Bioactivity of Natural and Engineered Antimicrobial Peptides from Venom of the Scorpions Urodacus yaschenkoi and U. manicatus
Abstract
:1. Introduction
2. Results
2.1. Peptide Engineering
2.2. Antimicrobial Assays
2.3. Hemolytic Assays
2.4. Cytotoxicity Assays
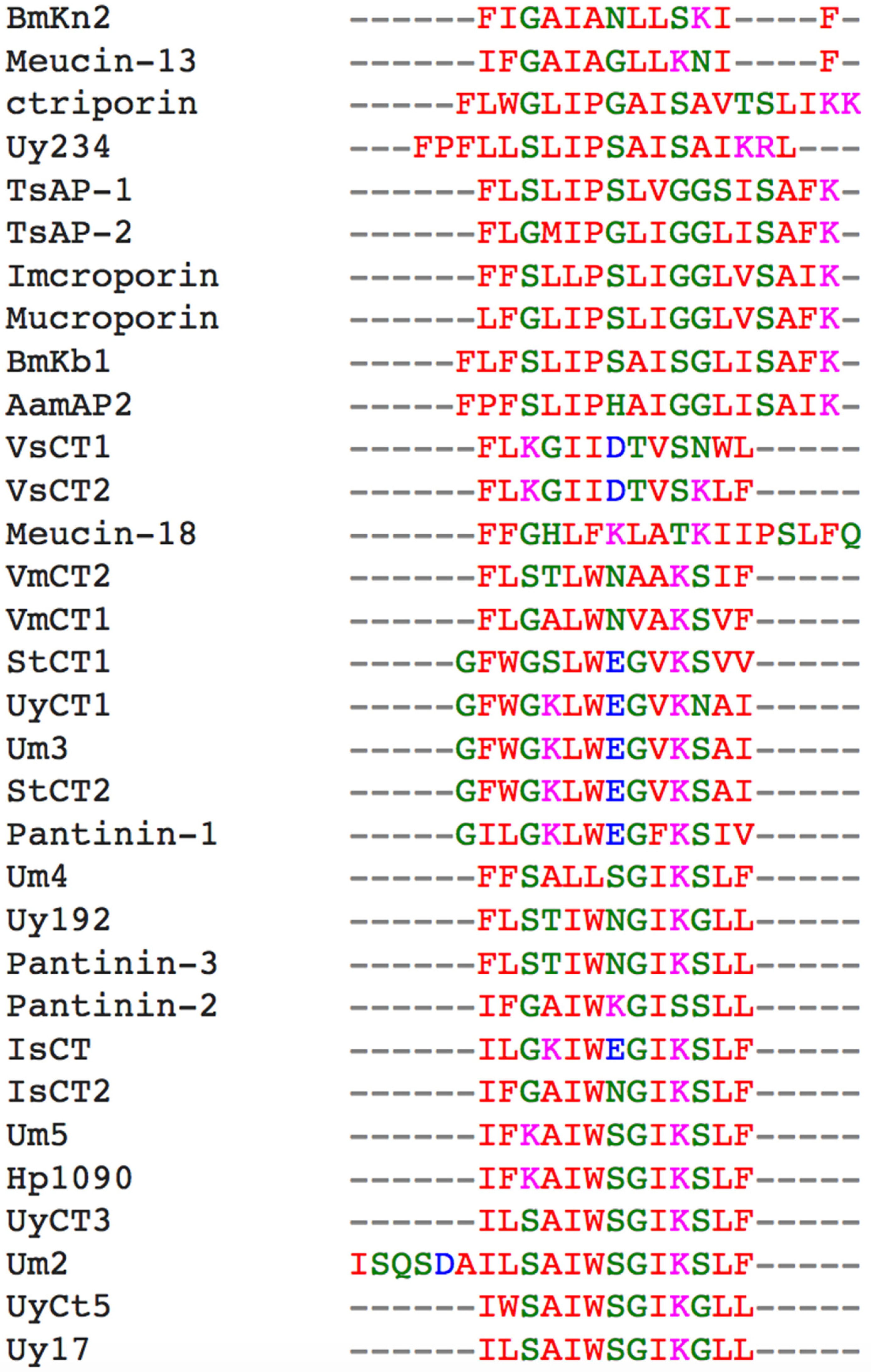
2.5. Phylogenetic Analysis
3. Discussion
4. Materials and Methods
4.1. Natural and Engineered AMPs
4.2. Antimicrobial Activity
4.3. Antifungal Assays
4.4. Hemolytic Activity
4.5. Cytotoxic Activity
4.6. Phylogenetic Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflict of Interest
References
- Wiesner, J.; Vilcinskas, A. Antimicrobial peptides: The ancient arm of the human immune system. Virulence 2010, 1, 440–464. [Google Scholar] [CrossRef] [PubMed]
- Mylonakis, E.; Podsiadlowski, L.; Muhammed, M.; Vilcinskas, A. Diversity, evolution and medical applications of insect antimicrobial peptides. Phil. Trans. R. Soc. B 2016, 371, 20150290. [Google Scholar] [CrossRef] [PubMed]
- Bolouri Moghaddam, M.R.; Tonk, M.; Schreiber, C.; Salzig, D.; Czermak, P.; Vilcinskas, A.; Rahnamaeian, M. The potential of the Galleria mellonella innate immune system is maximized by the co-presentation of diverse antimicrobial peptides. Biol. Chem. 2016, 397, 939–945. [Google Scholar] [CrossRef] [PubMed]
- Bolouri Moghaddam, M.R.; Vilcinskas, A.; Rahnamaeian, M. Cooperative interaction of antimicrobial peptides with the interrelated immune pathways in plants. Mol. Plant Pathol 2016, 17, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Vilcinskas, A. Evolutionary plasticity of insect immunity. J. Insect Physiol. 2013, 59, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Al Souhail, Q.; Hiromasa, Y.; Rahnamaeian, M.; Giraldo, M.C.; Takahashi, D.; Valent, B.; Vilcinskas, A.; Kanost, M.R. Characterization and regulation of expression of an antifungal peptide from hemolymph of an insect, Manduca sexta. Dev. Comp. Immunol. 2016, 61, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Tonk, M.; Knorr, E.; Cabezas-Cruz, A.; Valdes, J.J.; Kollewe, C.; Vilcinskas, A. Tribolium castaneum defensins are primarily active against gram-positive bacteria. J. Invertebr. Pathol. 2015, 132, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Tonk, M.; Cabezas-Cruz, A.; Valdes, J.J.; Rego, R.O.; Chrudimska, T.; Strnad, M.; Sima, R.; Bell-Sakyi, L.; Franta, Z.; Vilcinskas, A.; et al. Defensins from the tick Ixodes scapularis are effective against phytopathogenic fungi and the human bacterial pathogen listeria grayi. Parasit. Vectors 2014, 7, 554–561. [Google Scholar] [CrossRef] [PubMed]
