The Influence of the Toxin/Antitoxin mazEF on Growth and Survival of Listeria monocytogenes under Stress
Abstract
:1. Introduction
2. Results
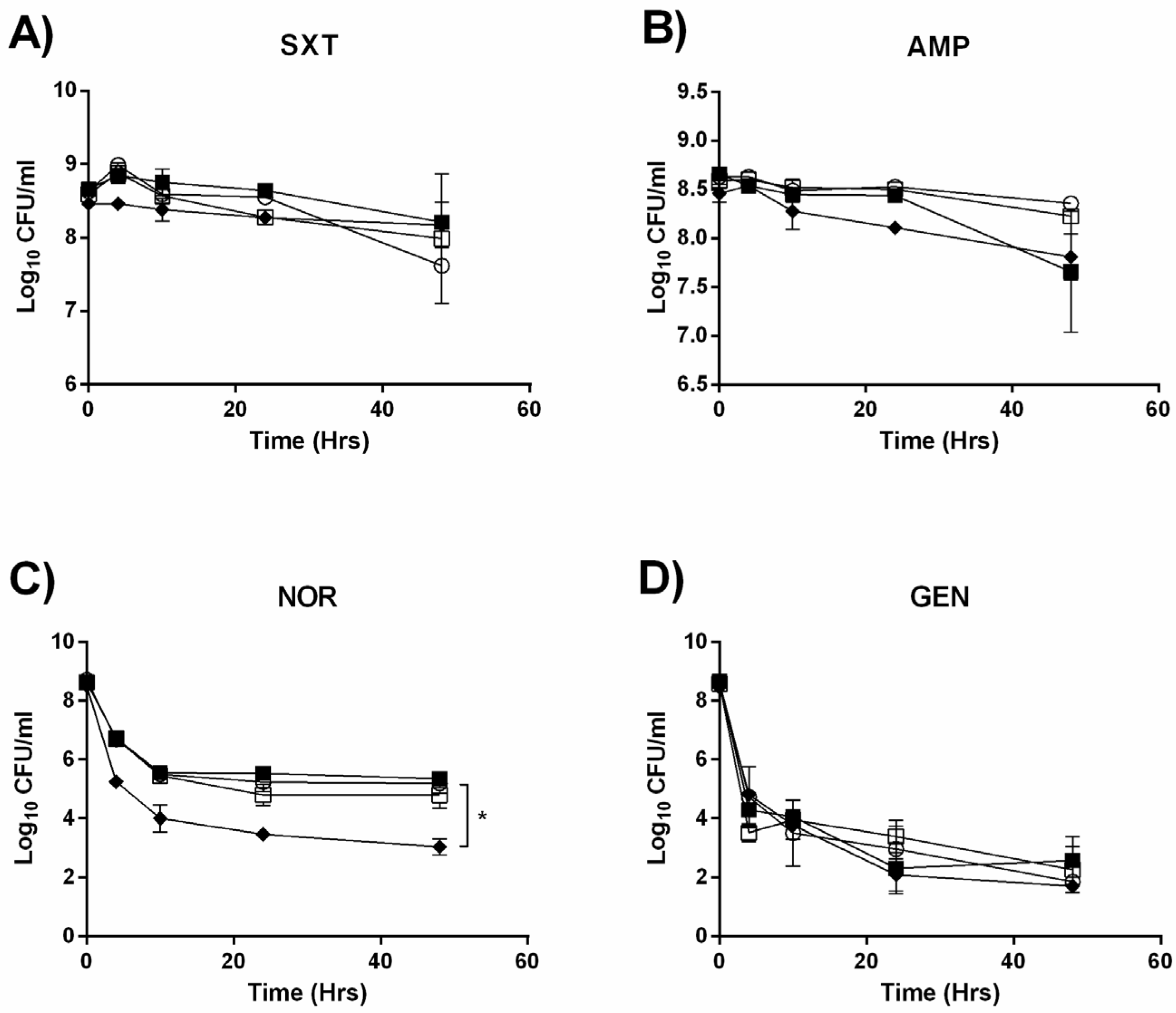
2.1. Deletion of the mazEF TA System does not Detectably Alter Persister Formation
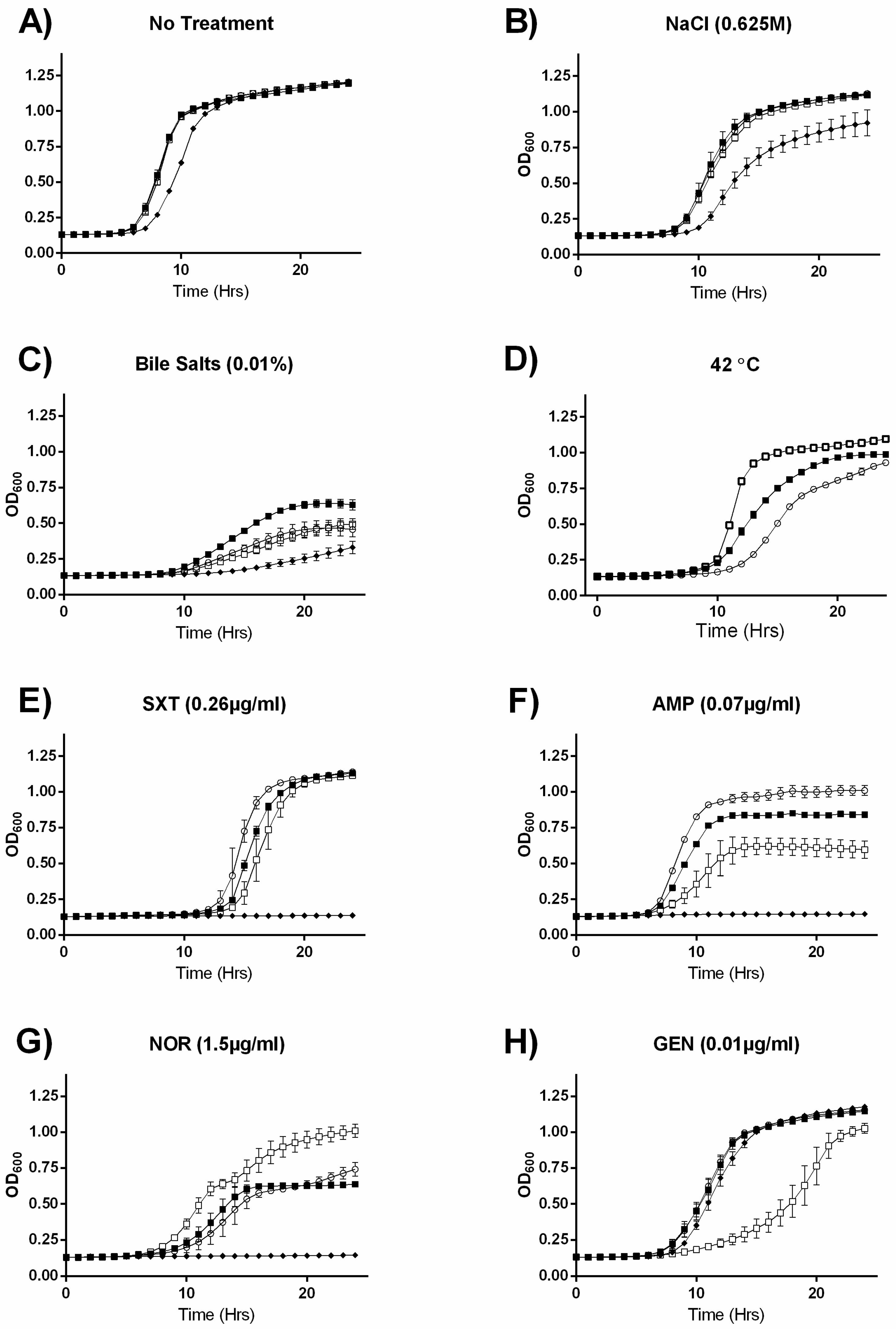
2.2. MazEF Affects Growth under Specific Sub-Inhibitory Growth Conditions
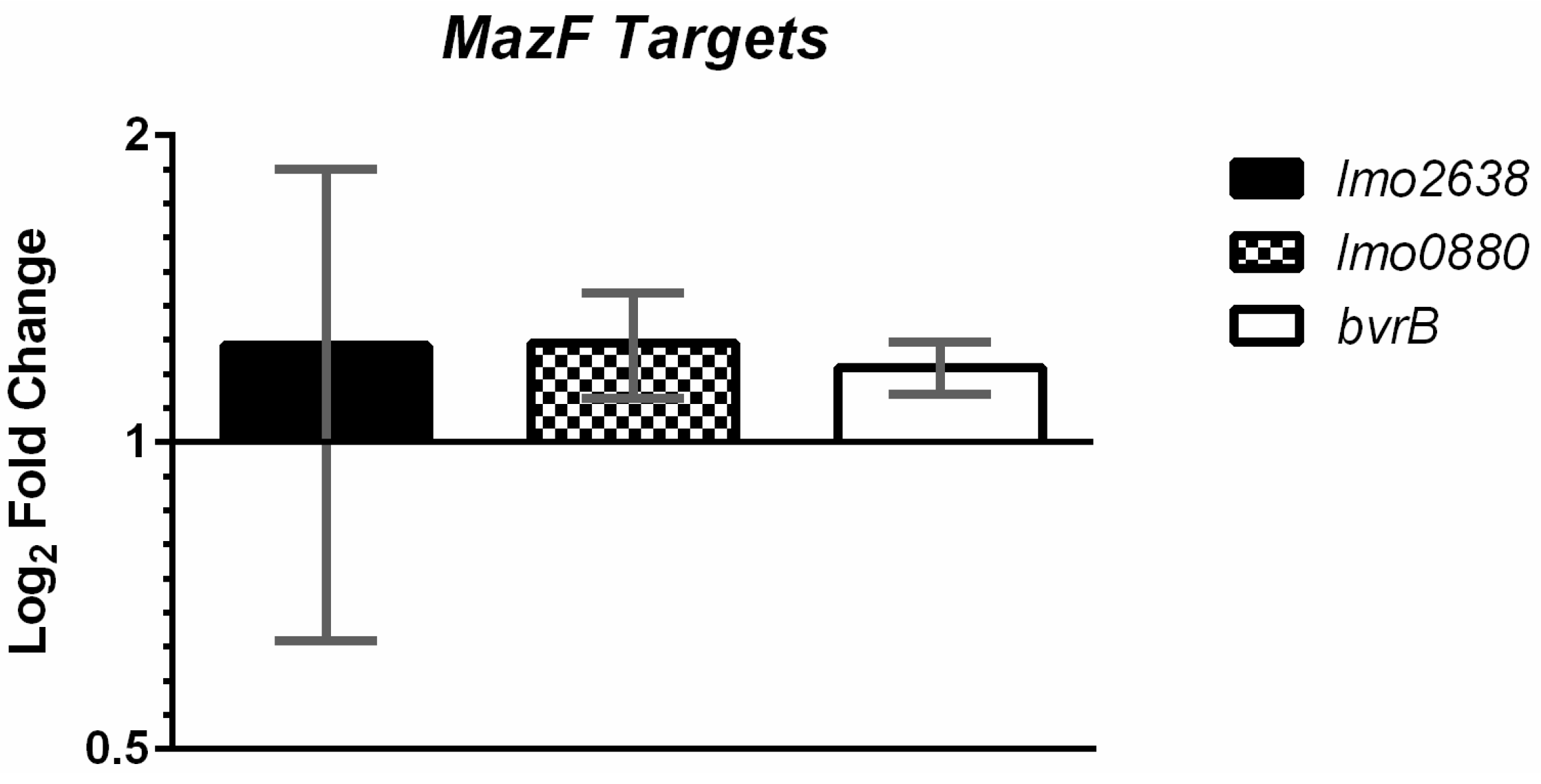
2.3. MazF Does not Detectably Degrade Putative Target Transcripts In Vivo
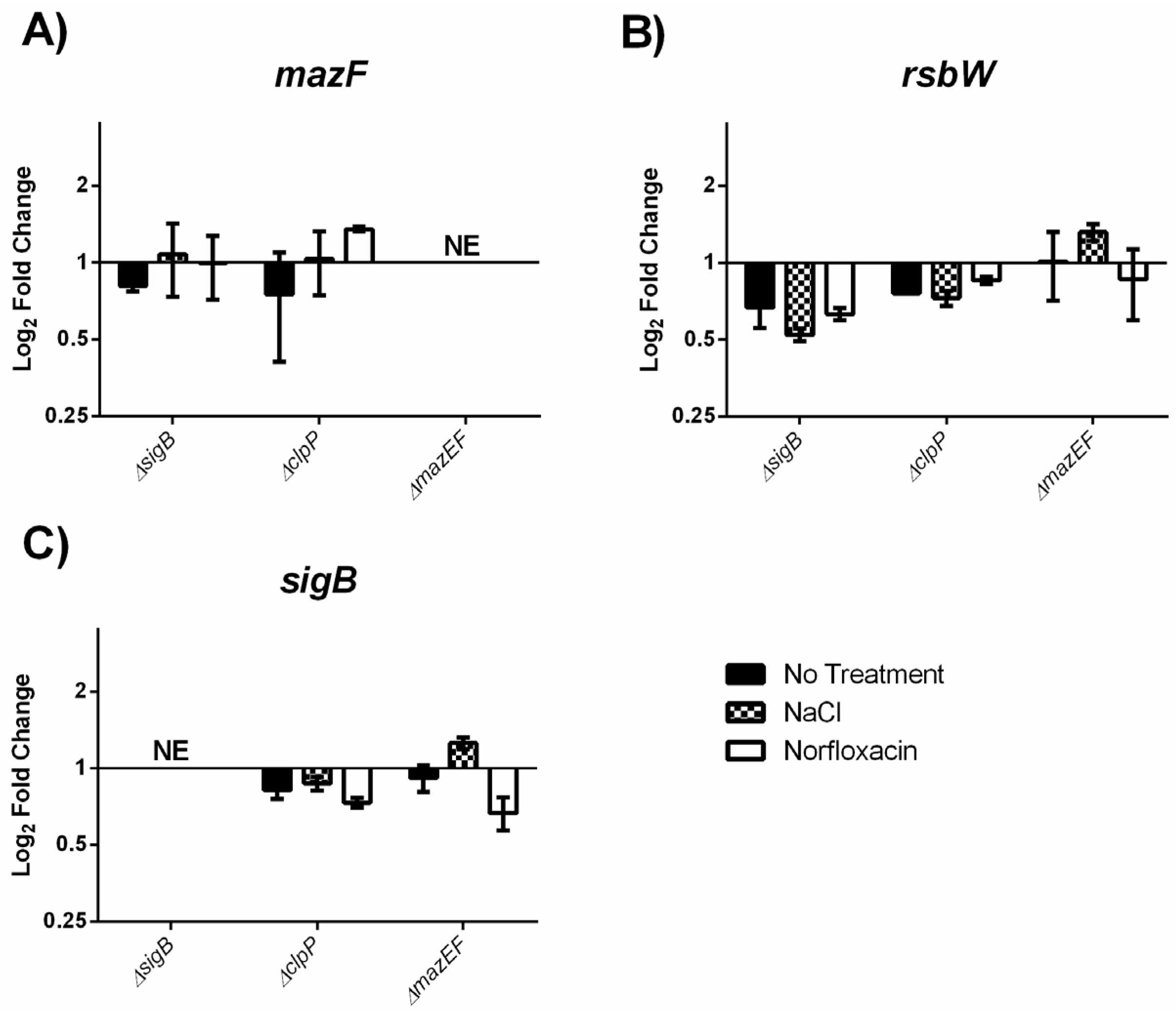
2.4. MazEF Affects Expression of sigB Dependent Genes, but not sigB Operon
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Bacterial Strains and Growth Conditions
5.2. Antibiotic Preparation
5.3. Constructions of Mutants
5.4. Killing Kinetics
5.5. Growth Kinetics
5.6. Sample Collection and RNA Isolation
5.7. Measuring Relative Gene Expression via Quantitative Reverse Transcription PCR (RT-qPCR)
5.8. Bioinformatic Analysis of MazF Cut Site Abundance in EDGe Genome
5.9. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Drevets, D.A.; Bronze, M.S. Listeria monocytogenes: Epidemiology, human disease, and mechanisms of brain invasion. FEMS Immunol. Med. Microbiol. 2008, 53, 151–165. [Google Scholar] [CrossRef] [PubMed]
- Freitag, N.E.; Port, G.C.; Miner, M.D. Listeria monocytogenes—From saprophyte to intracellular pathogen. Nat. Rev. Microbiol. 2009, 7, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Becker, L.A.; Cetin, M.S.; Hutkins, R.W.; Benson, A.K. Identification of the gene encoding the alternative sigma factor sigmaB from Listeria monocytogenes and its role in osmotolerance. J. Bacteriol. 1998, 180, 4547–4554. [Google Scholar] [PubMed]
- Wiedmann, M.; Arvik, T.J.; Hurley, R.J.; Boor, K.J. General stress transcription factor sigmaB and its role in acid tolerance and virulence of Listeria monocytogenes. J. Bacteriol. 1998, 180, 3650–3656. [Google Scholar] [PubMed]
- Tienungoon, S.; Ratkowsky, D.A.; McMeekin, T.A.; Ross, T. Growth limits of Listeria monocytogenes as a function of temperature, pH, NaCl, and lactic acid. Appl. Environ. Microbiol. 2000, 66, 4979–4987. [Google Scholar] [CrossRef] [PubMed]
