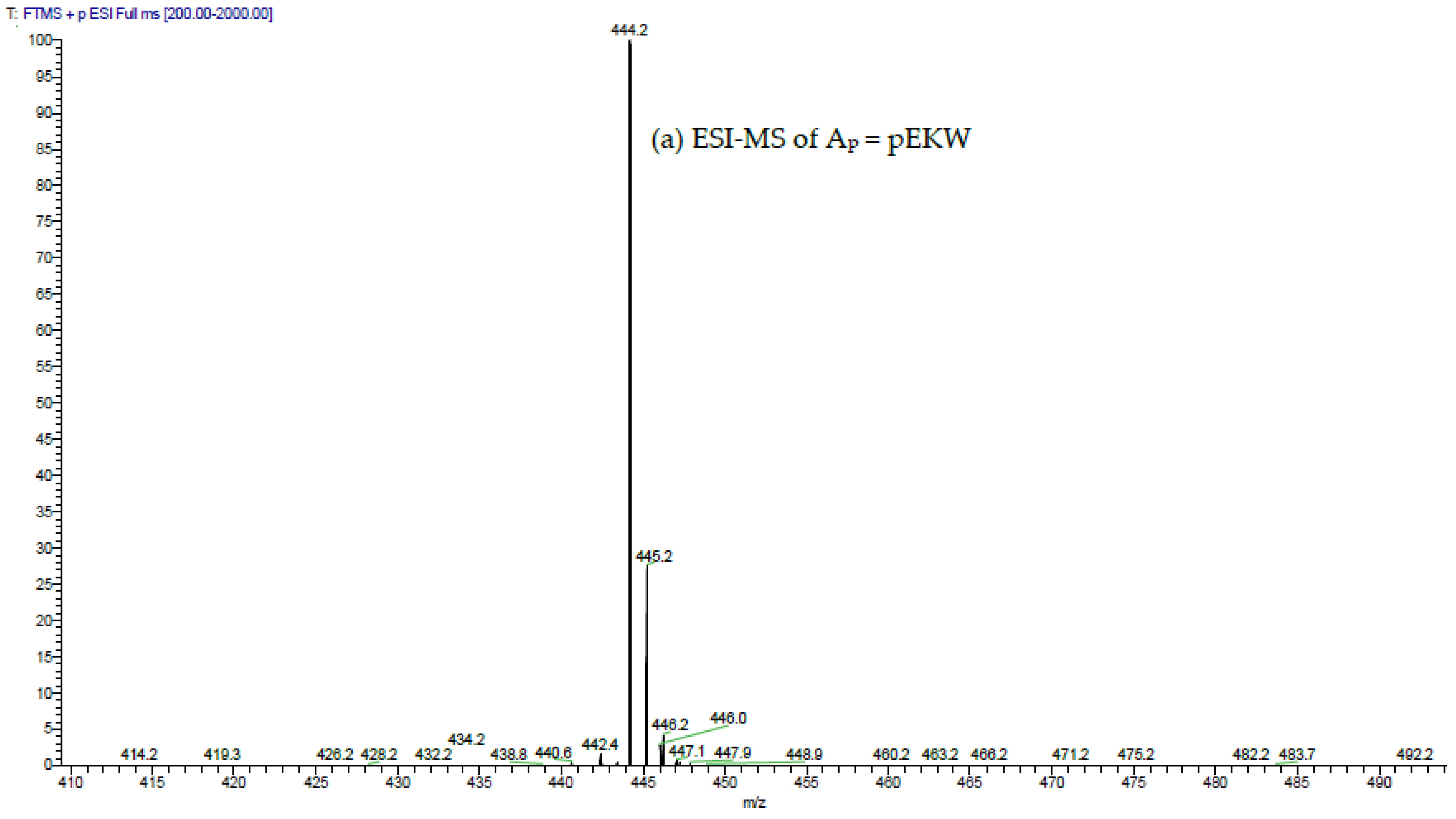
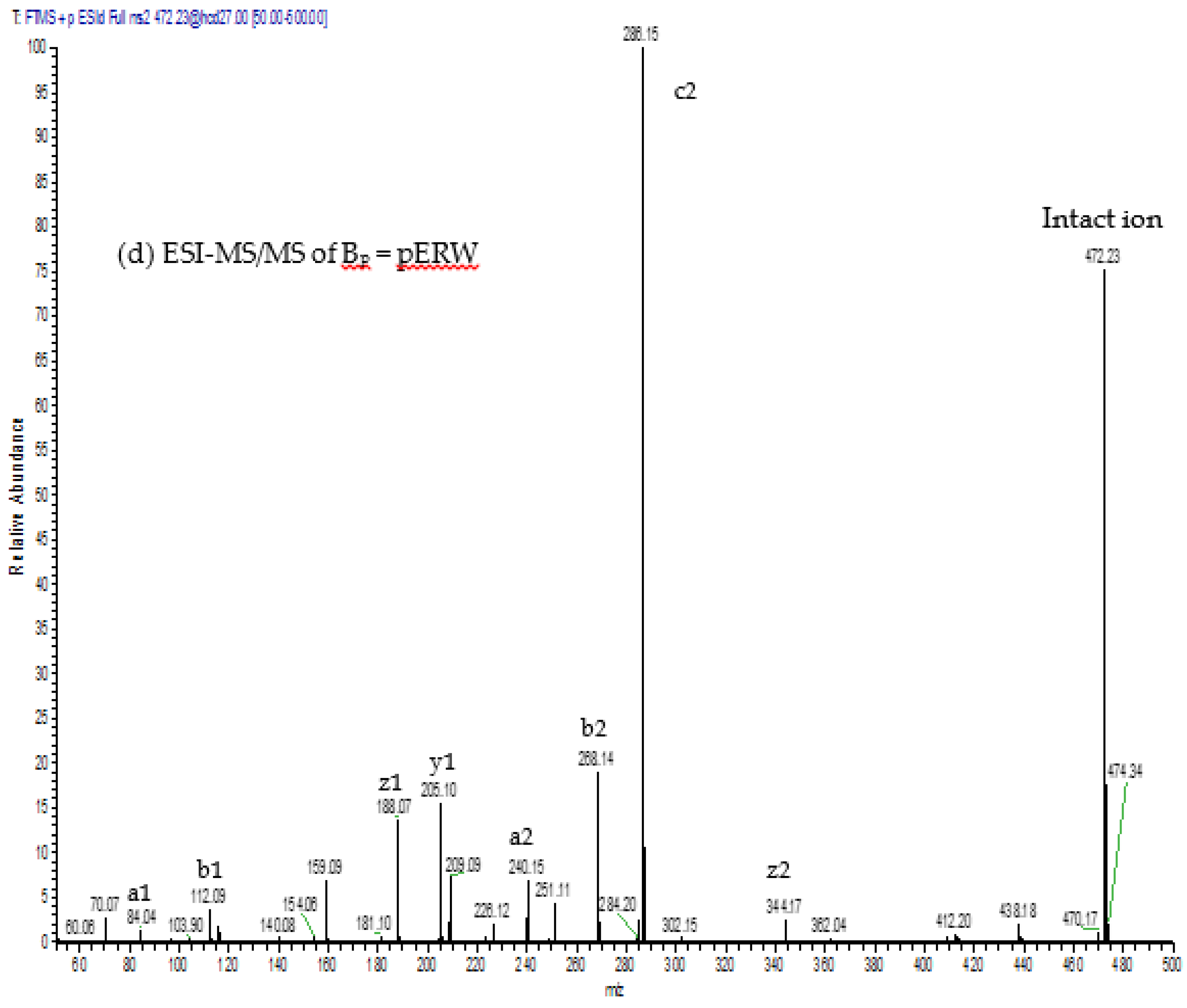
The aforementioned inhibitory tripeptides were purified from the MRV venom as their pyroglutamate forms, pEKW and pERW. These were identified using RP-HPLC (co-chromatography with synthetic peptides), LC-ESI-MS analysis of intact mass and LC-ESI-MS/MS sequencing. Although the tripeptide pEKW purified from MRV venom has been found in other snake species, such as
Trimeresurus mucrosquamatus [
10],
Echis ocellatus, and
Cerastes cerastes cerastes [
14], the tripeptide pERW purified here has not been found in the venom of any other snake species.
