Aflatoxin B1-Induced Developmental and DNA Damage in Caenorhabditis elegans
Abstract
:1. Introduction
2. Results
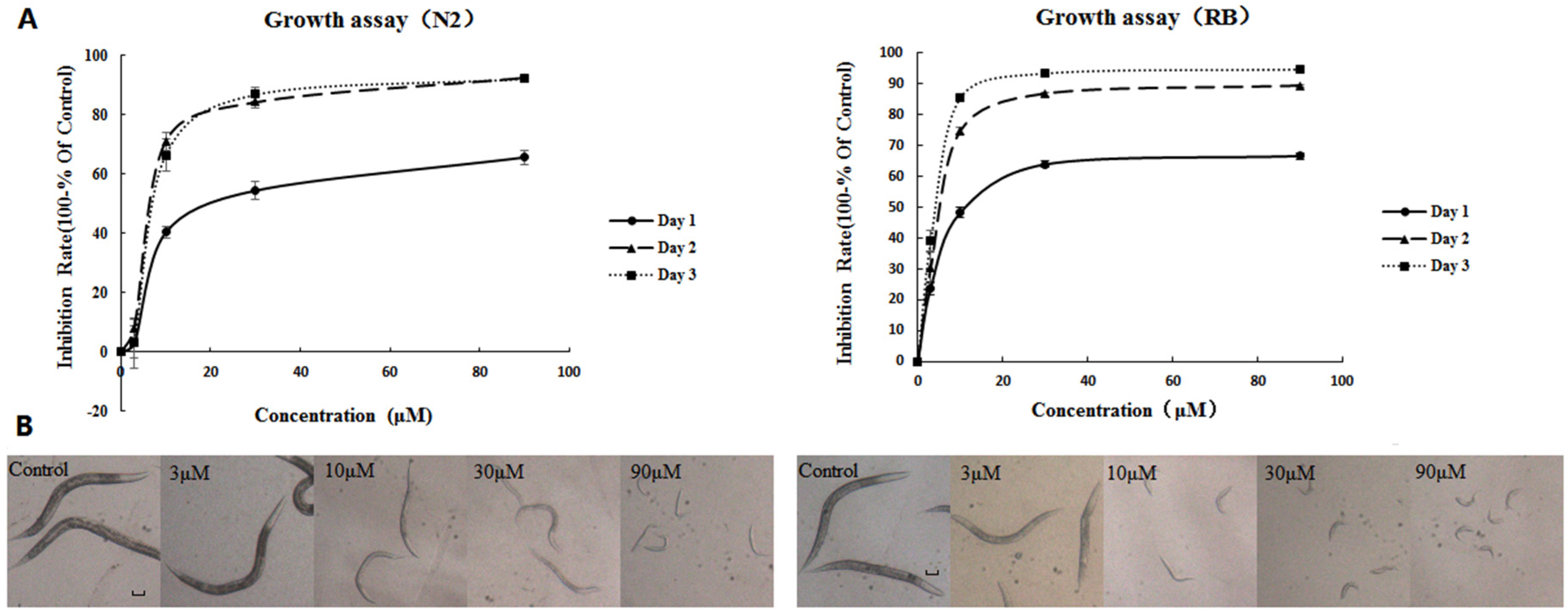
2.1. AFB1 Effects on the Growth of C. elegans
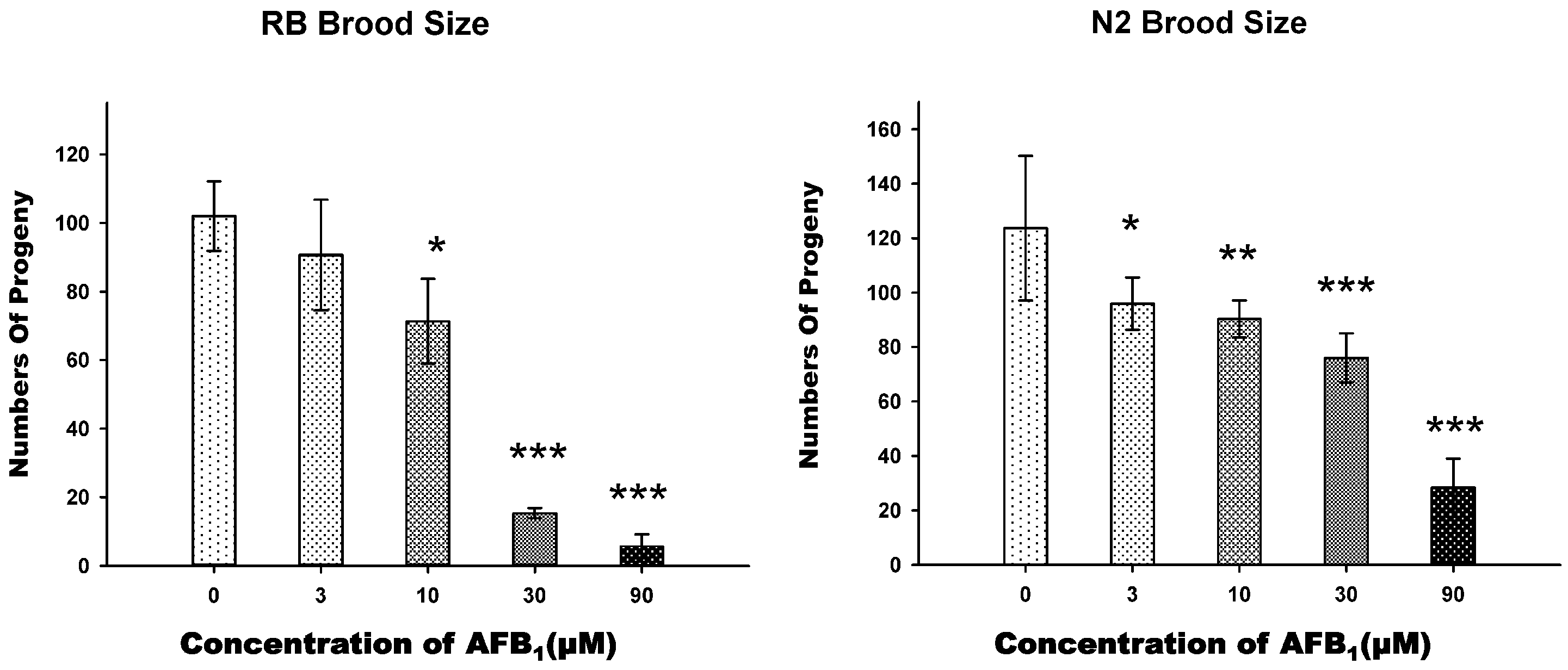
2.2. AFB1 Effects on the Brood Size of C. elegans
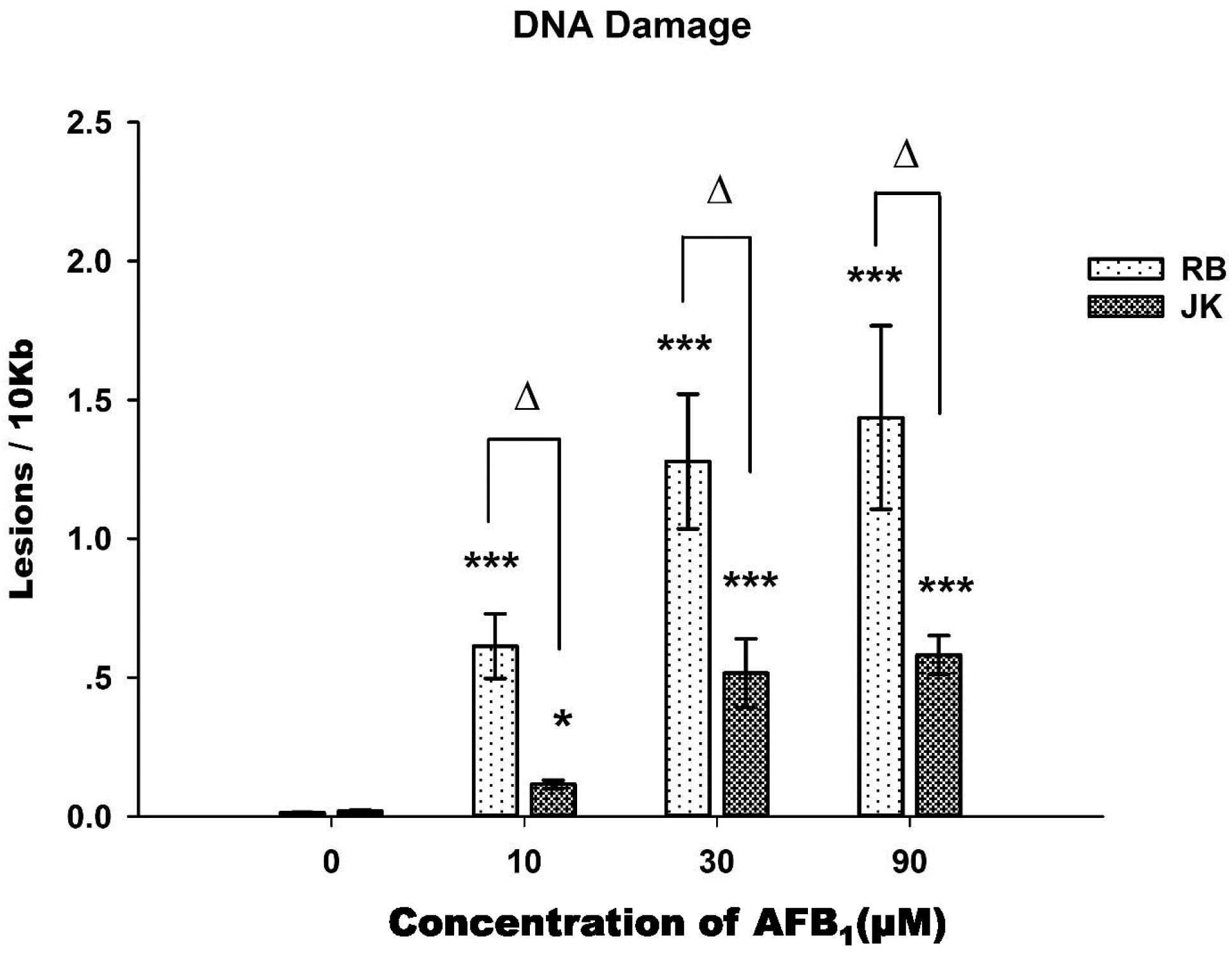
2.3. DNA Damage Assay
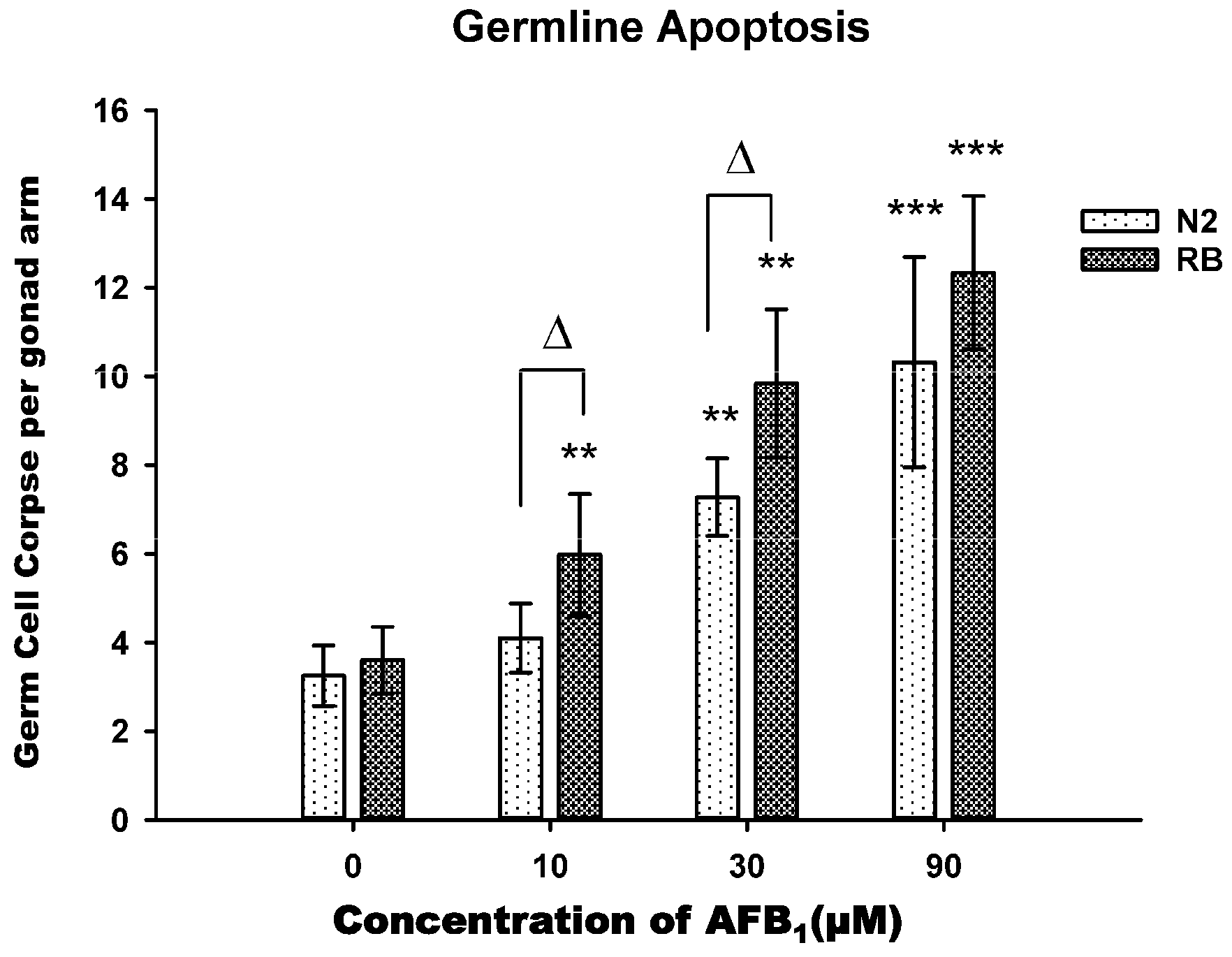
2.4. AFB1-Induced Germ Line Apoptosis
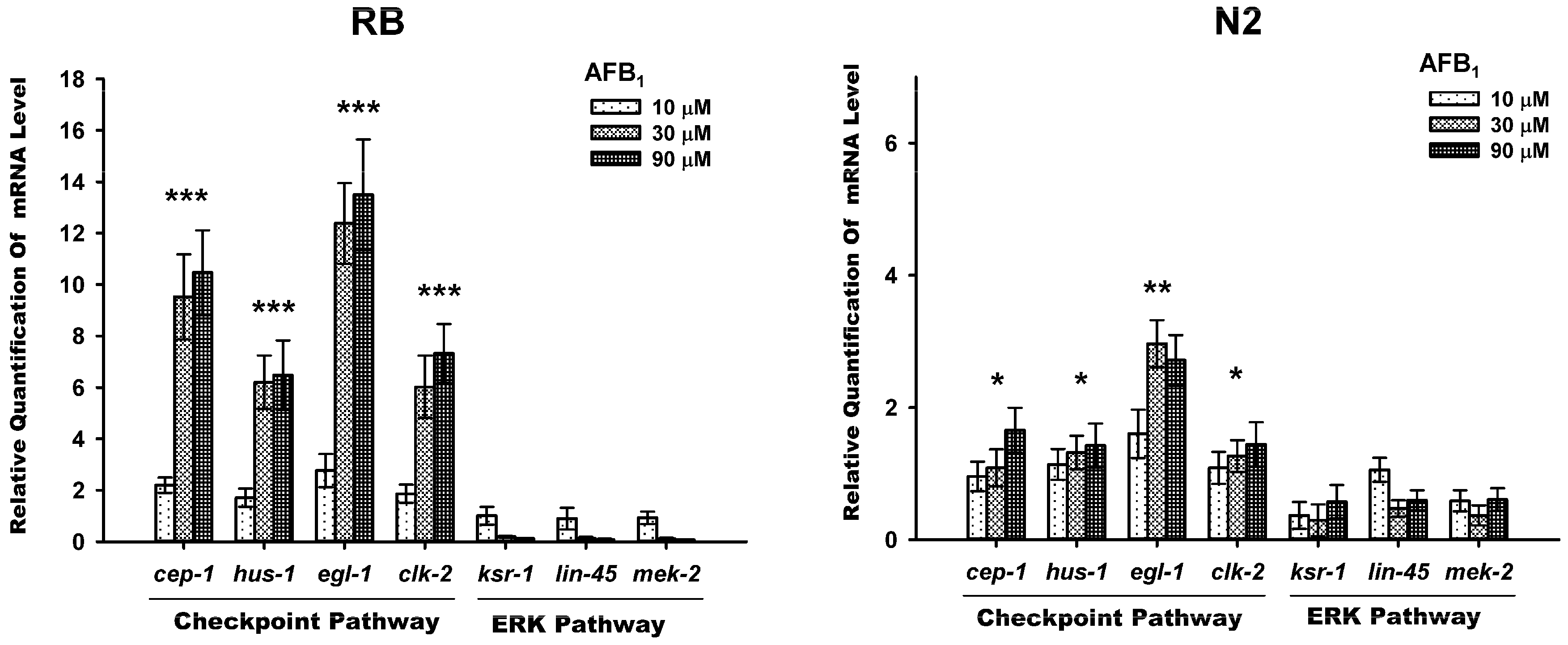
2.5. AFB1-Induced Gemline Apoptosis via the Checkpoint Pathway, Not ERK Pathway
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. C. elegans Culture
5.2. Chemicals Exposures
5.3. Growth Assay
5.4. Brood Size Assay
5.5. DNA Damage Assay
5.6. Real-Time PCR
5.7. Apoptosis Assay
5.8. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Busby, W.F.; Wogan, G.N. Aflatoxin. In Chemical Carcinogens, 2nd ed.; Searle, C.E., Ed.; American Chemical Society: Washington, DC, USA, 1984; pp. 945–1136. [Google Scholar]
- Creppy, E.E. Update of survey, regulation and toxic effects of mycotoxins in europe. Toxicol. Lett. 2002, 127, 19–28. [Google Scholar] [CrossRef]
- Dvorackova, I.; Stora, C.; Ayraud, N. Evidence of Aflatoxin B1 in two cases of lung cancer in man. J. Cancer Res. Clin. Oncol. 1981, 100, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Tam, A.S.; Foley, J.F.; Devereux, T.R.; Maronpot, R.R.; Massey, T.E. High frequency and heterogeneous distribution of p53 mutations in Aflatoxin B1-induced mouse lung tumors. Cancer Res. 1999, 59, 3634–3640. [Google Scholar] [PubMed]