- Tonk, M.; Cabezas-Cruz, A.; Valdes, J.J.; Rego, R.O.; Grubhoffer, L.; Estrada-Pena, A.; Vilcinskas, A.; Kotsyfakis, M.; Rahnamaeian, M. Ixodes ricinus defensins attack distantly-related pathogens. Dev. Comp. Immunol. 2015, 53, 358–365. [Google Scholar] [CrossRef] [PubMed]
- Harrison, P.L.; Abdel-Rahman, M.A.; Miller, K.; Strong, P.N. Antimicrobial peptides from scorpion venoms. Toxicon 2014, 88, 115–137. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Rahman, M.A.; Quintero-Hernandez, V.; Possani, L.D. Venom proteomic and venomous glands transcriptomic analysis of the egyptian scorpion Scorpio maurus palmatus (arachnida: Scorpionidae). Toxicon 2013, 74, 193–207. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.; Hou, X.; Wang, L.; Zhang, Y.; Xi, X.; Wang, H.; Zhou, M.; Duan, J.; Wei, M.; Chen, T.; et al. AaeAP1 and AaeAP2 : Novel antimicrobial peptides from the venom of the scorpion, Androctonus aeneas: Structural characterisation, molecular cloning of biosynthetic precursor-encoding cdnas and engineering of analogues with enhanced antimicrobial and anticancer activities. Toxins (Basel) 2015, 7, 219–237. [Google Scholar] [PubMed]
- Giangaspero, A.; Sandri, L.; Tossi, A. Amphipathic α helical antimicrobial peptides. Eur. J. Biochem. 2001, 268, 5589–5600. [Google Scholar] [CrossRef] [PubMed]
- Zasloff, M. Preface. Philos. Trans. R. Soc. B 2016, 371. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Kim, H.N.; Park, S.N.; Jang, S.H.; Choi, C.H.; Lim, H.T.; Hahm, K.S. Design of novel analogues with potent antibiotic activity based on the antimicrobial peptide, HP(2-9)-ME(1-12). Biotechnol. Lett. 2004, 26, 493–498. [Google Scholar] [CrossRef] [PubMed]
- Rahnamaeian, M.; Vilcinskas, A. Short antimicrobial peptides as cosmetic ingredients to deter dermatological pathogens. Appl. Microbiol. Biotechnol. 2015, 99, 8847–8855. [Google Scholar] [CrossRef] [PubMed]
- Luna-Ramirez, K.; Quintero-Hernandez, V.; Juarez-Gonzalez, V.R.; Possani, L.D. Whole transcriptome of the venom gland from Urodacus yaschenkoi scorpion. PLoS ONE 2015, 10, e0127883. [Google Scholar] [CrossRef] [PubMed]