The synthetic tripeptides pERW, pEKW and pEEW showed complete inhibition of the gelatinolytic activity of RVV-X and of the fibrinogenolytic activity of Daborhagin at 5 mM concentration of each inhibitor. Non-selective inhibition of all three synthetic peptides on biological activities of SVMPs reflects the importance of the first pyroGlu and the final tryptophan residue in the blocking mechanism at the active site of SVMP. The crystal structure of TM-3 (a SVMP from
Trimeresurus mucrosquamatus) bound to tripeptide inhibitors (a proteinase and inhibitors model from Taiwan habu) revealed that the inhibitor Trp residue deeply inserts into the S-1 pocket of the protease and provides a greater inhibition than other smaller amino acids. Similarly, the pyro-ring of the inhibitor is required for fitting into the S-3 position of the protease and the activity of inhibitor becomes weaker in the absence of pyro-ring. The native middle residue is also position-specific to the S-2 site [
11]. Tripeptides from different species share same first (pyroGlu-) and third (tryptophan) residues. The variability of the middle residue may be dependent on species variation of the SVMPs.
5. Materials and Methods
5.1. Venoms and Venom Glands
Lyophilised crude venom was obtained from No. 1, Myanmar Pharmaceutical Factory, Yangon, Myanmar. The salivary glands from Myanmar Russell’s viper (Daboia russelii siamensis) were dissected in the Snake Farm, MPF, Yangon, Myanmar and used for RNA-Seq (RNA sequencing). The experimental plan was approved by the Animal Care and Use Committee, Chulalongkorn University (CU-ACUC) (No. 17/2558). Synthetic tripeptides (98% purity) were purchased from Severn Biotech Ltd., Worcestershire, UK and supplied with data from MS analysis to confirm their masses to be within 0.4% of the predicted values.
5.2. Transcripts Analysis of SVMPIs
Next-generation sequencing of mRNA from Myanmar Russell’s viper (2 adult males and 2 adult females) venom glands was performed on an Illumina HiSeq2000 platform. De novo assembly was performed using Trinity (r20140717). Annotation of SVMPI transcripts were archived through Blastn searches against the collected NCBI nucleotide database with search words “venom” and “serpents”. The annotations with a high score at the top hit list were picked up.
The SVMPI transcripts were further analysed with Blastx and ORF finder for final best annotation and identification of the full-length transcript. The alignment of translated SVMPI sequences with those from other snakes were performed by using Clustal Omega followed by shading with BOXSHADE 3.21 (K. Hofmann, Koeln, Germany & M. Baron, Surrey, UK).
5.3. Protein Concentration
Protein concentration was determined by using Bradford reagent (BioRad, Hemel Hempstead, UK). The absorbance was measured at 595 nm and the calibration curve was prepared with a bovine gamma globulin standard (0–1.5 mg/mL).
5.4. Purification of SVMPs
All chromatographic procedures were performed on either a Bio-Cad Vision Workstation (GFC) or a GE Healthcare AKTA System (anion-exchange, HIC and RP-HPLC).
5.4.1. Gel Filtration Chromatography (GFC)
Crude venom (100 mg) dissolved in 5 mL of 0.01 M phosphate buffered saline (pH 7.4) was applied to a Superdex 200 column (5 × 160 cm) pre-equilibrated with the same buffer. Elution was carried out with the same buffer. The flow rate was 2 mL/min and 6 mL fractions were collected. The fractions having metalloproteinase activity (fractions 15–18) were combined for further purification. The fractions eluting near the total volume were analysed for tripeptides using RP-HPLC with subsequent MS analysis.
5.4.2. Anion-Exchange Chromatography
The SVMP-containing sample obtained from GFC was applied to a Resource Q anion-exchange column (6 mL) pre-equilibrated with 0.05 M Tris-Cl buffer (pH 8.0). Elution was achieved with a linear NaCl gradient from 0 to 0.5 M in the same buffer at a flow rate of 0.6 mL/min and 1.8 mL fractions were collected. Elution was monitored at 280 nm.
5.4.3. Hydrophobic Interaction Chromatography (HIC)
To further purify the SVMPs for activity measurements, fractions from Resource Q were loaded onto a Phenyl Superose column (1 mL) equilibrated in 2.5 M NaCl, 50 mM Tris-Cl, pH 7.8. Samples in Tris-Cl were adjusted to 2.5 M in NaCl and were centrifuged at 10,000× g for 5 min before loading onto the column. Separation was achieved by a 30 min-gradient of 2.5–0 M NaCl in 50 mM Tris-Cl, pH 7.0, using a flow rate of 0.25 mL/min. Elution was monitored at 280 nm and 0.25 mL fractions were collected.
