Variation and Distribution of L-A Helper Totiviruses in Saccharomyces sensu stricto Yeasts Producing Different Killer Toxins
Abstract
:1. Introduction
2. Results
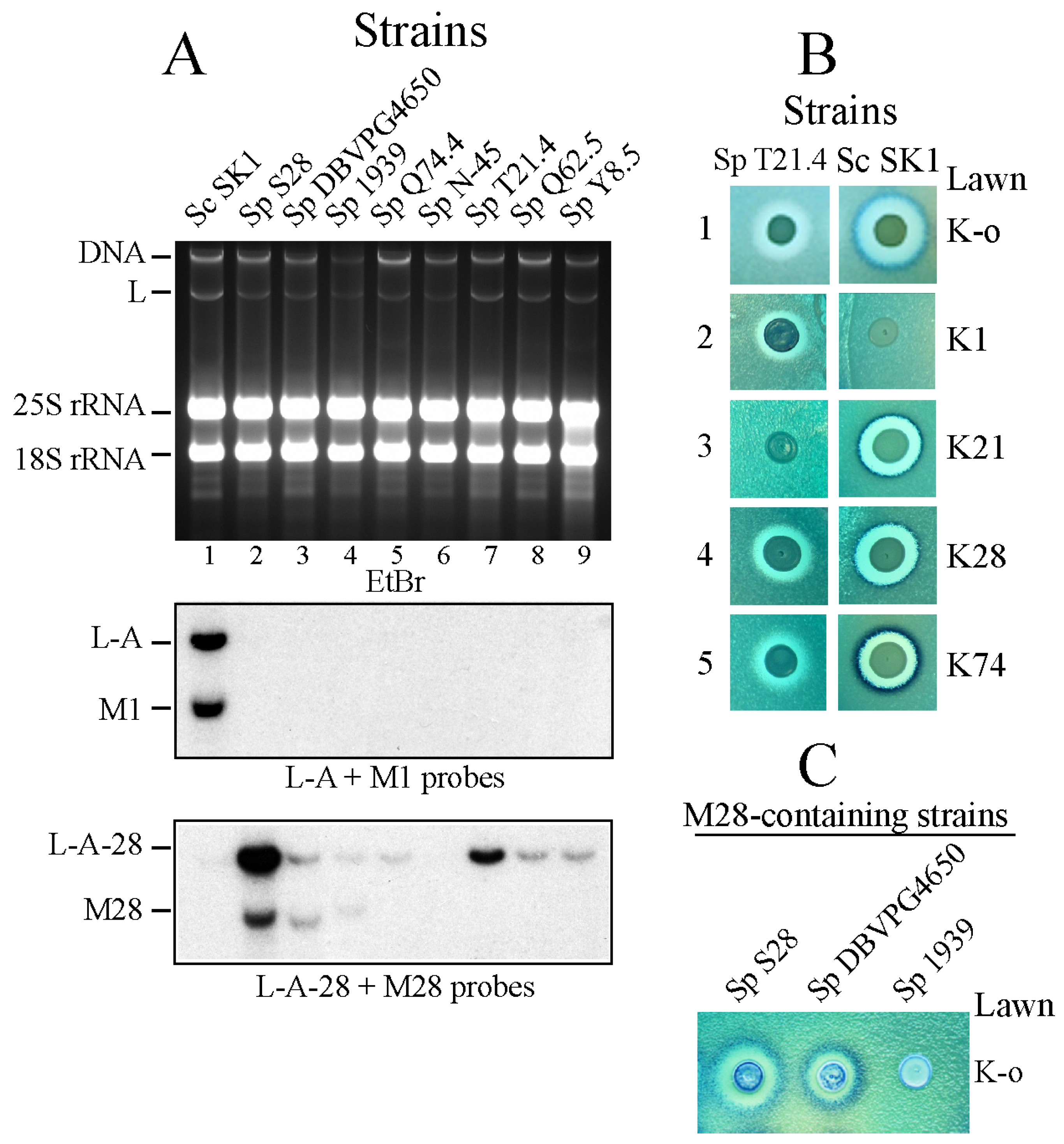
2.1. Killer Viruses from S. paradoxus Are Different from Those from S. cerevisiae
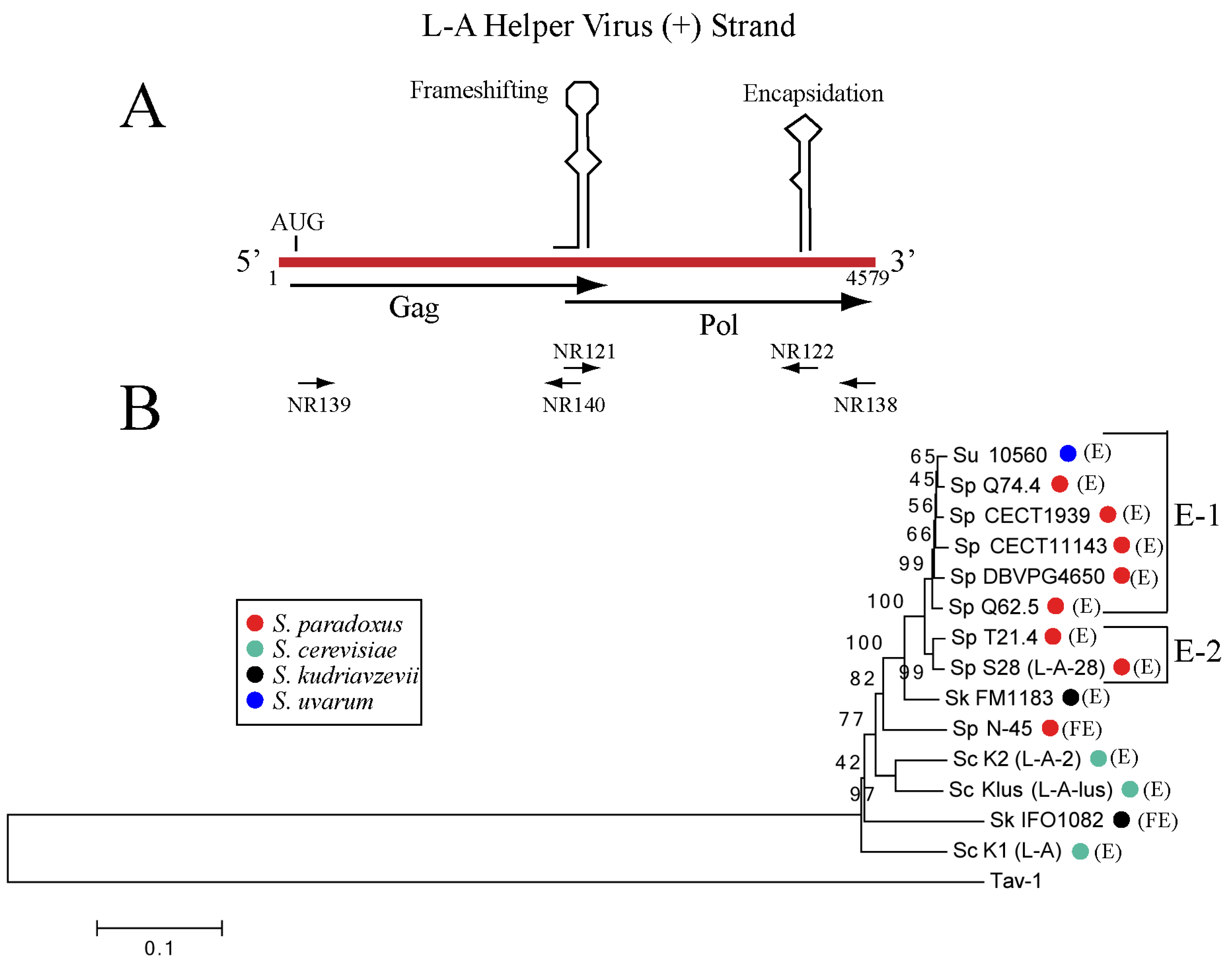
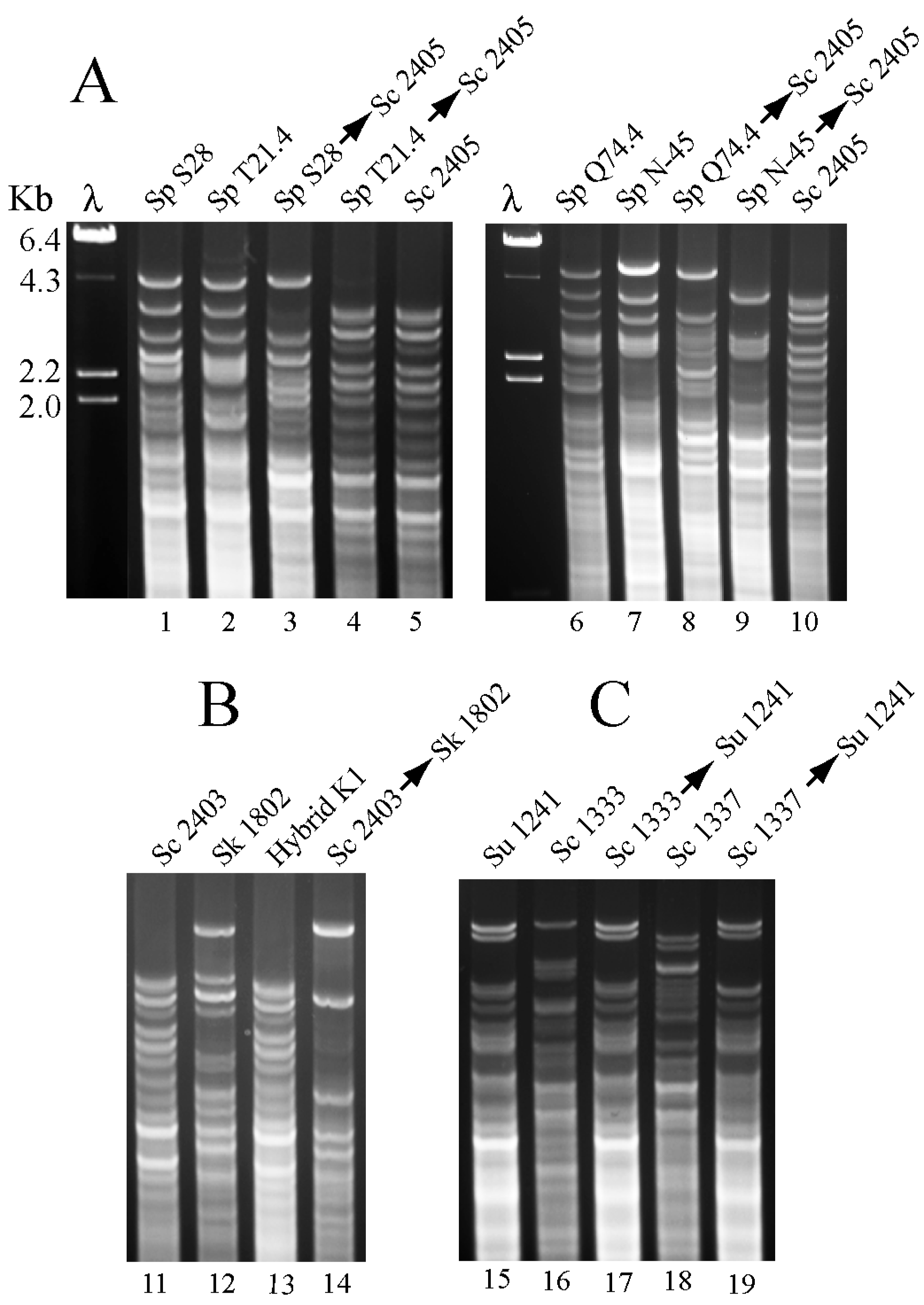
2.2. Different L-A Variants in S. paradoxus Strains
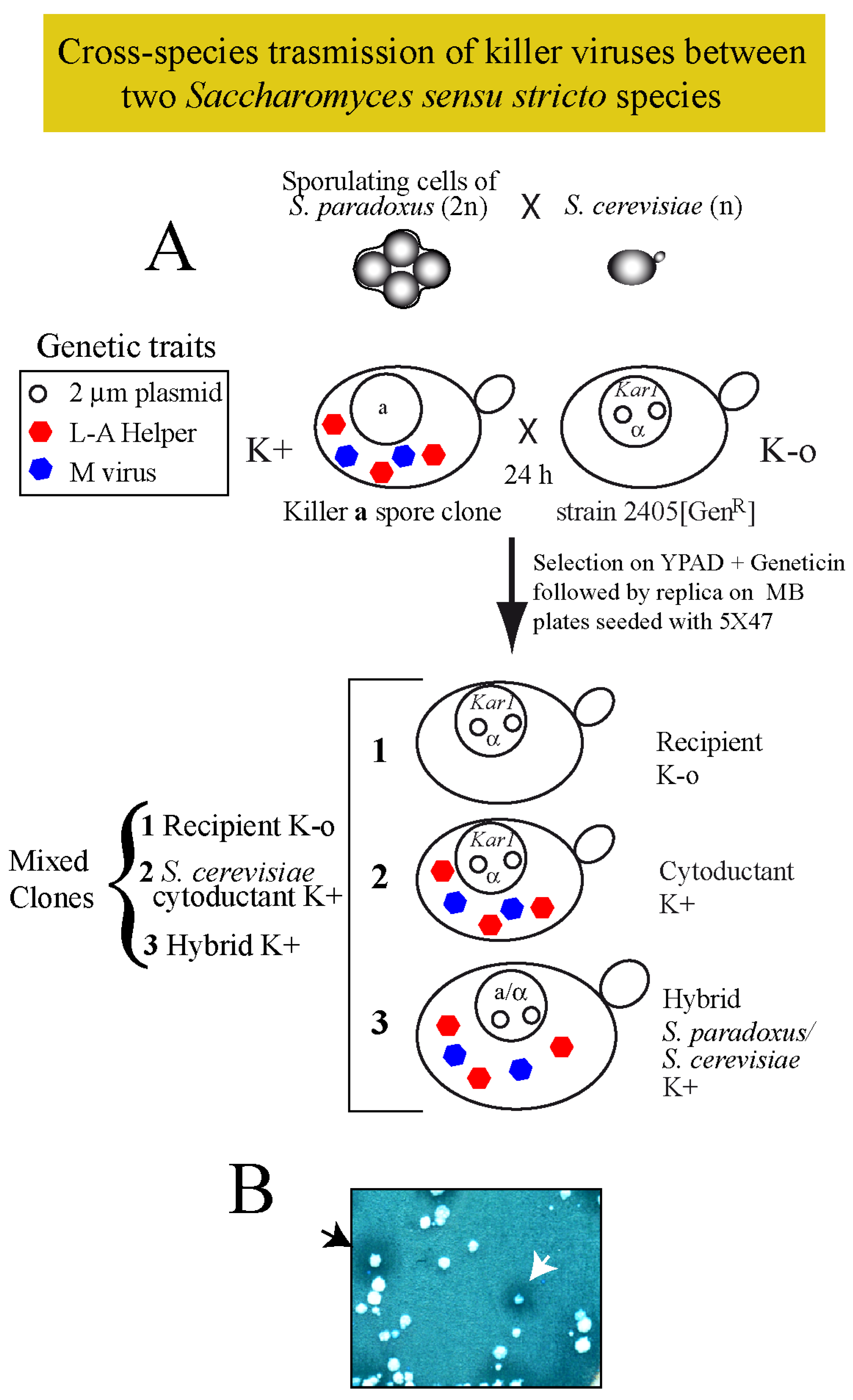
2.3. Killer Viruses Cross-Transmission between Saccharomyces Sensu Stricto Species
2.3.1. Maintenance of S. paradoxus Viruses in S. cerevisiae
2.3.2. Killer Viruses Cross-Transmission between Other Saccharomyces sensu stricto Species
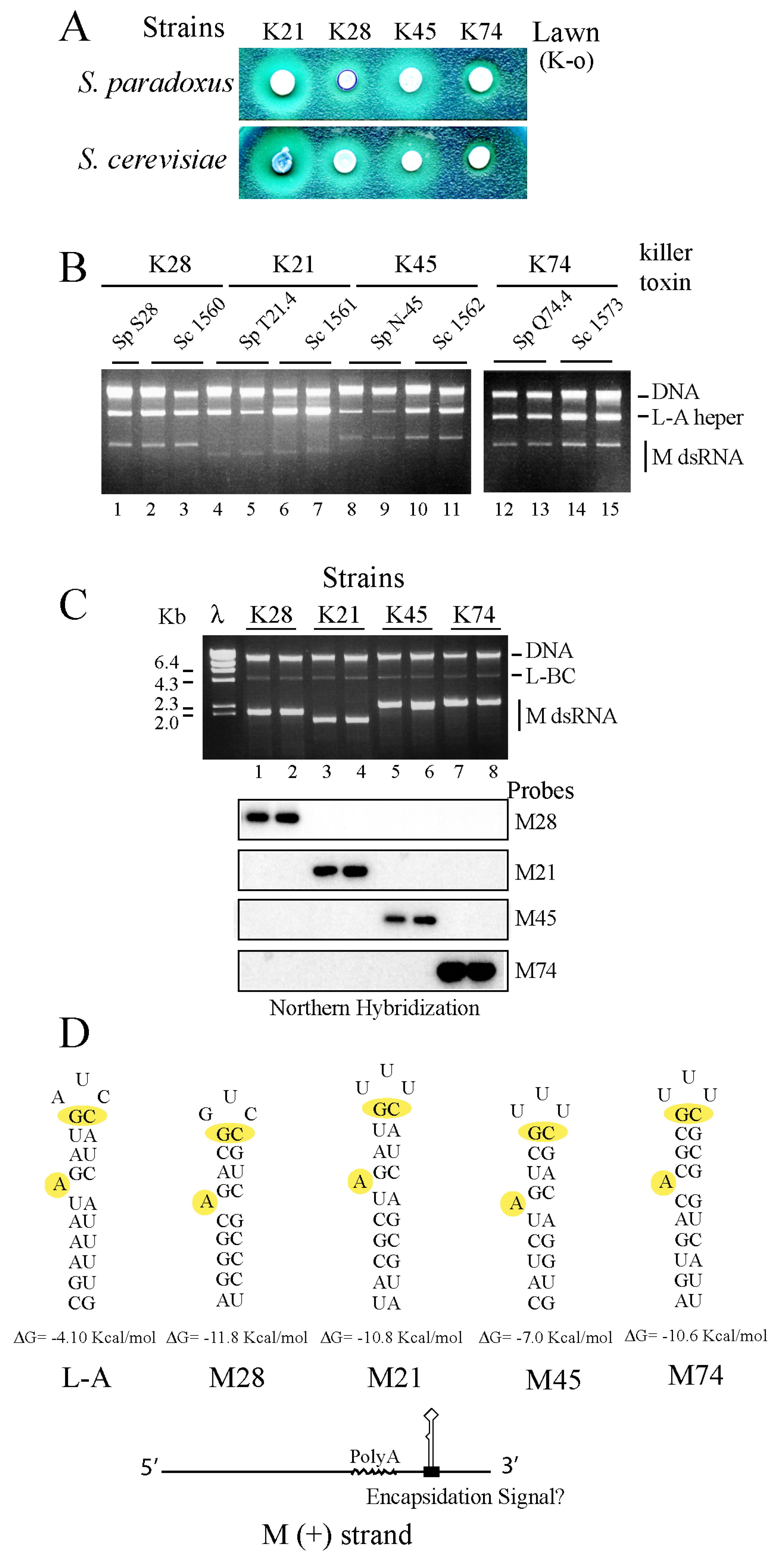
2.4. Mitochondria Incompatibility as the Main Barrier for Killer Viruses Spreading in the Wild?
2.5. M28, M21, M45 and M74 Can Be Maintained by L-A Proteins Expressed from a Vector
3. Discussion
3.1. Variation of L-A Helper Sequences Depend Mostly on the Geographical Location of the Host
3.2. Host–Virus Adaptation Previous to Speciation
3.3. Origins of Toxin-Encoding M Satellites
4. Materials and Methods
4.1. Yeast Strains and Media
4.2. Cytoduction (Cytoplasm Mixing)
4.3. Killer Assay
4.4. cDNA Synthesis and Sequencing of L-A Variants from Saccharomyces sensu stricto
4.5. Northern Hybridization and Plasmids
4.6. Other Procedures
4.7. GenBank Accession Numbers
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wickner, R.B.; Ghabrial, S.A.; Nibert, M.L.; Patterson, J.L.; Wang, C.C. Family Totiviridae. In Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowits, E.J., Eds.; Elsevier Academic Press: Tokyo, Japan, 2011; pp. 639–650. [Google Scholar]
- Esteban, R.; Fujimura, T.; Wickner, R.B. Internal and terminal cis-acting sites are necessary for in vitro replication of the L-A double-stranded RNA virus of yeast. EMBO J. 1989, 8, 947–954. [Google Scholar] [PubMed]
- Icho, T.; Wickner, R.B. The double-stranded RNA genome of yeast virus L-A encodes its own putative RNA polymerase by fusing two open reading frames. J. Biol. Chem. 1989, 264, 6716–6723. [Google Scholar] [PubMed]