- Guengerich, F.P.; Johnson, W.W.; Shimada, T.; Ueng, Y.F.; Yamazaki, H.; Langouet, S. Activation and detoxication of Aflatoxin B1. Mutat. Res. 1998, 402, 121–128. [Google Scholar] [CrossRef]
- Wang, J.S.; Groopman, J.D. DNA damage by mycotoxins. Mutat. Res. 1999, 424, 167–181. [Google Scholar] [CrossRef]
- Madrigal-Santillan, E.; Morales-Gonzalez, J.A.; Vargas-Mendoza, N.; Reyes-Ramirez, P.; Cruz-Jaime, S.; Sumaya-Martinez, T.; Perez-Pasten, R.; Madrigal-Bujaidar, E. Antigenotoxic studies of different substances to reduce the DNA damage induced by Aflatoxin B1 and ochratoxin A. Toxins 2010, 2, 738–757. [Google Scholar] [CrossRef] [PubMed]
- Kensler, T.W.; Roebuck, B.D.; Wogan, G.N.; Groopman, J.D. Aflatoxin: A 50-year odyssey of mechanistic and translational toxicology. Toxicol. Sci. 2011, 120 (Suppl. 1), S28–S48. [Google Scholar] [CrossRef] [PubMed]
- Faisal, K.; Periasamy, V.S.; Sahabudeen, S.; Radha, A.; Anandhi, R.; Akbarsha, M.A. Spermatotoxic effect of Aflatoxin B1 in rat: Extrusion of outer dense fibres and associated axonemal microtubule doublets of sperm flagellum. Reproduction 2008, 135, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Leung, M.C.; Williams, P.L.; Benedetto, A.; Au, C.; Helmcke, K.J.; Aschner, M.; Meyer, J.N. Caenorhabditis elegans: An emerging model in biomedical and environmental toxicology. Toxicol. Sci. 2008, 106, 5–28. [Google Scholar] [CrossRef] [PubMed]