5.4.4. Reversed Phase High Performance Liquid Chromatography (RP-HPLC)
For MS analysis, SVMPs were purified using RP-HPLC rather than HIC. Fractions from Resource Q chromatography were made up to 0.2% (v/v) in TFA, centrifuged at 10,000× g for 5 min and then applied to Phenomenex Aeris C4 column (150 × 2.1 mm, 5 micron). The proteins were separated in a two-part acetonitrile gradient in 0.08% TFA: 0%–40% over 25 min then 40%–65% over 5 min and elution was monitored at 280 nm. The flow rate was 0.15 mL/min and 0.25 mL fractions were collected.
5.5. Purification of Tripeptides
Fractions from GFC suspected to contain small molecular weight components were made up to 0.2% (v/v) in TFA, centrifuged at 10,000× g for 5 min and applied to Phenomenex Luna C18 RP-HPLC column (100 × 2.1 mm) equilibrated in 0.08% TFA. The components were separated at 0.15 mL/min with a three-part acetonitrile gradient in 0.08% TFA: 0%–12% over 5 min, 12%–28% over 50 min and then 28%–65% over 10 min. Elution was monitored at 280 nm.
5.6. Mass Spectrometric Analyses
Putative RVV-X (10 μg of R1 from RP-HPLC) was reduced, treated with iodoacetamide and digested with 1.0 μg trypsin in the presence of 2 M urea. The resulting peptides were analysed by LC-ESI-MS/MS using an Acquity UPLC CSH Peptide C18 RP column (Waters, Milford, MA, USA) connected to a Q-Exactive (ThermoFisher, Northumberland, UK) MS instrument. Peaks Studio 8.0 (BSI, Waterloo, Canada) was used to analyse the resulting MS/MS data against the sequences for Eastern Russell’s viper RVV-X H chain VM3CX_DABSI (Q7LZ61) and light chains LC1 SLLC1_DABSI (Q4PRD1) and LC2 SLLC2_DABSI (Q4PRD2).
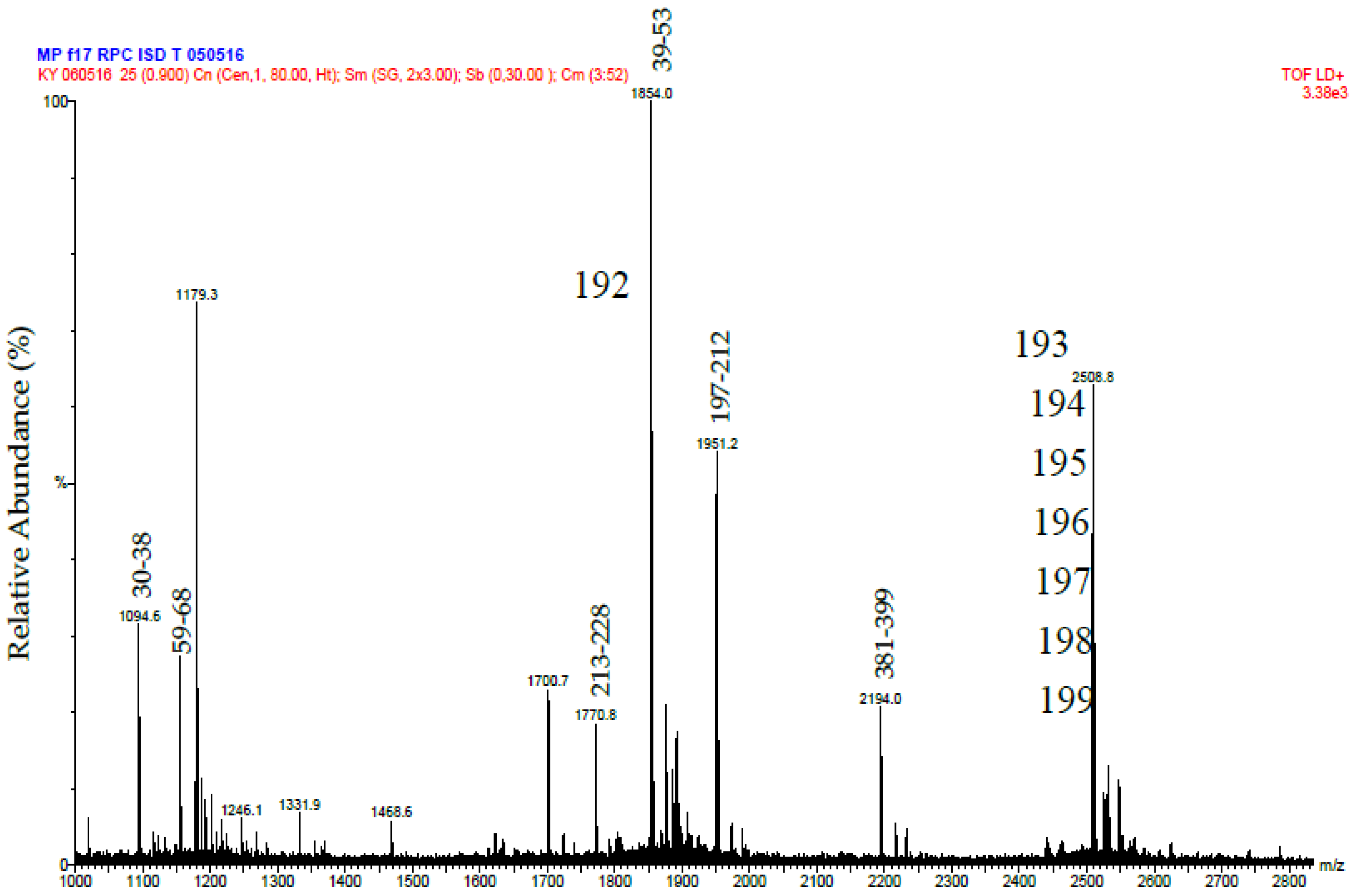