- Dinman, J.D.; Icho, T.; Wickner, R.B. A -1 ribosomal frame shift in a double-stranded RNA virus of yeast forms a gag–pol fusion protein. Proc. Natl. Acad. Sci. USA 1991, 88, 174–178. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.J.; Breinig, F. Yeast viral killer toxins: Lethality and self-protection. Nat. Rev. Microbiol. 2006, 4, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Wickner, R.B.; Fujimura, T.; Esteban, R. Viruses and prions of Saccharomyces cerevisiae. Adv. Virus Res. 2013, 86, 1–36. [Google Scholar] [CrossRef] [PubMed]
- Bevan, E.A.; Herring, A.J.; Mitchell, D.J. Preliminary characterization of two species of dsRNA in yeast and their relationship to the “killer” character. Nature 1973, 245, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Hannig, E.M.; Leibowitz, M.J. Structure and expression of the M2 genomic segment of a type 2 killer virus of yeast. Nucleic Acids Res. 1985, 13, 4379–4400. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.J.; Tipper, D.J. K28, a unique double-stranded RNA killer virus of Saccharomyces cerevisiae. Mol. Cell. Biol. 1990, 10, 4807–4815. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Cousiño, N.; Maqueda, M.; Ambrona, J.; Zamora, E.; Esteban, R.; Ramírez, M. A new wine Saccharomyces cerevisiae killer toxin (Klus), encoded by a double-stranded RNA virus, with broad antifungal activity is evolutionarily related to a chromosomal host gene. Appl. Environ. Microbiol. 2011, 77, 1822–1832. [Google Scholar] [CrossRef] [PubMed]
- De la Peña, P.; Barros, F.; Gascón, S.; Lazo, P.S.; Ramos, S. Effect of yeast killer toxin on sensitive yeast cells of Saccharomyces cerevisiae. J. Biol. Chem. 1981, 256, 10420–10425. [Google Scholar] [PubMed]
- Martinac, B.; Zhu, H.; Kubalski, A.; Zhou, X.L.; Culbertson, M.; Bussey, H.; Kung, C. Yeast K1 killer toxin forms ion channels in sensitive yeast spheroplasts and in artificial liposomes. Proc. Natl. Acad. Sci. USA 1990, 87, 6228–6232. [Google Scholar] [CrossRef] [PubMed]
- Lukša, J.; Podoliankaitė, M.; Vepštaitė, I.; Strazdaitė-Žielienė, Ž.; Urbonavičius, J.; Servienė, E. Yeast β-1,6-glucan is a primary target for the Saccharomyces cerevisiae K2 toxin. Eukaryot Cell 2015, 14, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Orentaite, I.; Poranen, M.M.; Oksanen, H.M.; Daugelavicius, R.; Bamford, D.H. K2 killer toxin-induced physiological changes in the yeast Saccharomyces cerevisiae. FEMS Yeast Res. 2016, 16, fow003. [Google Scholar] [CrossRef] [PubMed]