- Stergiou, L.; Hengartner, M.O. Death and more: DNA damage response pathways in the nematode C. elegans. Cell Death Differ. 2004, 11, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Leung, M.C.; Goldstone, J.V.; Boyd, W.A.; Freedman, J.H.; Meyer, J.N. Caenorhabditis elegans generates biologically relevant levels of genotoxic metabolites from Aflatoxin B1 but not benzo[a]pyrene in vivo. Toxicol. Sci. 2010, 118, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J.N.; Boyd, W.A.; Azzam, G.A.; Haugen, A.C.; Freedman, J.H.; Van Houten, B. Decline of nucleotide excision repair capacity in aging Caenorhabditis elegans. Genome Biol. 2007, 8, R70. [Google Scholar] [CrossRef] [PubMed]
- Wood, R.D. DNA damage recognition during nucleotide excision repair in mammalian cells. Biochimie 1999, 81, 39–44. [Google Scholar] [CrossRef]
- Wilson, D.M., 3rd; Thompson, L.H. Life without DNA repair. Proc. Natl. Acad. Sci. USA 1997, 94, 12754–12757. [Google Scholar] [CrossRef] [PubMed]
- Kaletta, T.; Hengartner, M.O. Finding function in novel targets: C. elegans as a model organism. Nat. Rev. Drug Discov. 2006, 5, 387–398. [Google Scholar] [CrossRef] [PubMed]
- Rajini, P.S.; Melstrom, P.; Williams, P.L. A comparative study on the relationship between various toxicological endpoints in Caenorhabditis elegans exposed to organophosphorus insecticides. J. Toxicol. Environ. Health Part A 2008, 71, 1043–1050. [Google Scholar] [CrossRef] [PubMed]
- Sprando, R.L.; Olejnik, N.; Cinar, H.N.; Ferguson, M. A method to rank order water soluble compounds according to their toxicity using Caenorhabditis elegans, a complex object parametric analyzer and sorter, and axenic liquid media. Food Chem. Toxicol. 2009, 47, 722–728. [Google Scholar] [CrossRef] [PubMed]