- Maisonneuve, E.; Gerdes, K. Molecular mechanisms underlying bacterial persisters. Cell 2014, 157, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, A.; Dewan, P.C.; Siddique, S.A.; Varadarajan, R. MazF-induced growth inhibition and persister generation in Escherichia coli. J. Biol. Chem. 2014, 289, 4191–4205. [Google Scholar] [CrossRef] [PubMed]
- Donegan, N.P.; Thompson, E.T.; Fu, Z.; Cheung, A.L. Proteolytic regulation of toxin-antitoxin systems by ClpPC in Staphylococcus aureus. J. Bacteriol. 2010, 192, 1416–1422. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.K.; Pedersen, K.; Hansen, F.G.; Gerdes, K. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 2003, 332, 809–819. [Google Scholar] [CrossRef]
- Zhu, L.; Inoue, K.; Yoshizumi, S.; Kobayashi, H.; Zhang, Y.; Ouyang, M.; Kato, F.; Sugai, M.; Inouye, M. Staphylococcus aureus MazF specifically cleaves a pentad sequence, UACAU, which is unusually abundant in the mRNA for pathogenic adhesive factor SraP. J. Bacteriol. 2009, 191, 3248–3255. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, G.M.; Ng, Y.; Gram, L. Survival of bactericidal antibiotic treatment by a persister subpopulation of Listeria monocytogenes. Appl. Environ. Microbiol. 2013, 79, 7390–7397. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Harrison, E.M.; Bi, D.; Tai, C.; He, X.; Ou, H.-Y.; Rajakumar, K.; Deng, Z. TADB: A web-based resource for Type 2 toxin-antitoxin loci in bacteria and archaea. Nucleic Acids Res. 2011, 39, D606–D611. [Google Scholar] [CrossRef] [PubMed]
- Pandey, D.P.; Gerdes, K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005, 33, 966–976. [Google Scholar] [CrossRef] [PubMed]
- Schmitter, S. Role of a Toxin-Antitoxin System in L. monocytogenes. Doctoral and Habilitation Thesis, Eidgenossische Technische Hochschule ETH Zurich, Zürich, Switzerland, 2014. [Google Scholar]
- Christensen, S.K.; Gerdes, K. Delayed-relaxed response explained by hyperactivation of RelE. Mol. Microbiol. 2004, 53, 587–597. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez Barrios, A.F.; Zuo, R.; Hashimoto, Y.; Yang, L.; Bentley, W.E.; Wood, T.K. Autoinducer 2 Controls Biofilm Formation in Escherichia coli through a Novel Motility Quorum-Sensing Regulator (MqsR, B3022). J. Bacteriol. 2006, 188, 305–316. [Google Scholar] [CrossRef] [PubMed]
- Metzger, S.; Dror, I.B.; Aizenman, E.; Schreiber, G.; Toone, M.; Friesen, J.D.; Cashel, M.; Glaser, G. The nucleotide sequence and characterization of the relA gene of Escherichia coli. J. Biol. Chem. 1988, 263, 15699–15704. [Google Scholar] [PubMed]
