The Aromatic Head Group of Spider Toxin Polyamines Influences Toxicity to Cancer Cells
Abstract
:1. Introduction
2. Results
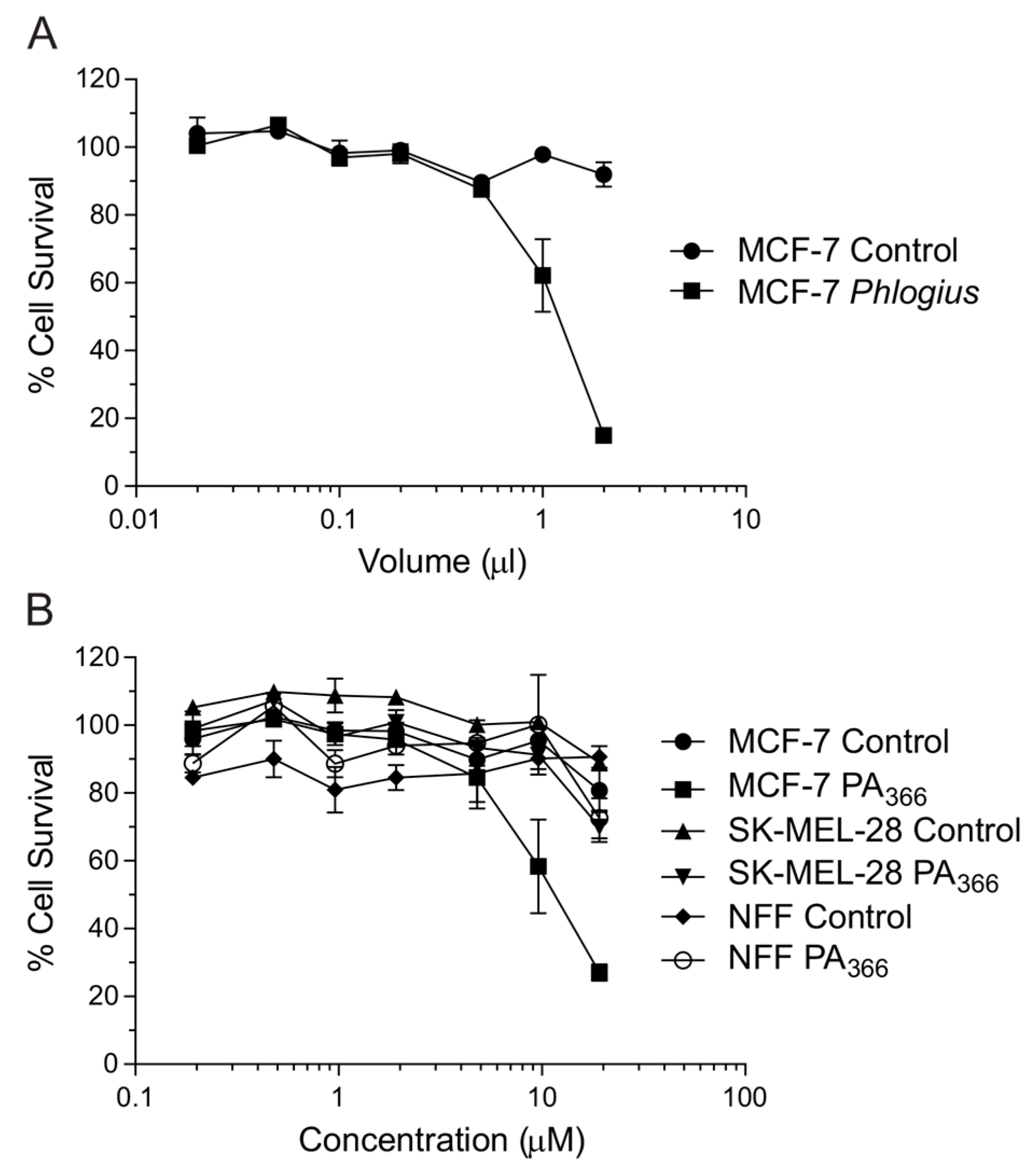
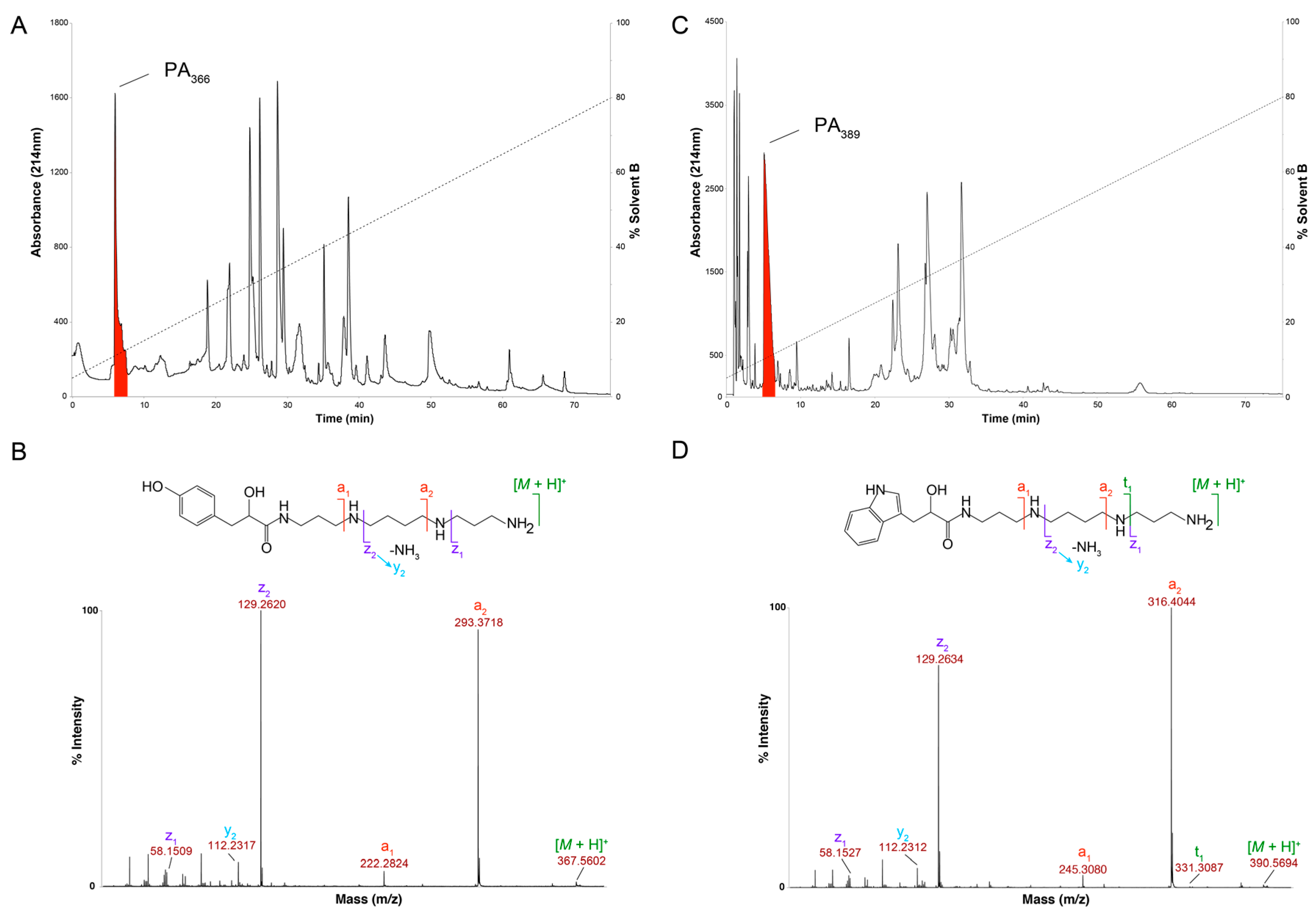
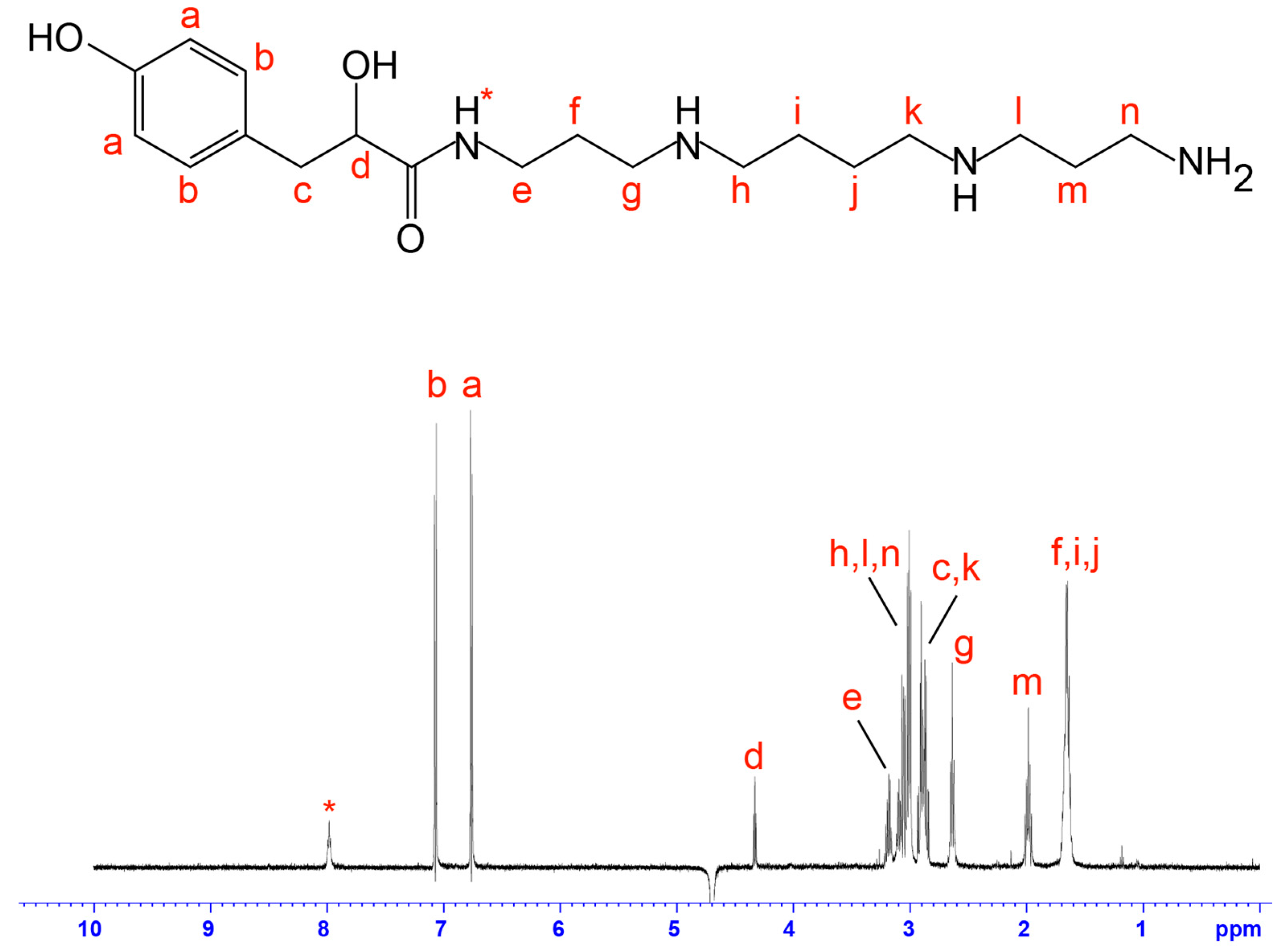
2.1. Cytotoxicity Assays and Characterisation of Bioactive Compound
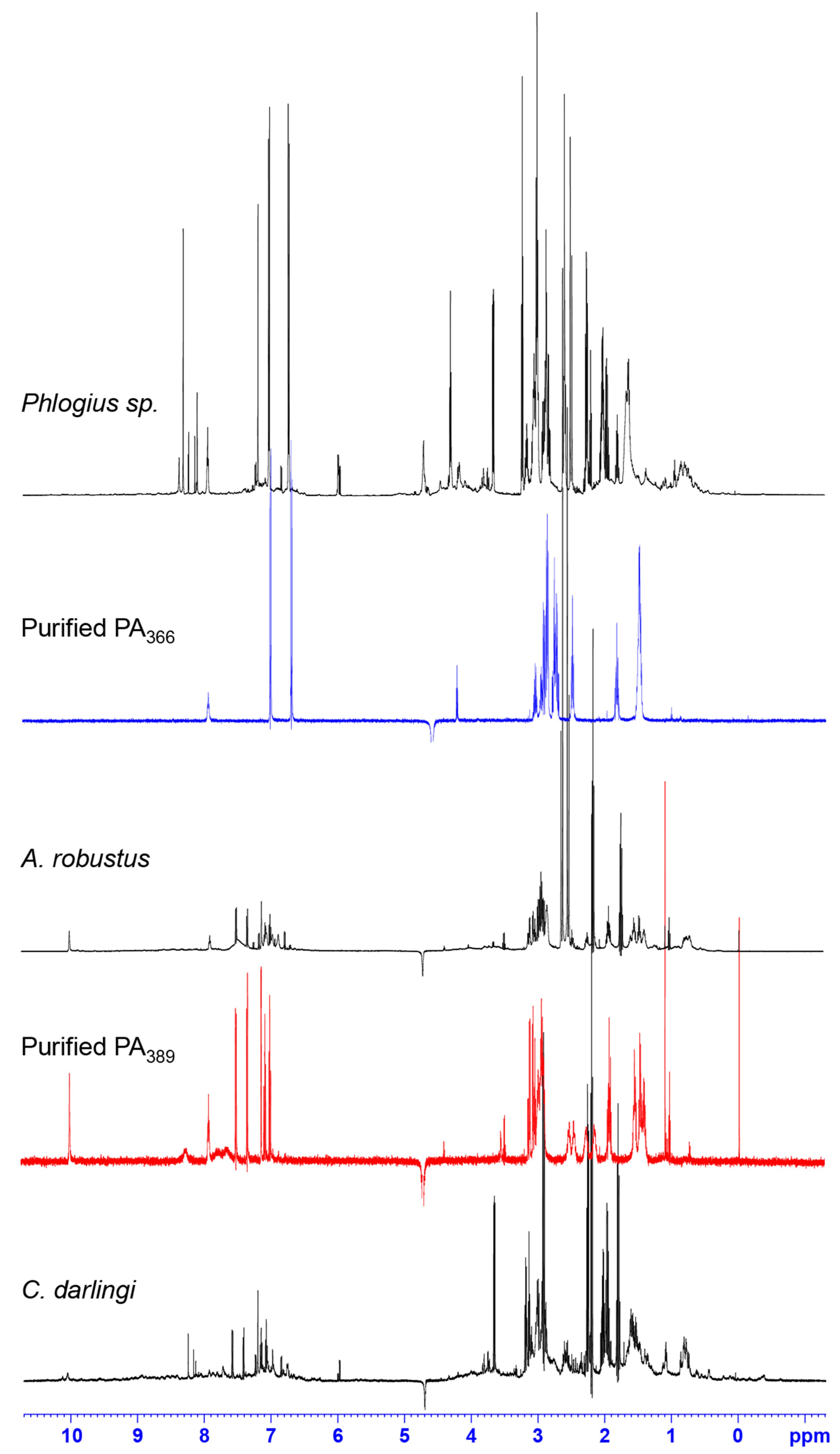
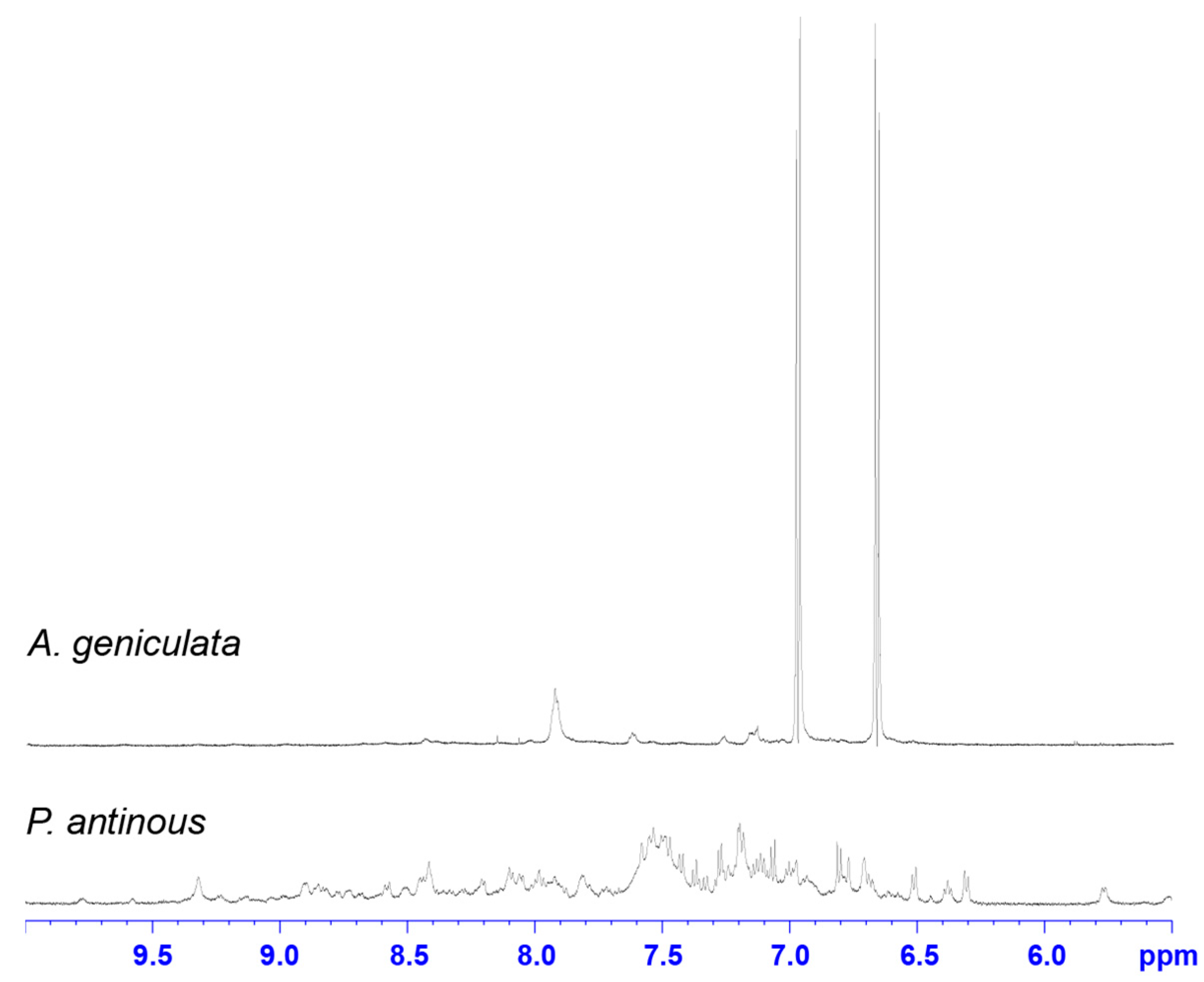