- Luna-Ramírez, K.; Quintero-Hernández, V.; Vargas-Jaimes, L.; Batista, C.V.F.; Winkel, K.D.; Possani, L.D. Characterization of the venom from the australian scorpion Urodacus yaschenkoi: Molecular mass analysis of components, cdna sequences and peptides with antimicrobial activity. Toxicon 2013, 63, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Luna-Ramírez, K.; Sani, M.-A.; Silva-Sanchez, J.; Jiménez-Vargas, J.M.; Reyna-Flores, F.; Winkel, K.D.; Wright, C.E.; Possani, L.D.; Separovic, F. Membrane interactions and biological activity of antimicrobial peptides from Australian scorpion. BBA-Biomembranes 2014, 1838, 2140–2148. [Google Scholar] [CrossRef] [PubMed]
- Sunagar, K.; Undheim, E.A.B.; Chan, A.H.C.; Koludarov, I.; Muñoz-Gómez, S.A.; Antunes, A.; Fry, B.G. Evolution stings: The origin and diversification of scorpion toxin peptide scaffolds. Toxins 2013, 5, 2456–2487. [Google Scholar] [CrossRef] [PubMed]
- Quintero-Hernández, V.; Ramírez-Carreto, S.; Romero-Gutiérrez, M.T.; Valdez-Velázquez, L.L.; Becerril, B.; Possani, L.D.; Ortiz, E. Transcriptome analysis of scorpion species belonging to the Vaejovis genus. PLoS ONE 2015, 10, e0117188. [Google Scholar] [CrossRef] [PubMed]
- Luan, N.; Shen, W.; Liu, J.; Wen, B.; Lin, Z.; Yang, S.; Lai, R.; Liu, S.; Rong, M. A combinational strategy upon RNA sequencing and peptidomics unravels a set of novel toxin peptides in scorpion Mesobuthus martensii. Toxins (Basel) 2016, 8, 286–299. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.A.; Alam, M.; Abbasi, A.; Undheim, E.A.; Fry, B.G.; Kalbacher, H.; Voelter, W. Structure-activity relationship of chlorotoxin-like peptides. Toxins (Basel) 2016, 8, 36–53. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Ma, C.; Du, Q.; Wei, R.; Wang, L.; Zhou, M.; Chen, T.; Shaw, C. Two peptides, TsAP-1 and TsAP-2, from the venom of the brazilian yellow scorpion, Tityus serrulatus: Evaluation of their antimicrobial and anticancer activities. Biochimie 2013, 95, 1784–1794. [Google Scholar] [CrossRef] [PubMed]
- Almaaytah, A.; Albalas, Q. Scorpion venom peptides with no disulfide bridges: A review. Peptides 2014, 51, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Deslouches, B.; Steckbeck, J.D.; Craigo, J.K.; Doi, Y.; Mietzner, T.A.; Montelaro, R.C. Rational design of engineered cationic antimicrobial peptides consisting exclusively of arginine and tryptophan, and their activity against multidrug-resistant pathogens. Antimicrob Agents Ch. 2013, 57, 2511–2521. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Li, Z.; Zhang, R.; Wu, Y.; Li, W.; Cao, Z. Stct2, a new antibacterial peptide characterized from the venom of the scorpion Scorpiops tibetanus. Peptides 2012, 36, 213–220. [Google Scholar] [CrossRef]