- Donegan, N.P.; Cheung, A.L. Regulation of the mazEF toxin-antitoxin module in Staphylococcus aureus and its impact on sigB expression. J. Bacteriol. 2009, 191, 2795–2805. [Google Scholar] [CrossRef] [PubMed]
- Pellegrini, O.; Mathy, N.; Gogos, A.; Shapiro, L.; Condon, C. The Bacillus subtilis ydcDE operon encodes an endoribonuclease of the MazF/PemK family and its inhibitor. Mol. Microbiol. 2005, 56, 1139–1148. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Tamber, S.; Memmi, G.; Donegan, N.P.; Cheung, A.L. Overexpression of MazFsa in Staphylococcus aureus induces bacteriostasis by selectively targeting mRNAs for cleavage. J. Bacteriol. 2009, 191, 2051–2059. [Google Scholar] [CrossRef] [PubMed]
- Schuster, C.F.; Mechler, L.; Nolle, N.; Krismer, B.; Zelder, M.-E.; Gotz, F.; Bertram, R. The MazEF Toxin-Antitoxin System Alters the β-Lactam Susceptibility of Staphylococcus aureus. PLoS ONE 2015, 10. [Google Scholar] [CrossRef]
- Park, J.H.; Yamaguchi, Y.; Inouye, M. Bacillus subtilis MazF-bs (EndoA) is a UACAU-specific mRNA interferase. FEBS Lett. 2011, 585, 2526–2532. [Google Scholar] [CrossRef] [PubMed]
- Schuster, C.F.; Park, J.H.; Prax, M.; Herbig, A.; Nieselt, K.; Rosenstein, R.; Inouye, M.; Bertrama, R. Characterization of a MazEF toxin-antitoxin homologue from Staphylococcus equorum. J. Bacteriol. 2013, 195, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Wurtzel, O.; Sesto, N.; Mellin, J.R.; Karunker, I.; Edelheit, S.; Becavin, C.; Archambaud, C.; Cossart, P.; Sorek, R. Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol. Syst. Biol. 2012, 8. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, G.M.; Fromberg, A.; Ng, Y.; Gram, L. Sublethal Concentrations of Antibiotics Cause Shift to Anaerobic Metabolism in Listeria monocytogenes and Induce Phenotypes Linked to Antibiotic Tolerance. Front. Microbiol. 2016, 7, 1091. [Google Scholar] [CrossRef] [PubMed]