2.2. NMR Screening of Crude Spider Venoms
2.3. Protein Array Analysis
3. Discussion
4. Materials and Methods
4.1. Venom Collection and Purification
4.2. Cytotoxicity Assays
4.3. Mass Spectrometry and NMR Analysis
4.4. Protein Array Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Deyrup, S.T.; Eckman, L.E.; McCarthy, P.H.; Smedley, S.R.; Meinwald, J.; Schroeder, F.C. 2D NMR-spectroscopic screening reveals polyketides in ladybugs. Proc. Natl. Acad. Sci. USA 2011, 108, 9753–9758. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, M.K.; Lee, J.H.; Chai, E.Z.; Kanchi, M.M.; Kar, S.; Arfuso, F.; Dharmarajan, A.; Kumar, A.P.; Ramar, P.S.; Looi, C.Y.; et al. Cancer prevention and therapy through the modulation of transcription factors by bioactive natural compounds. Semin. Cancer Biol. 2016, 40–41, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Doroghazi, J.R.; Albright, J.C.; Goering, A.W.; Ju, K.S.; Haines, R.R.; Tchalukov, K.A.; Labeda, D.P.; Kelleher, N.L.; Metcalf, W.W. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat. Chem. Biol. 2014, 10, 963–968. [Google Scholar] [CrossRef] [PubMed]
- Wickenden, A.; Priest, B.; Erdemli, G. Ion channel drug discovery: Challenges and future directions. Future Med. Chem. 2012, 4, 661–679. [Google Scholar] [CrossRef] [PubMed]
- Escoubas, P.; Sollod, B.; King, G.F. Venom landscapes: Mining the complexity of spider venoms via a combined cdna and mass spectrometric approach. Toxicon 2006, 47, 650–663. [Google Scholar] [CrossRef] [PubMed]
- World Spider Catalog v18.0. Available online: http://www.wsc.nmbe.ch/ (accessed on 10 April 2017).