- Eisfeld, K.; Riffer, F.; Mentges, J.; Schmitt, M.J. Endocytotic uptake and retrograde transport of a virally encoded killer toxin in yeast. Mol. Microbiol. 2000, 37, 926–940. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.J.; Tipper, D.J. Sequence of the M28 dsRNA: Preprotoxin is processed to an α/β heterodimeric protein. Virology 1995, 213, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Cousiño, N.; Gómez, P.; Esteban, R. L-A-lus, a new variant of L-A totivirus in wine yeasts associated to Klus killer toxin-producing Mlus dsRNA. Possible role of satellite RNAs encoding killer toxins on the evolution of their helper viruses. Appl. Environ. Microbiol. 2013, 79, 4661–4674. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Cousiño, N.; Esteban, R. Relationships and Evolution of Double-Stranded RNA Totiviruses of Yeasts Inferred from Analysis of L-A-2 and L-BC Variants in Wine Yeast Strain Populations. Appl. Environ. Microbiol. 2017, 83, e02991-16. [Google Scholar] [CrossRef] [PubMed]
- Konovalovas, A.; Serviené, E.; Serva, S. Genome Sequence of Saccharomyces cerevisiae Double-Stranded RNA Virus L-A-28. Genome Announc. 2016, 4, e00549-16. [Google Scholar] [CrossRef] [PubMed]
- Park, C.M.; Bruenn, J.A.; Ganesa, C.; Flurkey, W.F.; Bozarth, R.F.; Koltin, Y. Structure and heterologous expression of the Ustilago maydis viral toxin KP4. Mol. Microbiol. 1994, 11, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.J.; Neuhausen, F. Killer toxing-secreting double-stranded RNA mycoviruses in the yeasts Hanseniaspora uvarum and Zygosaccharomyces bailii. J. Virol. 1994, 68, 1765–1772. [Google Scholar] [PubMed]
- Ramírez, M.; Velázquez, R.; Maqueda, M.; López-Piñeiro, A.; Ribas, J.C. A new wine Torulaspora delbrueckii killer strain with broad antifungal activity and its toxin-encoding double-stranded RNA virus. Front. Microbiol. 2015, 6, 983. [Google Scholar] [CrossRef] [PubMed]
- Ivannikova, Y.V.; Naumova, E.S.; Naumov, G.I. Viral dsRNA in the wine yeast Saccharomyces bayanus var. uvarum. Res. Microbiol. 2007, 158, 638–643. [Google Scholar] [CrossRef] [PubMed]