- Silva, E.C.; Camargos, T.S.; Maranhao, A.Q.; Silva-Pereira, I.; Silva, L.P.; Possani, L.D.; Schwartz, E.F. Cloning and characterization of cDNA sequences encoding for new venom peptides of the brazilian scorpion Opisthacanthus cayaporum. Toxicon 2009, 54, 252–261. [Google Scholar] [CrossRef] [PubMed]
- Uggerhoj, L.E.; Poulsen, T.J.; Munk, J.K.; Fredborg, M.; Sondergaard, T.E.; Frimodt-Moller, N.; Hansen, P.R.; Wimmer, R. Rational design of alpha-helical antimicrobial peptides: Do's and don'ts. Chembiochem 2015, 16, 242–253. [Google Scholar] [CrossRef] [PubMed]
- Sakai, N.; Matile, S. Anion-mediated transfer of polyarginine across liquid and bilayer membranes. J. Am. Chem. Soc. 2003, 125, 14348–14356. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.I.; Prenner, E.J.; Vogel, H.J. Tryptophan- and arginine-rich antimicrobial peptides: Structures and mechanisms of action. Biochim. Biophys. Acta 2006, 1758, 1184–1202. [Google Scholar] [CrossRef] [PubMed]
- Albada, H.B.; Prochnow, P.; Bobersky, S.; Langklotz, S.; Bandow, J.E.; Metzler-Nolte, N. Short antibacterial peptides with significantly reduced hemolytic activity can be identified by a systematic L-to-D exchange scan of their amino acid residues. ACS Comb. Sci. 2013, 15, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Bao, A.; Zhong, J.; Zeng, X.C.; Nie, Y.; Zhang, L.; Peng, Z.F. A novel cysteine-free venom peptide with strong antimicrobial activity against antibiotics-resistant pathogens from the scorpion Opistophthalmus glabrifrons. J. Pept. Sci. 2015, 21, 758–764. [Google Scholar] [CrossRef] [PubMed]
- Rahnamaeian, M.; Cytrynska, M.; Zdybicka-Barabas, A.; Vilcinskas, A. The functional interaction between abaecin and pore-forming peptides indicates a general mechanism of antibacterial potentiation. Peptides 2016, 78, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Rahnamaeian, M.; Langen, G.; Imani, J.; Khalifa, W.; Altincicek, B.; von Wettstein, D.; Kogel, K.H.; Vilcinskas, A. Insect peptide metchnikowin confers on barley a selective capacity for resistance to fungal ascomycetes pathogens. J. Exp. Bot. 2009, 60, 4105–4114. [Google Scholar] [CrossRef] [PubMed]
- Ponce, D.; Brinkman, D.L.; Luna-Ramirez, K.; Wright, C.E.; Dorantes-Aranda, J.J. Comparative study of the toxic effects of Chrysaora quinquecirrha (cnidaria: Scyphozoa) and Chironex fleckeri (cnidaria: Cubozoa) venoms using cell-based assays. Toxicon 2015, 106, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Dorantes-Aranda, J.J.; Waite, T.D.; Godrant, A.; Rose, A.L.; Tovar, C.D.; Woods, G.M.; Hallegraeff, G.M. Novel application of a fish gill cell line assay to assess ichthyotoxicity of harmful marine microalgae. Harmful Algae 2011, 10, 366–373. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, Scalable generation of high-quality protein multiple sequence alignments using Clustal omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]