Putative Daborhagin (5 μg of R2 from RP-HPLC) was reduced, treated with iodoacetamide and digested with 0.5 μg trypsin in the presence of 2 M urea. The resulting peptides were desalted and mass spectrometric analysis was performed using a MALDI-TOF instrument (Waters-Micromass, Milford, MA, USA). Samples were analysed by mixing a 1 μL solution of the tryptic peptides with an equal volume of 5.7 mg/mL α-cyano-4-hydroxycinnamic acid in 60% acetonitrile/0.1% trifluoroacetic acid and laying this onto a dried bed of 1 μL of 25 mg/mL α-cyano-4-hydroxycinnamic acid. Laser energy was set at 25% and detector voltage 1800 V. Ion spectra were collected in the mass range of 1000–3000 Da. Data analysis was performed using MassLynx (Waters, Milford, MA, USA). The tryptic peptide masses obtained were matched manually with those predicted (using ExPASy Peptide Mass) of a sequence for Daborhagin-K (VM3DK_DABRR; B8K1W0) retrieved from UniprotKB [
27] using search word ‘Daborhagin’.
The purified tripeptides were analysed by ESI-MS and ESI-MS/MS using the same instrument and conditions as used to analyse the tryptic peptides from RVV-X.
5.7. Analysis by SDS-PAGE
Protein purity was determined by SDS-PAGE [
28] on a 12% or 15% resolving gel and 4% stacking gel using a Mini-PROTEAN 3 electrophoresis system (BioRad, Hemel Hemstead, UK). Samples were loaded in either reduced or non-reduced form. Gels were run at 200 V, 30 mA per gel, for 50 min. Proteins were visualised with Coomassie Brilliant Blue R250 V followed by destaining with methanol: water: acetic acid (30:60:10). Alternatively, proteins were visualised by silver staining as performed by method of Heukeshoven & Dernick [
29].