- Pieczynska, M.; de Visser, J.A.G.M.; Korona, R. Incidence of symbiotic dsRNA ‘killer’ viruses in wild and domesticated yeast. FEMS Yeast Res. 2013, 13, 856–859. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.L.; Leu, J.Y.; Chang, T.H. A population study of killer viruses reveals different evolutionary histories of two closely related Saccharomyces sensu stricto yeasts. Mol. Ecol. 2015, 24, 4312–4322. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P. Phylogenetic circumscription of Saccharomyces, Kluyveromyces and other members of the Saccharomycetaceae, and the proposal of the new genera Lachancea, Nakaseomyces, Naumovia, Vanderwaltozyma and Zygotorulaspora. FEMS Yeast Res. 2003, 4, 233–245. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Robnett, C.J. Phylogenetic relationships among yeasts of the ‘Saccharomyces complex’ determined from multigene sequence analyses. FEMS Yeast Res. 2003, 3, 417–432. [Google Scholar] [CrossRef]
- Nguyen, H.V.; Gaillardin, C. Evolutionary relationships between the former species Saccharomyces uvarum and the hybrids Saccharomyces bayanus and Saccharomyces pastorianus; reinstatement of Saccharomyces uvarum (Beijerinck) as a distinct species. FEMS Yeast Res. 2005, 5, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Naumov, G.I.; Naumova, E.S.; Masneuf, I.; Aigle, M.; Kondratieva, V.I.; Dubourdieu, D. Natural polyploidization of some cultured yeast Saccharomyces sensu stricto: Auto- and allotetraploidy. Syst. Appl. Microbiol. 2000, 23, 442–449. [Google Scholar] [CrossRef]
- Wang, S.A.; Bai, F.Y. Saccharomyces arboricolus sp. nov., a yeast species from tree bark. Int. J. Syst. Evol. Microbiol. 2008, 58, 510–514. [Google Scholar] [CrossRef] [PubMed]
- Libkind, D.; Hittinger, C.T.; Valério, E.; Gonçalves, C.; Dover, J.; Johnston, M.; Gonçalves, P.; Sampaio, J.P. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc. Natl. Acad. Sci. USA 2011, 108, 14539–14544. [Google Scholar] [CrossRef] [PubMed]
- Hittinger, C.T. Saccharomyces diversity and evolution: A budding model genus. Trends Genet. 