- Leelaja, B.C.; Rajini, P.S. Biochemical and physiological responses in Caenorhabditis elegans exposed to sublethal concentrations of the organophosphorus insecticide, monocrotophos. Ecotoxicol. Environ. Saf. 2013, 94, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Xue, K.S.; Sun, X.; Tang, L.; Wang, J.S. Multi-toxic endpoints of the foodborne mycotoxins in nematode Caenorhabditis elegans. Toxins 2015, 7, 5224–5235. [Google Scholar] [CrossRef] [PubMed]
- Dhanapal, J.; Ravindrran, M.B.; Baskar, S.K. Toxic effects of Aflatoxin B1 on embryonic development of zebrafish (danio rerio): Potential activity of piceatannol encapsulated chitosan/poly (lactic acid) nanoparticles. Anti-Cancer Agents Med. Chem. 2015, 15, 248–257. [Google Scholar] [CrossRef]
- McKean, C.; Tang, L.; Billam, M.; Tang, M.; Theodorakis, C.W.; Kendall, R.J.; Wang, J.S. Comparative acute and combinative toxicity of Aflatoxin B1 and T-2 toxin in animals and immortalized human cell lines. J. Appl. Toxicol. 2006, 26, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Hoeijmakers, J.H. Genome maintenance mechanisms for preventing cancer. Nature 2001, 411, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Astin, J.W.; O’neil, N.J.; Kuwabara, P.E. Nucleotide excision repair and the degradation of RNA pol II by the Caenorhabditis elegans XPA and Rsp5 orthologues, RAD-3 and WWP-1. DNA Repair 2008, 7, 267–280. [Google Scholar] [CrossRef] [PubMed]