5.8. Caseinolytic Activity
The proteolytic activity was estimated by hydrolysis of heated casein using the Anson method [
30]. The reaction mixture, consisting of 500 μL casein (20 mg/mL) in 0.1 M Tris-Cl (pH 8.0), 20 μL venom was incubated for 30 min at 37 °C. The reaction was quenched by the addition of 500 μL of 5% trichloroacetic acid (TCA) at room temperature. After centrifugation at 10,000×
g for 5 min, the hydrolysed substrate un-precipitated with TCA was determined by Folin Ciocalteau method [
31]. Thus, 400 μL of the supernatant was mixed with 1 mL of 0.5M Na
2CO
3 and 200 μL of diluted (1:5) Folin & Ciocalteau’s phenol reagent. The mixture was then incubated at 37 °C for 30 min and the absorbance was measured at 660 nm. One enzyme unit is defined as the amount of enzyme which hydrolyses casein to produce color equivalent to 1.0 μmole of tyrosine per minute at pH 8.0 at 37 °C.
5.9. Gelatinolytic Activity
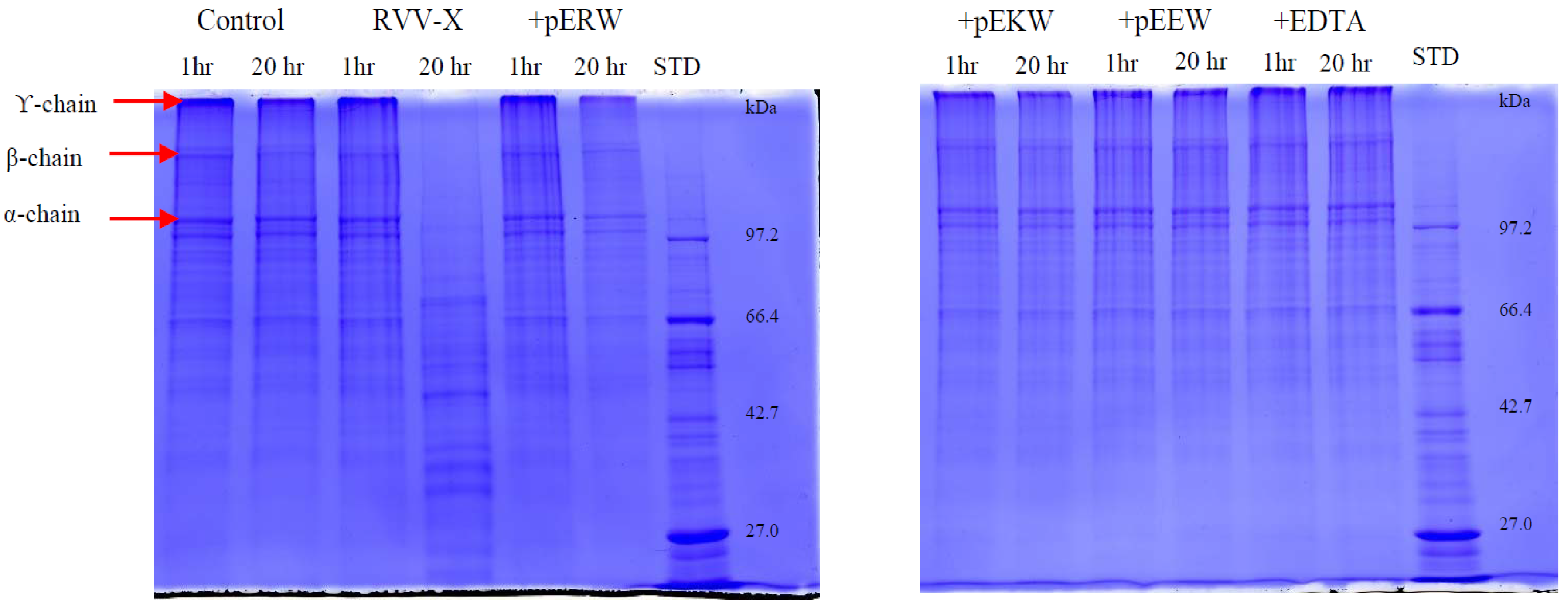
The gelatinolytic activity of the purified enzyme was analysed by zymography [
32]. The purified metalloproteinase was diluted in SDS sample buffer under non-reducing conditions and run on 10% SDS-polyacrylamide gels (0.75 mm) co-polymerised with 0.5 mg/mL of gelatin. After electrophoresis, the gels were washed in 2.5% Triton X-100 for 30 min and then washed three times in distilled water to remove any Triton. Gels were then incubated in developing buffer (50 mM Tris-Cl, pH 7.8, 200 mM NaCl, 5 mM CaCl
2, 0.02% Brij 35) for 18 h at 37 °C. The gels were stained with 0.5% Coomassie blue R-250 in methanol: acetic acid: water (5:10:85) solution and subsequently destained in methanol: acetic acid: water (10:5:85). The presence of gelatinolytic activity was defined as clear bands on the dark blue background.