- Merfa, M.V.; Niza, B.; Takita, M.A.; De Souza, A.A. The MqsRA Toxin-Antitoxin System from Xylella fastidiosa Plays a Key Role in Bacterial Fitness, Pathogenicity, and Persister Cell Formation. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Lobato-Marquez, D.; Moreno-Cordoba, I.; Figueroa, V.; Diaz-Orejas, R.; Garcia-del Portillo, F. Distinct type I and type II toxin-antitoxin modules control Salmonella lifestyle inside eukaryotic cells. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, K.; Christensen, S.K.; Lobner-Olesen, A. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 2005, 3, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Lemos, J.A.C.; Brown, T.A.; Abranches, J.; Burne, R.A. Characteristics of Streptococcus mutans strains lacking the MazEF and RelBE toxin-antitoxin modules. FEMS Microbiol. Lett. 2005, 253, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Silva-Herzog, E.; McDonald, E.M.; Crooks, A.L.; Detweiler, C.S. Physiologic Stresses Reveal a Salmonella Persister State and TA Family Toxins Modulate Tolerance to These Stresses. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Page, R.; Peti, W. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 2016, 12, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, K.; Christensen, S.K.; Gerdes, K. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol. Microbiol. 2002, 45, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Zhang, J.; Liu, M.; Woychik, N.A.; Inouye, M. Single protein production in living cells facilitated by an mRNA interferase. Mol. Cell 2005, 18, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Bukowski, M.; Rojowska, A.; Wladyka, B. Prokaryotic toxin-antitoxin systems--the role in bacterial physiology and application in molecular biology. Acta Biochim. Pol. 2011, 58, 1–9. [Google Scholar] [PubMed]
- Maisonneuve, E.; Gerdes, K. Supporting Information: Bacterial persistence by RNA endonucleases. Proc. Natl. Acad. Sci. USA 2011, 108, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, P.; Arora, G.; Singh, M.; Kidwai, S.; Narayan, O.P.; Singh, R. MazF ribonucleases promote Mycobacterium tuberculosis drug tolerance and virulence in guinea pigs. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Conlon, B.P.; Rowe, S.E.; Gandt, A.B.; Nuxoll, A.S.; Donegan, N.P.; Zalis, E.A.; Clair, G.; Adkins, J.N.; Cheung, A.L.; Lewis, K. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat. Microbiol. 2016, 1. [Google Scholar] [CrossRef] [PubMed]
- Gaillot, O.; Pellegrini, E.; Bregenholt, S.; Nair, S.; Berche, P. The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol. Microbiol. 2000, 35, 1286–1294. [Google Scholar] [CrossRef] [PubMed]