2013, 29, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Liti, G.; Carter, D.M.; Moses, A.M.; Warringer, J.; Parts, L.; James, S.A.; Davey, R.P.; Roberts, I.N.; Burt, A.; Koufopanou, V.; et al. Population genomics of domestic and wild yeasts. Nature 2009, 458, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Sniegowski, P.D.; Dombrowski, P.G.; Fingerman, E. Saccharomyces cerevisiae and Saccharomyces paradoxus coexist in a natural woodland site in North America and display different levels of reproductive isolation from European conspecifics. FEMS Yeast Res. 2002, 1, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Sampaio, J.P.; Gonçalves, P. Natural populations of Saccharomyces kudriavzevii in Portugal are associated with oak bark and are sympatric with S. cerevisiae and S. paradoxus. Appl. Environ. Microbiol. 2008, 74, 2144–2152. [Google Scholar] [CrossRef] [PubMed]
- Stefanini, I.; Dapporto, L.; Legras, J.L.; Calabretta, A.; Di Paola, M.; De Filippo, C.; Viola, R.; Capretti, P.; Polsinelli, M.; Turillazzi, S.; et al. Role of social wasps in Saccharomyces cerevisiae ecology and evolution. Proc. Natl. Acad. Sci. USA 2012, 109, 13398–13403. [Google Scholar] [CrossRef] [PubMed]
- Stefanini, I.; Dapporto, L.; Berná, L.; Polsinelli, M.; Turillazzi, S.; Cavalieri, D. Social wasps are a Saccharomyces mating nest. Proc. Natl. Acad. Sci. USA 2016, 113, 2247–2251. [Google Scholar] [CrossRef] [PubMed]
- Pieczynska, M.D.; Wloch-Salamon, D.; Korona, R.; de Visser, J.A. Rapid multiple-level coevolution in experimental populations of yeast killer and nonkiller strains. Evolution 2016, 70, 1342–1353. [Google Scholar] [CrossRef] [PubMed]
- Bevan, E.A.; Makower, M. The physiological basis of the killer character in yeast. In Genetics Today; Proceedings of the XIth International Congress of Genetics, The Hague, The Netherlands, September 1963; Geerts, S.J., Ed.; Pergamon Press: Oxford, UK, 1963; pp. 202–203. [Google Scholar]
- Philliskirk, G.; Young, T.W. The occurrence of killer character in yeasts of various genera. Antonie Van Leeuwenhoek 1975, 41, 147–151. [Google Scholar] [CrossRef] [PubMed]
- Diamond, M.E.; Dowhanick, J.J.; Nemeroff, M.E.; Pietras, D.F.; Tu, C.L.; Bruenn, J.A. Overlapping genes in a yeast double-stranded RNA virus. J. Virol. 1989, 63, 3983–3990. [Google Scholar] [PubMed]
- Pfeiffer, P.; Radler, F. Comparison of the killer toxin of several yeasts and the purification of a toxin of type K2. Arch. Microbiol. 1984, 137, 357–361. [Google Scholar] [CrossRef] [PubMed]