- Gumienny, T.L.; Lambie, E.; Hartwieg, E.; Horvitz, H.R.; Hengartner, M.O. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development 1999, 126, 1011–1022. [Google Scholar] [PubMed]
- Vermezovic, J.; Stergiou, L.; Hengartner, M.O.; d’Adda di Fagagna, F. Differential regulation of DNA damage response activation between somatic and germline cells in Caenorhabditis elegans. Cell Death Differ. 2012, 19, 1847–1855. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.G.; Yang, Z.B.; Wang, Y.; Yang, W.R.; Zhou, H.J. Effects of dietary supplementation of multi-enzyme on growth performance, nutrient digestibility, small intestinal digestive enzyme activities, and large intestinal selected microbiota in weanling pigs. J. Anim. Sci. 2014, 92, 2063–2069. [Google Scholar] [CrossRef] [PubMed]
- Gartner, A.; Milstein, S.; Ahmed, S.; Hodgkin, J.; Hengartner, M.O. A conserved checkpoint pathway mediates DNA damage—induced apoptosis and cell cycle arrest in C. elegans. Mol. Cell 2000, 5, 435–443. [Google Scholar] [CrossRef]
- Reddy, L.; Odhav, B.; Bhoola, K. Aflatoxin B1-induced toxicity in HepG2 cells inhibited by carotenoids: Morphology, apoptosis and DNA damage. Biol. Chem. 2006, 387, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Raj, H.G.; Kohli, E.; Rohil, V.; Dwarakanath, B.S.; Parmar, V.S.; Malik, S.; Adhikari, J.S.; Tyagi, Y.K.; Goel, S.; Gupta, K.; et al. Acetoxy-4-methylcoumarins confer differential protection from Aflatoxin B1-induced micronuclei and apoptosis in lung and bone marrow cells. Mutat. Res. 2001, 494, 31–40. [Google Scholar] [CrossRef]