- Springer, M.T.; Singh, V.K.; Cheung, A.L.; Donegan, N.P.; Chamberlain, N.R. Effect of clpP and clpC deletion on persister cell number in Staphylococcus aureus. J. Med. Microbiol. 2016, 65, 848–857. [Google Scholar] [CrossRef] [PubMed]
- Page, M. The Chemistry of β-Lactams; Page, M.I., Ed.; Springer: Dordrecht, The Netherlands, 1992. [Google Scholar]
- Blondeau, J.M. Fluoroquinolones: Mechanism of action, classification, and development of resistance. Surv. Ophthalmol. 2004, 49 (Suppl. 2), S73–S78. [Google Scholar] [CrossRef] [PubMed]
- Kazmierczak, M.J.; Wiedmann, M.; Boor, K.J. Contributions of Listeria monocytogenes SigmaB and PrfA to expression of virulence and stress response genes during extra- and intracellular growth. Microbiology 2006, 152, 1827–1838. [Google Scholar] [CrossRef] [PubMed]
- Sue, D.; Boor, K.J.; Wiedmann, M. Sigma(B)-dependent expression patterns of compatible solute transporter genes opuCA and lmo1421 and the conjugated bile salt hydrolase gene bsh in Listeria monocytogenes. Microbiology 2003, 149, 3247–3256. [Google Scholar] [CrossRef] [PubMed]
- McGann, P.; Ivanek, R.; Wiedmann, M.; Boor, K.J. Temperature-dependent expression of Listeria monocytogenes internalin and internalin-like genes suggests functional diversity of these proteins among the listeriae. Appl. Environ. Microbiol. 2007, 73, 2806–2814. [Google Scholar] [CrossRef] [PubMed]
- Muthuramalingam, M.; White, J.C.; Bourne, C.R. Toxin-Antitoxin Modules Are Pliable Switches Activated by Multiple Protease Pathways. Toxins 2016, 8, 214. [Google Scholar] [CrossRef] [PubMed]
- Gerth, U.; Kruger, E.; Derre, I.; Msadek, T.; Hecker, M. Stress induction of the Bacillus subtilis clpP gene encoding a homologue of the proteolytic component of the Clp protease and the involvement of ClpP and ClpX in stress tolerance. Mol. Microbiol. 1998, 28, 787–802. [Google Scholar] [CrossRef] [PubMed]
- Prax, M.; Bertram, R. Metabolic aspects of bacterial persisters. Front. Cell. Infect. Microbiol. 2014, 4, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Syed, M.A.; Koyanagi, S.; Sharma, E.; Jobin, M.C.; Yakunin, A.F.; Levesque, C.M. The chromosomal mazEF locus of Streptococcus mutans encodes a functional type II toxin-antitoxin addiction system. J. Bacteriol. 2011, 193, 1122–1130. [Google Scholar] [CrossRef] [PubMed]