- Saez, N.J.; Senff, S.; Jensen, J.E.; Er, S.Y.; Herzig, V.; Rash, L.D.; King, G.F. Spider-venom peptides as therapeutics. Toxins (Basel) 2010, 2, 2851–2871. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, F.C.; Taggi, A.E.; Gronquist, M.; Malik, R.U.; Grant, J.B.; Eisner, T.; Meinwald, J. NMR-spectroscopic screening of spider venom reveals sulfated nucleosides as major components for the brown recluse and related species. Proc. Natl. Acad. Sci. USA 2008, 105, 14283–14287. [Google Scholar] [CrossRef] [PubMed]
- Mortari, M.R.; Cunha, A.O.; Ferreira, L.B.; dos Santos, W.F. Neurotoxins from invertebrates as anticonvulsants: From basic research to therapeutic application. Pharmacol. Ther. 2007, 114, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Rhee, H.J.; Kim, E.J.; Lee, J.K. Physiological polyamines: Simple primordial stress molecules. J. Cell. Mol. Med. 2007, 11, 685–703. [Google Scholar] [CrossRef] [PubMed]
- Linsalata, M.; Orlando, A.; Russo, F. Pharmacological and dietary agents for colorectal cancer chemoprevention: Effects on polyamine metabolism (review). Int. J. Oncol. 2014, 45, 1802–1812. [Google Scholar] [CrossRef] [PubMed]
- Stromgaard, K.; Jensen, L.S.; Vogensen, S.B. Polyamine toxins: Development of selective ligands for ionotropic receptors. Toxicon 2005, 45, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.A.; Vullo, D.; Supuran, C.T.; Poulsen, S.A. Natural product polyamines that inhibit human carbonic anhydrases. Biomed. Res. Int. 2014, 2014, 374079. [Google Scholar] [CrossRef] [PubMed]
- Kostopoulou, O.N.; Kouvela, E.C.; Magoulas, G.E.; Garnelis, T.; Panagoulias, I.; Rodi, M.; Papadopoulos, G.; Mouzaki, A.; Dinos, G.P.; Papaioannou, D.; et al. Conjugation with polyamines enhances the antibacterial and anticancer activity of chloramphenicol. Nucleic Acids Res. 2014, 42, 8621–8634. [Google Scholar] [CrossRef] [PubMed]
- Quistad, G.B.; Suwanrumpha, S.; Jarema, M.A.; Shapiro, M.J.; Skinner, W.S.; Jamieson, G.C.; Lui, A.; Fu, E.W. Structures of paralytic acylpolyamines from the spider agelenopsis aperta. Biochem. Biophys. Res. Commun. 1990, 169, 51–56. [Google Scholar] [CrossRef]