5.10. Fibrinogenolytic Activity
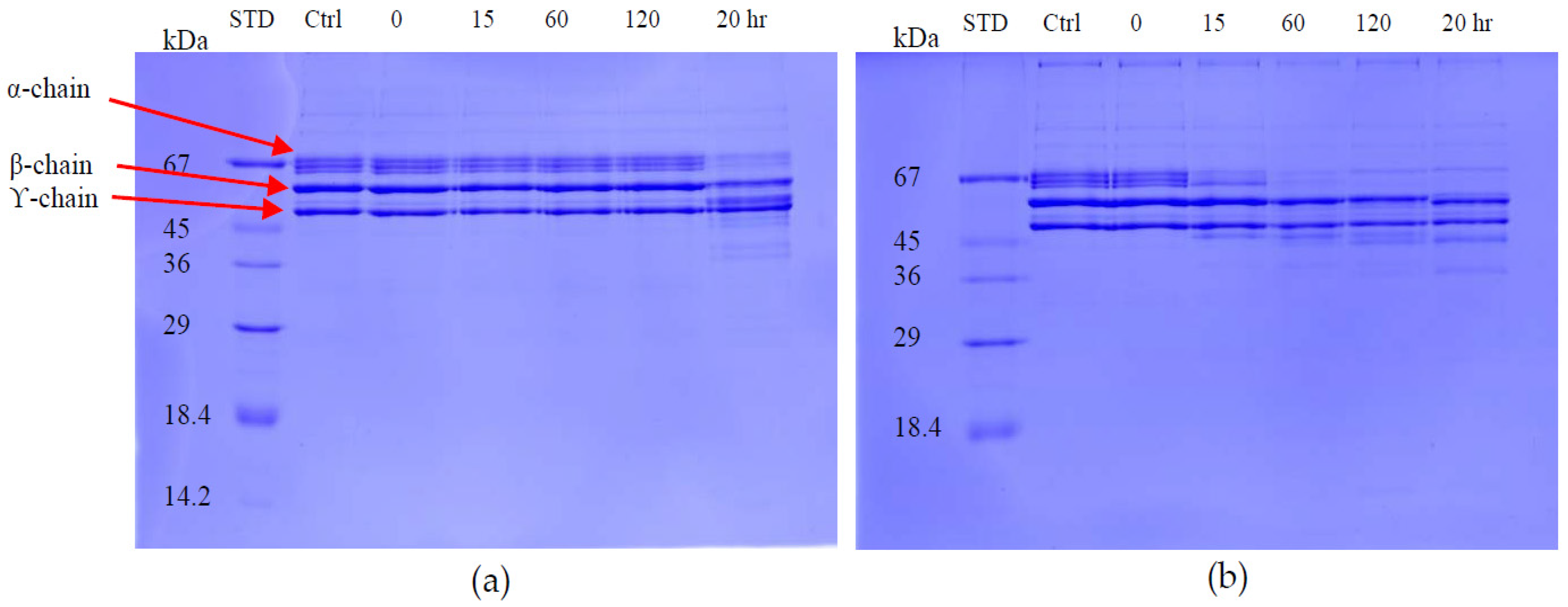
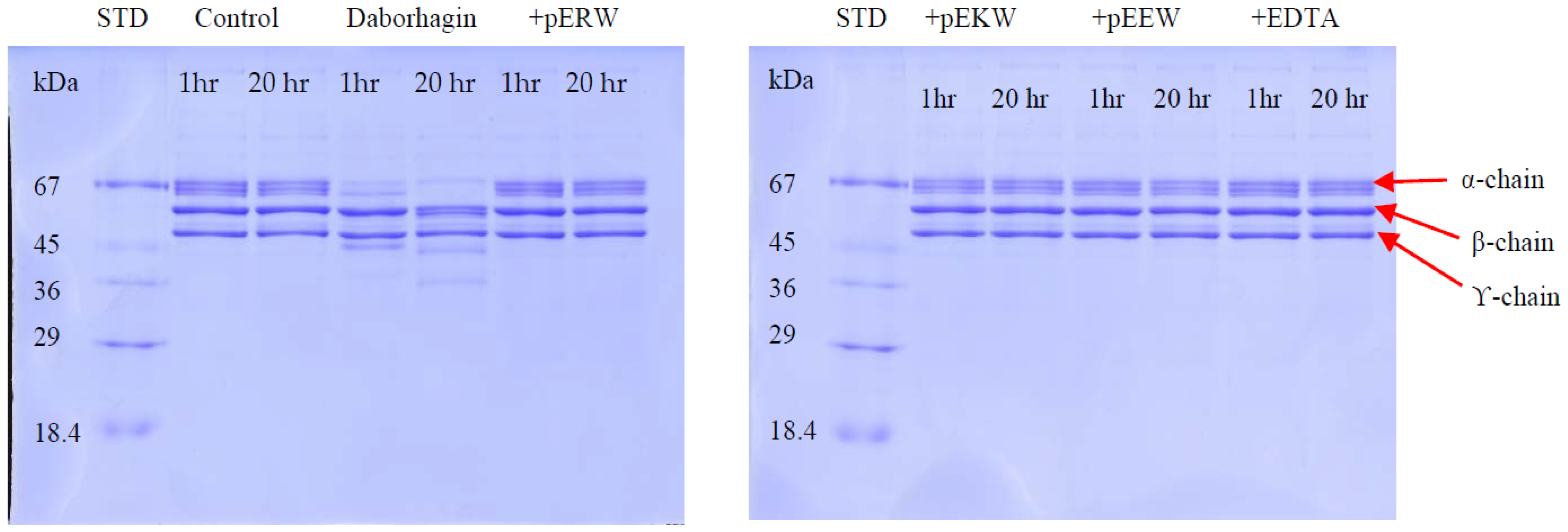
The fibrinogenolytic activity was assayed by SDS-PAGE (4% stacking/12% resolving gel) as described by Ouyang & Teng [
33]. Equal volumes of fibrinogen (1 mg/mL in 0.05 M Tris-Cl, pH 8.5) and 20 μg/mL of enzyme were incubated at 37 °C for various times intervals. At 0, 5, 15, 30, 60 and 120 min, 200 μL of the incubated solution was mixed with 400 μL of denaturing buffer containing 0.2 M Tris-Cl (pH6.8), 20% glycerol, 10% sodium dodecyl sulfate (SDS), 0.05% bromophenol blue and 10 mM β-mercaptoethanol and heated at 100 °C for 10 min to stop the digestion. Proteolytic activity was determined on the Coomassie blue-stained gel after electrophoresis by observing the cleavage patterns of purified fibrinogen chains.
5.11. Inhibition of Gelatinolytic Activity
The effect of synthetic tripeptides and EDTA on purified protein was assayed using SDS-PAGE (4% stacking/10% resolving gel) to determine gelatin degradation. The purified protein (10 ng/μL) was incubated firstly with synthetic tripeptide (5 mM) or EDTA (100 μM) at 37 °C for 10 min. Then, 10 μL of gelatin solution (2 mg/mL in distilled water) was added and 20 μL of this incubated solution was taken out at 1 h and 20 h, mixed with 5 μL of 5× denaturing buffer and heated at 95 °C for 2 min. The cleavage patterns on gelatin by the enzyme was observed on Coomassie blue-stained gels after electrophoresis.
5.12. Inhibition of Fibrinogenolytic Activity
The effect of synthetic tripeptides or EDTA on purified protein was assayed using SDS-PAGE (4% stacking/12% resolving gel) to determine fibrinogen degradation. The purified protein (32 ng/μL) was incubated firstly with synthetic tripeptide (5 mM) or EDTA (100 μM) at 37 °C for 10 min. Then, 10 μL of fibrinogen solution (2 mg/mL in distilled water) was added and 20 μL of this incubated solution was taken out at 1 h and 20 h, mixed with 5 μL of 5× denaturing buffer and heated at 95 °C for 2 min. The cleavage effect on fibrinogen chains by the SVMPs were observed on Coomassie blue-stained gel following electrophoresis.