- Mok, W.W.K.; Park, J.O.; Rabinowitz, J.D.; Brynildsen, M.P. RNA Futile Cycling in Model Persisters Derived from MazF Accumulation. MBio 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Mingeot-Leclercq, M.-P.; Glupczynski, Y.; Tulkens, P.M. Aminoglycosides: Activity and Resistance. Antimicrob. Agents Chemother. 1999, 43, 727–737. [Google Scholar] [PubMed]
- Scortti, M.; Monzo, H.J.; Lacharme-Lora, L.; Lewis, D.A.; Vazquez-Boland, J.A. The PrfA virulence regulon. Microbes Infect. 2007, 9, 1196–1207. [Google Scholar] [CrossRef] [PubMed]
- Oliver, H.F.; Orsi, R.H.; Ponnala, L.; Keich, U.; Wang, W.; Sun, Q.; Cartinhour, S.W.; Filiatrault, M.J.; Wiedmann, M.; Boor, K.J. Deep RNA sequencing of L. monocytogenes reveals overlapping and extensive stationary phase and sigma B-dependent transcriptomes, including multiple highly transcribed noncoding RNAs. BMC Genom. 2009, 10. [Google Scholar] [CrossRef] [PubMed]
- Hain, T.; Hossain, H.; Chatterjee, S.S.; Machata, S.; Volk, U.; Wagner, S.; Brors, B.; Haas, S.; Kuenne, C.T.; Billion, A.; et al. Temporal transcriptomic analysis of the Listeria monocytogenes EGD-e sigmaB regulon. BMC Microbiol. 2008, 8. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, T.; Leimeister-Wachter, M.; Domann, E.; Hartl, M.; Goebel, W.; Nichterlein, T.; Notermans, S. Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J. Bacteriol. 1992, 174, 568–574. [Google Scholar] [CrossRef] [PubMed]
- Horton, R.M.; Cai, Z.; Ho, S.N.; Pease, L.R. Gene splicing by overlap extension: Tailor-made genes using the polymerase chain reaction. Biotechniques 2013, 54, 528–535. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001; p. 999. [Google Scholar]
- Guzman, C.A.; Rohde, M.; Chakraborty, T.; Domann, E.; Hudel, M.; Wehland, J.; Timmis, K.N. Interaction of Listeria monocytogenes with mouse dendritic cells. Infect. Immun. 1995, 63, 3665–3673. [Google Scholar] [PubMed]
- Monk, I.R.; Gahan, C.G.M.; Hill, C. Tools for functional postgenomic analysis of Listeria monocytogenes. Appl. Environ. Microbiol. 2008, 74, 3921–3934. [Google Scholar] [CrossRef] [PubMed]
- Primer3. Available online: http://frodo.wi.mit.edu/primer3/input.htm (accessed on 14 May 2016).