- Traynelis, S.F.; Wollmuth, L.P.; McBain, C.J.; Menniti, F.S.; Vance, K.M.; Ogden, K.K.; Hansen, K.B.; Yuan, H.; Myers, S.J.; Dingledine, R. Glutamate receptor ion channels: Structure, regulation, and function. Pharmacol. Rev. 2010, 62, 405–496. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.; Alewood, P.F. Taxonomy of australian funnel-web spiders using rp-hplc/esi-ms profiling techniques. Toxicon 2006, 47, 614–627. [Google Scholar] [CrossRef] [PubMed]
- Tzouros, M.; Chesnov, S.; Bigler, L.; Bienz, S. A template approach for the characterization of linear polyamines and derivatives in spider venom. Eur. J. Mass Spectrom. (Chichester) 2013, 19, 57–69. [Google Scholar] [CrossRef]
- Skinner, W.S.; Dennis, P.A.; Lui, A.; Carney, R.L.; Quistad, G.B. Chemical characterization of acylpolyamine toxins from venom of a trap-door spider and two tarantulas. Toxicon 1990, 28, 541–546. [Google Scholar] [CrossRef]
- Pineda, S.S.; Undheim, E.A.; Rupasinghe, D.B.; Ikonomopoulou, M.P.; King, G.F. Spider venomics: Implications for drug discovery. Future Med. Chem. 2014, 6, 1699–1714. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Zhang, S.; Li, M.; Wu, X.; Weng, H.; Ding, Q.; Cao, Y.; Bao, R.; Shu, Y.; Mu, J.; et al. Regulation of cell proliferation and migration in gallbladder cancer by zinc finger x-chromosomal protein. Gene 2013, 528, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Ladomery, M.; Dellaire, G. Multifunctional zinc finger proteins in development and disease. Ann. Hum. Genet. 2002, 66, 331–342. [Google Scholar] [CrossRef] [PubMed]
- Thomas, T.; Kiang, D.T. Modulation of the binding of progesterone receptor to DNA by polyamines. Cancer Res. 1988, 48, 1217–1222. [Google Scholar] [PubMed]
- Slattery, M.L.; Lundgreen, A.; John, E.M.; Torres-Mejia, G.; Hines, L.; Giuliano, A.R.; Baumgartner, K.B.; Stern, M.C.; Wolff, R.K. Mapk genes interact with diet and lifestyle factors to alter risk of breast cancer: The breast cancer health disparities study. Nutr. Cancer 2015, 67, 292–304. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Ramaswamy, B. Mechanisms and therapeutic advances in the management of endocrine-resistant breast cancer. World J. Clin. Oncol. 2014, 5, 248–262. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; Lu, Q.; Li, Q.; Ji, Y.; Chen, W.; Xue, X. Astragaloside iv inhibits breast cancer cell invasion by suppressing vav3 mediated rac1/mapk signaling. Int. Immunopharmacol. 2017, 42, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Thomas, T.; Thomas, T.J. Polyamines in cell growth and cell death: Molecular mechanisms and therapeutic applications. Cell. Mol. Life Sci. 2001, 58, 244–258. [Google Scholar] [CrossRef] [PubMed]