- Al-Hammadi, S.; Marzouqi, F.; Al-Mansouri, A.; Shahin, A.; Al-Shamsi, M.; Mensah-Brown, E.; Souid, A.K. The cytotoxicity of Aflatoxin B1 in human lymphocytes. Sultan Qaboos Univ. Med. J. 2014, 14, e65–e71. [Google Scholar] [CrossRef] [PubMed]
- Cagnol, S.; Chambard, J.C. ERK and cell death: Mechanisms of ERK-induced cell death—Apoptosis, autophagy and senescence. FEBS J. 2010, 277, 2–21. [Google Scholar] [CrossRef] [PubMed]
- Brenner, S. The genetics of Caenorhabditis elegans. Genetics 1974, 77, 71–94. [Google Scholar] [PubMed]
- Kodoyianni, V.; Maine, E.M.; Kimble, J. Molecular basis of loss-of-function mutations in the glp-1 gene of Caenorhabditis elegans. Mol. Biol. Cell 1992, 3, 1199–1213. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.A.; Fleming, J.T. Basic culture methods. Methods Cell Biol. 1995, 48, 3–29. [Google Scholar] [PubMed]
- Williams, P.L.; Dusenbery, D.B. Aquatic toxicity testing using the nematode, Caenorhabditis elegans. Environ. Toxicol. Chem. 1990, 9, 1285–1290. [Google Scholar] [CrossRef]
- Bischof, L.J.; Huffman, D.L.; Aroian, R.V. Assays for toxicity studies in C. elegans with Bt crystal proteins. Methods Mol. Biol. 2006, 351, 139–154. [Google Scholar] [PubMed]
- Hunter, S.E.; Jung, D.; Di Giulio, R.T.; Meyer, J.N. The QPCR assay for analysis of mitochondrial DNA damage, repair, and relative copy number. Methods 2010, 51, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J.N. Qpcr: A tool for analysis of mitochondrial and nuclear DNA damage in ecotoxicology. Ecotoxicology 2010, 19, 804–811. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Kelly, K.O.; Dernburg, A.F.; Stanfield, G.M.; Villeneuve, A.M. Caenorhabditis elegans msh-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics 2000, 156, 617–630. [Google Scholar] [PubMed]
- Wang, S.; Zhao, Y.; Wu, L.; Tang, M.; Su, C.; Hei, T.K.; Yu, Z. Induction of germline cell cycle arrest and apoptosis by sodium arsenite in Caenorhabditis elegans. Chem. Res. Toxicol. 2007, 20, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Bretz, F. An extension of the williams trend test to general unbalanced linea models. Comput. Stat. Data Anal. 2006, 50, 1735–1748. [Google Scholar] [CrossRef]
| Time (h) | N2 (95% CI) | xpa-1 (95% CI) |
|---|---|---|
| 24 | 7.87 (6.93–8.93) | 5.02 (4.38–5.76) |
| 48 | 6.36 (5.91–6.86) | 4.25 (3.92–4.62) |
| 72 | 7.40 (6.79–8.01) | 3.54 (3.37–3.71) |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, W.-H.; Xue, K.S.; Tang, L.; Williams, P.L.; Wang, J.-S. Aflatoxin B1-Induced Developmental and DNA Damage in Caenorhabditis elegans. Toxins 2017, 9, 9. https://doi.org/10.3390/toxins9010009
Feng W-H, Xue KS, Tang L, Williams PL, Wang J-S. Aflatoxin B1-Induced Developmental and DNA Damage in Caenorhabditis elegans. Toxins. 2017; 9(1):9. https://doi.org/10.3390/toxins9010009
Chicago/Turabian StyleFeng, Wei-Hong, Kathy S. Xue, Lili Tang, Phillip L. Williams, and Jia-Sheng Wang. 2017. "Aflatoxin B1-Induced Developmental and DNA Damage in Caenorhabditis elegans" Toxins 9, no. 1: 9. https://doi.org/10.3390/toxins9010009