- Tasara, T.; Stephan, R. Evaluation of housekeeping genes in Listeria monocytogenes as potential internal control references for normalizing mRNA expression levels in stress adaptation models using real-time PCR. FEMS Microbiol. Lett. 2007, 269, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCt Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- GitHub. Available online: https://github.com/thocu/MazF_Targets.git (accessed on 7 October 2016).


| Gene Name | Length | UACMU Sites | p | Functional Description |
|---|---|---|---|---|
| lmo2638 | 1887 | 10 | 0.9738 | NADH dehydrogenase |
| secY | 1296 | 8 | 0.9436 | preprotein translocase subunit SecY |
| lmo1310 | 1305 | 8 | 0.9413 | hypothetical protein |
| bvrB | 1923 | 8 | 0.9303 | beta-glucoside-specific phosphotransferase enzyme II ABC component |
| lmo1911 | 1137 | 7 | 0.8988 | histidine kinase |
| lmo0835 | 1005 | 6 | 0.8770 | peptidoglycan binding protein |
| lmo1090 | 984 | 6 | 0.8698 | glycosyltransferase |
| lmo1030 | 1029 | 6 | 0.8600 | LacI family transcriptional regulator |
| lmo0880 | 1389 | 6 | 0.8261 | wall associated protein precursor |
| pgi | 1353 | 6 | 0.8259 | glucose-6-phosphate isomerase |
| Strain or Plasmid | Genotype and Relevant Characteristics | Source or Reference |
|---|---|---|
| E. coli DH5α | Plasmid construction and cloning | Lab stock |
| L. monocytogenes strains | ||
| EGDe | L. monocytogenes virulent wild-type BUG1600, Lineage ΙΙ, serotype 1/2a, MLST ST35 | O. Dussurget |
| ∆clpP | clpP deletion mutant derived from L .monocytogenes EDGe | This study |
| ∆mazEF | lmo0887 and lmo0888 (mazEF antitoxin/toxin) deletion mutant derived from L. monocytogenes EDGe | This study |
| ∆sigB | sigB deletion mutant derived from L. monocytogenes EDGe | This study |
| Plasmids | ||
| pAUL-A | Temperature sensitive origin of replication, lacZa’ multiple cloning site, erythromycin resistance marker | [54] |
| Primer | Purpose | Sequence (5′- > 3′) a,b |
|---|---|---|
| Mutagenesis | ||
| ClpP_UpSt_A | A primer: EcoRΙ | AAAGAATTCCAGTTAATGGGCCAGATT |
| ClpP_UpSt_B | B primer | ACGAATGGTCAAACTAGG |
| ClpP_DnSt_C | C primer | CCTAGTTTGACCATTCGTGCACAAAATGCAAAACCTCT |
| ClpP_DnSt_D | D primer: BamHΙ | AAAGGATCCCGTGACGGATTATTACCA |
| ClpP_IC_Fw | Integration and deletion check | TGGCTCTAACGATGATCTTG |
| ClpP_IC_Rv | TTGATGTTAGTGCACCTGTTG | |