- Mellor, I.R.; Usherwood, P.N. Targeting ionotropic receptors with polyamine-containing toxins. Toxicon 2004, 43, 493–508. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, S.K. Isolation, mode of action and properties of the major toxin (atraxotoxin) in the venom of the sydney funnel web spider (atrax robustus). Proc. Aust. Soc. Med. Res. 1973, 3, 172. [Google Scholar]
- Ogbourne, S.M.; Suhrbier, A.; Jones, B.; Cozzi, S.J.; Boyle, G.M.; Morris, M.; McAlpine, D.; Johns, J.; Scott, T.M.; Sutherland, K.P.; et al. Antitumor activity of 3-ingenyl angelate: Plasma membrane and mitochondrial disruption and necrotic cell death. Cancer Res. 2004, 64, 2833–2839. [Google Scholar] [CrossRef] [PubMed]
- Cozzi, S.J.; Parsons, P.G.; Ogbourne, S.M.; Pedley, J.; Boyle, G.M. Induction of senescence in diterpene ester-treated melanoma cells via protein kinase c-dependent hyperactivation of the mitogen-activated protein kinase pathway. Cancer Res. 2006, 66, 10083–10091. [Google Scholar] [CrossRef] [PubMed]

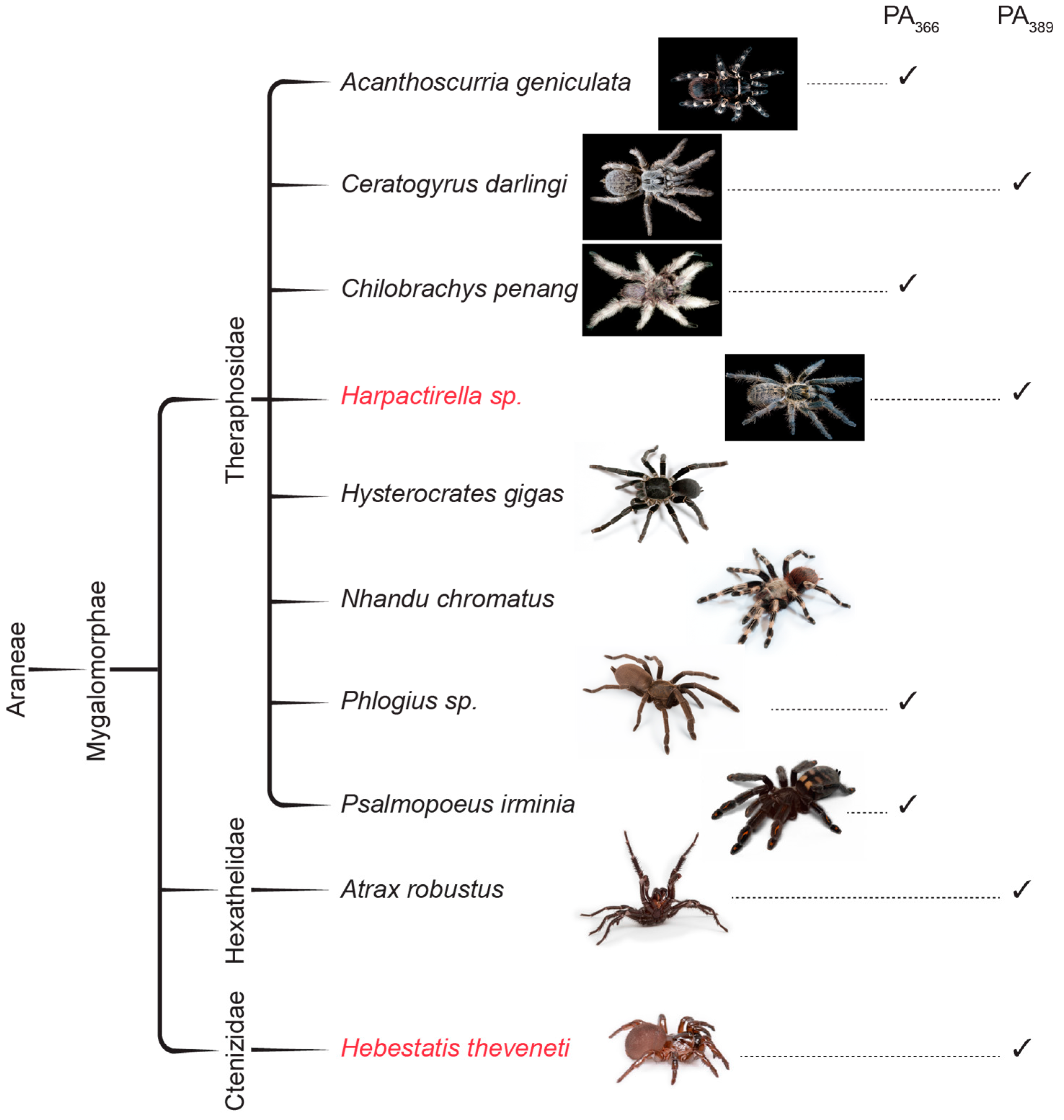
| Species | PA366 | PA389 |
|---|---|---|
| Acanthoscuria geniculata | × | |
| Ceratogyrus darlingi | × | |
| Chilobrachy penang | × | |
| Histerocrastes gigas | ||
| Nhandu chromatus | ||
| Phlogius sp. | × | |
| Psalmopoeus irminia | × | |
| Atrax robustus | × |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wilson, D.; Boyle, G.M.; McIntyre, L.; Nolan, M.J.; Parsons, P.G.; Smith, J.J.; Tribolet, L.; Loukas, A.; Liddell, M.J.; Rash, L.D.; et al. The Aromatic Head Group of Spider Toxin Polyamines Influences Toxicity to Cancer Cells. Toxins 2017, 9, 346. https://doi.org/10.3390/toxins9110346
Wilson D, Boyle GM, McIntyre L, Nolan MJ, Parsons PG, Smith JJ, Tribolet L, Loukas A, Liddell MJ, Rash LD, et al. The Aromatic Head Group of Spider Toxin Polyamines Influences Toxicity to Cancer Cells. Toxins. 2017; 9(11):346. https://doi.org/10.3390/toxins9110346
Chicago/Turabian StyleWilson, David, Glen M. Boyle, Lachlan McIntyre, Matthew J. Nolan, Peter G. Parsons, Jennifer J. Smith, Leon Tribolet, Alex Loukas, Michael J. Liddell, Lachlan D. Rash, and et al. 2017. "The Aromatic Head Group of Spider Toxin Polyamines Influences Toxicity to Cancer Cells" Toxins 9, no. 11: 346. https://doi.org/10.3390/toxins9110346