| AMP | Amino Acid Sequence | MW (Da) | Length | Net Charge | GRAVY+ | Accession No. |
|---|---|---|---|---|---|---|
| UyCT1 | GFWGKLWEGVKNAI | 1603.9 | 14 | +2 | −0.050 | AGA82754 |
| UyCT3 | ILSAIWSGIKSLF | 1433.7 | 13 | +2 | 1.392 | AGA82755 |
| UyCT5 | IWSAIWSGIKGLL | 1442.7 | 13 | +2 | 1.138 | AGA82756 |
| Uy17 | ILSAIWSGIKGLL | 1369.43 | 13 | +2 | 1.500 | SRP045734 * |
| Uy192 | FLSTIWNGIKGLL | 1459.98 | 13 | +2 | 0.969 | SRP045734 * |
| Uy234 | FPFLLSLIPSAISAIKRL | 1986.19 | 18 | +3 | 1.328 | SRP045734 * |
| Um2 | ISQSDAILSAIWSGIKSLF | 2034.56 | 19 | +1 | 0.832 | JAA98072 |
| Um3 | GFWGKLWEGVKSAI | 1577.23 | 14 | +2 | 0.143 | JAA98071 |
| Um4 | FFSALLSGIKSLF | 1428.58 | 13 | +2 | 1.492 | JAA98071 |
| Um5 | IFKAIWSGIKSLF | 1508.82 | 13 | +3 | 1.077 | JAA98069 |
| D1 | IFGAIWSGIKSLF | 1437.11 | 13 | +2 | 1.346 | – |
| D2 | FLSTIWNGIKSLF | 1524.00 | 13 | +2 | 0.862 | – |
| D4 | GFWGKLWKPVKKAI | 1657.87 | 14 | +5 | −0.193 | – |
| D5 | GFWGKLLEGVKKAI | 1544.52 | 14 | +3 | 0.257 | – |
| D10 | FPFLKLSLKIPKSAIKSAIKRL | 2497.71 | 22 | +7 | 0.377 | – |
| D11 | GFWGKLWEGVKNAIKKK | 1987.55 | 17 | +5 | −0.729 | – |
| Peptide | Sequence |
|---|---|
| UyCT3 | ILSAIWSGIKSLF |
| D1 | IFGAIWSGIKSLF |
| Uy192 | FLSTIWNGIKGLL |
| D2 | FLSTIWNGIKSLF |
| UyCT1 | GFWGKLWEGVKNAI |
| D4 | GFWGKLWKPVKKAI |
| D5 | GFWGKLLEGVKKAI |
| D11 | GFWGKLWEGVKNAIKKK |
| D12 | GFWKGKLWKEGVKNAIK |
| Uy234 | FPFL-LSL-IP-SAI-SAIKRL |
| D10 | FPFLKLSLKIPKSAIKSAIKRL |
| AMP | M. luteus | E. coli | S. aureus | L. grayi | L. fleischmannii | L. monocytogenes | EC10 | EC50 | EC90 | HC10 | HC50 | HC90 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UyCT1 | 1 | 4 | – | 4 | 4 | 4 | 0.65 | 17.37 | 52.12 | 31.5 | 142.50 | 644.46 |
| UyCT3 | 4 | 8 | – | 8 | 4 | 4 | 0.57 | 15.37 | 46.12 | 15.78 | 58.15 | 214.22 |
| UyCT5 | 4 | 15 | 8 | 8 | 8 | 4 | 0.91 | 24.55 | 73.65 | 2.39 | 20.59 | 177.49 |
| Uy17 | 15 | – | 30 | – | – | 15 | 2.73 | 73.63 | 220.80 | 26.65 | 138.40 | 718.55 |
| Uy192 | 15 | 15 | 15 | 15 | 4 | 8 | 1.92 | 51.95 | 155.90 | 35.85 | 155.6 | 675.22 |
| Uy234 | 2 | – | – | 4 | – | 2 | 0.23 | 6.29 | 18.86 | 55.14 | 104.50 | 198.04 |