| mazEF_A_Hind | A primer: Hind ΙΙΙ | ATGCAAGCTTTTAGTAGGCGGGGAACTTGCC |
| mazEF_B | B primer | TAACACGTGTCACACCCCCAA |
| mazEF _C | C primer | TTGGGGGTGTGACACGTGTTAGGTTAATGGCTGATGGTGAA |
| mazEF _D_Bam | D primer: BamHΙ | ATGCGGATCCTCAGACCCTTTTGCCCTGC |
| mazEF _IC_Fw | Integration and deletion check | CCTTCCACAGAAATCAAAAC |
| mazEF _IC_Rv | CCAACCTTTCTCCACTATT | |
| SigB_A_3 | A primer: EcoRΙ | AAAGAATTCAGCTGTAAGTGAAGCCATCAC |
| SigB_B_3 | B primer | CGCCTCTTTATCAGGTTGAGA |
| SigB_C_3 | C primer | TCTCAACCTGATAAAGAGGCGGTGTCTAGAATCCAACGTCAA |
| SigB_D_3 | D primer: BamHΙ | AAAGGATCCTAATAGCTATCGCAGCACC |
| SigB_IC_Fw | Integration and deletion check | CGTCAACGCCAAAGTGAA |
| SigB_IC_Rv | CACCTTTCAAACCATCGCTA | |
| qPCR | ||
| mazF_qPCR_Fw | Quantification of mazF | ACGGCCTGTTCTCATCATTC |
| mazF_qPCR_Rv | expression | CGTTGGCAATTTTGCTTTTT |
| sigB_qPCR_Fw | Quantification of sigB | GAAGCAATGGAAATGGGAAA |
| sigB_qPCR_Rv | expression | CCGTACCACCAACAACATCA |
| rsbW_qPCR_Fw | Quantification of rsbW | ATTACAACTTCCTGCCAAGC |
| rsbW_qPCR_Rv | expression | AATTGCTTCATAAGAAAATCCTG |
| opuCA_qPCR_Fw | Quantification of opuCA | ACATCGATAAAGGAGAATTTC |
| opuCA_qPCR_Rv | expression | GCCGGTTAATCATCTTCATTG |
| 2638_qPCR_Fw | Quantification of lmo2638 | CTGCTGCTACATCTGGTGCT |
| 2638_qPCR_Rv | expression | ACTGGAACCAACCAGGCATA |
| bvrB_qPCR_Fw | Quantification of bvrB | GCAATTGGCGCTAAAACTTC |
| bvrB_qPCR_Rv | expression | ATTGTAACGATGGCGGTTTC |
| 0880_qPCR_Fw | Quantification of lmo0880 | ATTCCGAACAAAATGGCAAA |
| 0880_qPCR_Rv | expression | TTTCTGCAACGGAGACATCA |
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Curtis, T.D.; Takeuchi, I.; Gram, L.; Knudsen, G.M. The Influence of the Toxin/Antitoxin mazEF on Growth and Survival of Listeria monocytogenes under Stress. Toxins 2017, 9, 31. https://doi.org/10.3390/toxins9010031
Curtis TD, Takeuchi I, Gram L, Knudsen GM. The Influence of the Toxin/Antitoxin mazEF on Growth and Survival of Listeria monocytogenes under Stress. Toxins. 2017; 9(1):31. https://doi.org/10.3390/toxins9010031
Chicago/Turabian StyleCurtis, Thomas D., Ippei Takeuchi, Lone Gram, and Gitte M. Knudsen. 2017. "The Influence of the Toxin/Antitoxin mazEF on Growth and Survival of Listeria monocytogenes under Stress" Toxins 9, no. 1: 31. https://doi.org/10.3390/toxins9010031