- Ferrè, F.; Clote, P. Disulfide connectivity prediction using secondary structure information and diresidue frequencies. Bioinformatics 2005, 21, 2336–2346. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Cousiño, N.; Gómez, P.; Esteban, R. Characterization of three new killer toxins from Saccharomyces paradoxus strains. Unpublished work. 2017. [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Zuckerkandl, E.; Pauling, L. Evolutionary divergence and convergence in proteins. In Evolving Genes and Proteins; Bryson, V., Vogel, H.J., Eds.; Academic Press: New York, NY, USA, 1965; pp. 97–166. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Esteve-Zarzoso, B.; Belloch, C.; Uruburu, F.; Querol, A. Identification of yeasts by RFLP analysis of the 5.8S rRNA gene and the two ribosomal internal transcribed spacers. Int. J. Syst. Bacteriol. 1999, 49, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Špírek, M.; Poláková, S.; Jatzová, K.; Sulo, P. Post-zygotic sterility and cytonuclear compatibility limits in S. cerevisiae xenomitochondrial cybrids. Front. Genet. 2015, 5, 454. [Google Scholar] [CrossRef] [PubMed]
- Querol, A.; Barrio, E.; Ramón, D. A comparative study of different methods of yeast strains characterization. Syst. Appl. Microbiol. 1992, 15, 439–446. [Google Scholar] [CrossRef]
- Greig, D.; Borts, R.H.; Louis, E.J.; Travisano, M. Epistasis and hybrid sterility in Saccharomyces. Proc. Biol. Sci. 2002, 269, 1167–1171. [Google Scholar] [CrossRef] [PubMed]
- Liti, G.; Peruffo, A.; James, S.A.; Roberts, I.N.; Louis, E.J. Inferences of evolutionary relationships from a population survey of LTR-retrotransposons and telomeric-associated sequences in the Saccharomyces sensu stricto complex. Yeast 2005, 22, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Skelton, A.; Gardner, R.C.; Goddard, M.R. Saccharomyces paradoxus and Saccharomyces cerevisiae reside on oak trees in New Zealand: Evidence for migration from Europe and interspecies hybrids. FEMS Yeast Res. 2010, 10, 941–947. [Google Scholar] [CrossRef] [PubMed]
- Wickner, R.B.; Icho, T.; Fujimura, T.; Widner, W.R. Expression of yeast L-A double-stranded RNA virus proteins produce derepressed replication: A ski- phenocopy. J. Virol. 1991, 65, 155–161. [Google Scholar] [PubMed]