| Um2 | 4 | – | – | – | – | – | 0.58 | 15.71 | 47.12 | 2.36 | 129 | 7033.22 |
| Um3 | 2 | 15 | 15 | 4 | 4 | 8 | 2.359 | 63.68 | 191.1 | 70.48 | 126.2 | 246.84 |
| Um4 | 8 | 8 | 15 | 15 | 4 | 8 | 3.22 | 87.01 | 261 | n.c. | n.c. (100) | n.c. |
| Um5 | 2 | 15 | – | 2 | 2 | 2 | 0.36 | 9.71 | 29.12 | 11.944 | 59.25 | 293.90 |
| D1 | 4 | 8 | – | 8 | 4 | 4 | 0.69 | 18.58 | 55.75 | 15.50 | 48 | 148.51 |
| D2 | 4 | – | 8 | – | 15 | 15 | 13.95 | 376.70 | 1130 | 2.94 | 24.48 | 200.83 |
| D4 | 2 | – | – | 8 | 15 | 30 | 36.30 | 980 | 2940 | n.c. | n.c. (100) | n.c. |
| D5 | 8 | 15 | – | 8 | 15 | 15 | 2.50 | 67.63 | 202.90 | 110.86 | 1636 | 24,141.72 |
| D10 | 0.25 | 8 | – | 8 | 30 | 30 | 5.24 | 141.40 | 424.30 | 149.75 | 1726 | 19,894.17 |
| D11 | 0.25 | 4 | – | 1 | 1 | 2 | 0.66 | 17.81 | 53.43 | 39.76 | 1110 | 30,984.53 |
| AMP | M. luteus | E. coli | S. aureus | L. grayi | L. fleischmannii | L. monocytogenes |
|---|---|---|---|---|---|---|
| UyCT1 | 142.50 | 35.62 | - | 35.62 | 35.62 | 35.62 |
| UyCT3 | 14.53 | 7.26 | - | 7.26 | 14.53 | 14.53 |
| UyCT5 | 5.14 | 1.37 | 2.57 | 2.57 | 2.57 | 5.14 |
| Uy17 | 9.22 | - | 4.61 | - | - | 9.22 |
| Uy192 | 10.37 | 10.37 | 10.37 | 10.37 | 38.90 | 19.45 |
| Uy234 | 52.25 | - | - | 26.12 | - | 52.25 |
| Um2 | 32.25 | - | - | - | - | - |
| Um3 | 63.10 | 8.41 | 8.41 | 31.55 | 31.55 | 15.77 |
| Um4 | - | - | - | - | - | - |
| Um5 | 29.62 | 3.95 | - | 29.62 | 29.62 | 29.62 |
| D1 | 11.99 | 5.99 | - | 5.99 | 11.99 | 11.99 |
| D2 | 6.12 | - | 3.06 | - | 1.63 | 1.63 |
| D4 | - | - | - | - | - | - |
| D5 | 204.50 | 109.06 | - | 204.50 | 109.06 | 109.06 |
| D10 | 6904 | 215.75 | - | 215.75 | 57.53 | 57.53 |
| D11 | 4440 | 277.5 | - | 1110 | 1110 | 555 |
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luna-Ramirez, K.; Tonk, M.; Rahnamaeian, M.; Vilcinskas, A. Bioactivity of Natural and Engineered Antimicrobial Peptides from Venom of the Scorpions Urodacus yaschenkoi and U. manicatus. Toxins 2017, 9, 22. https://doi.org/10.3390/toxins9010022
Luna-Ramirez K, Tonk M, Rahnamaeian M, Vilcinskas A. Bioactivity of Natural and Engineered Antimicrobial Peptides from Venom of the Scorpions Urodacus yaschenkoi and U. manicatus. Toxins. 2017; 9(1):22. https://doi.org/10.3390/toxins9010022
Chicago/Turabian StyleLuna-Ramirez, Karen, Miray Tonk, Mohammad Rahnamaeian, and Andreas Vilcinskas. 2017. "Bioactivity of Natural and Engineered Antimicrobial Peptides from Venom of the Scorpions Urodacus yaschenkoi and U. manicatus" Toxins 9, no. 1: 22. https://doi.org/10.3390/toxins9010022