- Rowley, P.A.; Ho, B.; Bushong, S.; Johnson, A.; Sawyer, S.L. XRN1 Is a Species-Specific Virus Restriction Factor in Yeasts. PLoS Pathog. 2016, 12, e1005890. [Google Scholar] [CrossRef] [PubMed]
- Fujimura, T.; Esteban, R. Cap-snatching mechanism in yeast L-A double-stranded RNA virus. Proc. Natl. Acad. Sci. USA 2011, 108, 17667–17671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McBride, R.C.; Boucher, N.; Park, D.S.; Turner, P.E.; Townsend, J.P. Yeast response to LA virus indicates coadapted global gene expression during mycoviral infection. FEMS Yeast Res. 2013, 13, 162–179. [Google Scholar] [CrossRef] [PubMed]
- Lukša, J.; Ravoitytė, B.; Konovalovas, A.; Aitmanaitė, L.; Butenko, A.; Yurchenko, V.; Serva, S.; Servienė, E. Different Metabolic Pathways Are Involved in Response of Saccharomyces cerevisiae to L-A and M Viruses. Toxins 2017, 9, E233. [Google Scholar] [CrossRef] [PubMed]
- Conde, J.; Fink, G.R. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc. Natl. Acad. Sci. USA 1976, 73, 3651–3655. [Google Scholar] [CrossRef] [PubMed]
- Chou, J.Y.; Hung, Y.S.; Lin, K.H.; Lee, H.Y.; Leu, J.Y. Multiple molecular mechanisms cause reproductive isolation between three yeast species. PLoS Biol. 2010, 8, e1000432. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Garrastacho, M.; Esteban, R. Yeast RNA viruses as indicators of exosome activity: Human exosome hCsl4p participates in RNA degradation in Saccharomyces cerevisiae. Yeast 2011, 28, 821–832. [Google Scholar] [CrossRef]
- Fujimura, T.; Esteban, R.; Esteban, L.M.; Wickner, R.B. Portable encapsidation signal of the L-A double-stranded RNA virus of S. cerevisiae. Cell 1990, 62, 819–828. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Zuker, M.; Mathews, D.H.; Turner, D.H. Algorithms and thermodynamics for RNA secondary structure prediction: A practical guide. In RNA Biochemistry and Biotechnology; Barciszewski, J., Clark, B.F.C., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1999; pp. 11–43. [Google Scholar]
| Strain | Description and or Genotype |
|---|---|
| S. paradoxus (Sp) | |
| S28 | Europe (wine); K28 |
| T21.4 | United Kingdom (oak trees); K21 |
| Q74.4 | United Kingdom (oak trees); K74 |
| DBVPG4650 | Italy (fossilized guano); K28 |
| Q62.5 | United Kingdom (oak trees); K62 |
| Y8.5 | United Kingdom (oak trees); K74 |
| CECT11143/(CBS5829) | Denmark (moor soil); K-o |
| CECT1939/(CBS432) | Russia (oak trees); K28 |
| N-45 | Far East (mongolian oak); K45 |
| S. uvarum (Su) | |
| CECT10560 | Spain (wine) |
| CECT1884 | Spain (wine) |
| 1241 | MATα ho∆::NatMX his3 lys2 ura3 |
| S. Kudriavzevii (Sk) | |
| CECT11825/(IFO1802) | Japan (decayed leaf) |
| S. cerevisiae (Sc) | |
| SK1 | Standard laboratory K1 killer strain |
| NCYC 232 | Ex-American Yeast Foam |
| 5X47 | Diploid tester strain sensitive for killer assay |
| 2928 | α ura3 his3 trp1 L-A-o |
| 2405 | α his4-1, kar1-1 L-A-o |
| 2403 | α his4-1, kar1-1 L-A, M1 |
| 1137 | α his4-1 kar1-1, L-A-2, M2 |
| 1332 | α leu1, trp1, kar1-1 L-A, M1 |
| 1333 | α leu1, trp1, kar1-1 L-A-lus |
| 1337 | α leu1, trp1, kar1-1 L-A-2, M2 |
| 1530 | 2405 with L-A-28 and M28 from Sp S28 (obtained by cytoduction) |
| 1531 | 2405 with L-A-T21.4 and M21 from Sp T21.4 (obtained by cytoduction) |
| 1539 | 2405 with L-A-Q74.4 and M74 from Sp Q74.4 (obtained by cytoduction) |
| 1546 | 2405 with L-A-N45 and M45 from Sp N-45 (obtained by cytoduction) |
| 1560 | diploid between 2928 and 1530 (L-A-28 and M28 from Sp S28) |
| 1561 | diploid between 2928 and 1531 (L-A-T21.4 and M21 from Sp T21.4) |
| 1562 | diploid between 2928 and 1546 (L-A-N45 and M45 from Sp N-45) |
| 1573 | diploid between 2928 and 1539 (L-A Q74.4 and M74 from Sp Q74.4) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Cousiño, N.; Gómez, P.; Esteban, R. Variation and Distribution of L-A Helper Totiviruses in Saccharomyces sensu stricto Yeasts Producing Different Killer Toxins. Toxins 2017, 9, 313. https://doi.org/10.3390/toxins9100313
Rodríguez-Cousiño N, Gómez P, Esteban R. Variation and Distribution of L-A Helper Totiviruses in Saccharomyces sensu stricto Yeasts Producing Different Killer Toxins. Toxins. 2017; 9(10):313. https://doi.org/10.3390/toxins9100313
Chicago/Turabian StyleRodríguez-Cousiño, Nieves, Pilar Gómez, and Rosa Esteban. 2017. "Variation and Distribution of L-A Helper Totiviruses in Saccharomyces sensu stricto Yeasts Producing Different Killer Toxins" Toxins 9, no. 10: 313. https://doi.org/10.3390/toxins9100313





