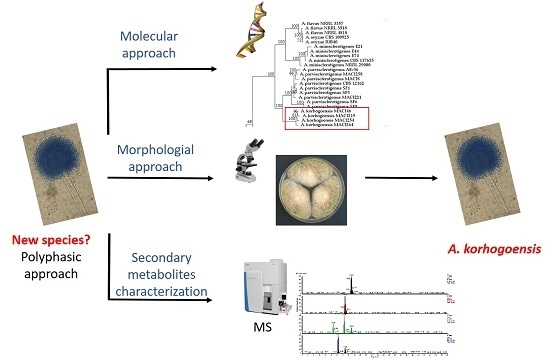
Aspergillus korhogoensis, a Novel Aflatoxin Producing Species from the Côte d’Ivoire
Abstract
:1. Introduction
2. Results
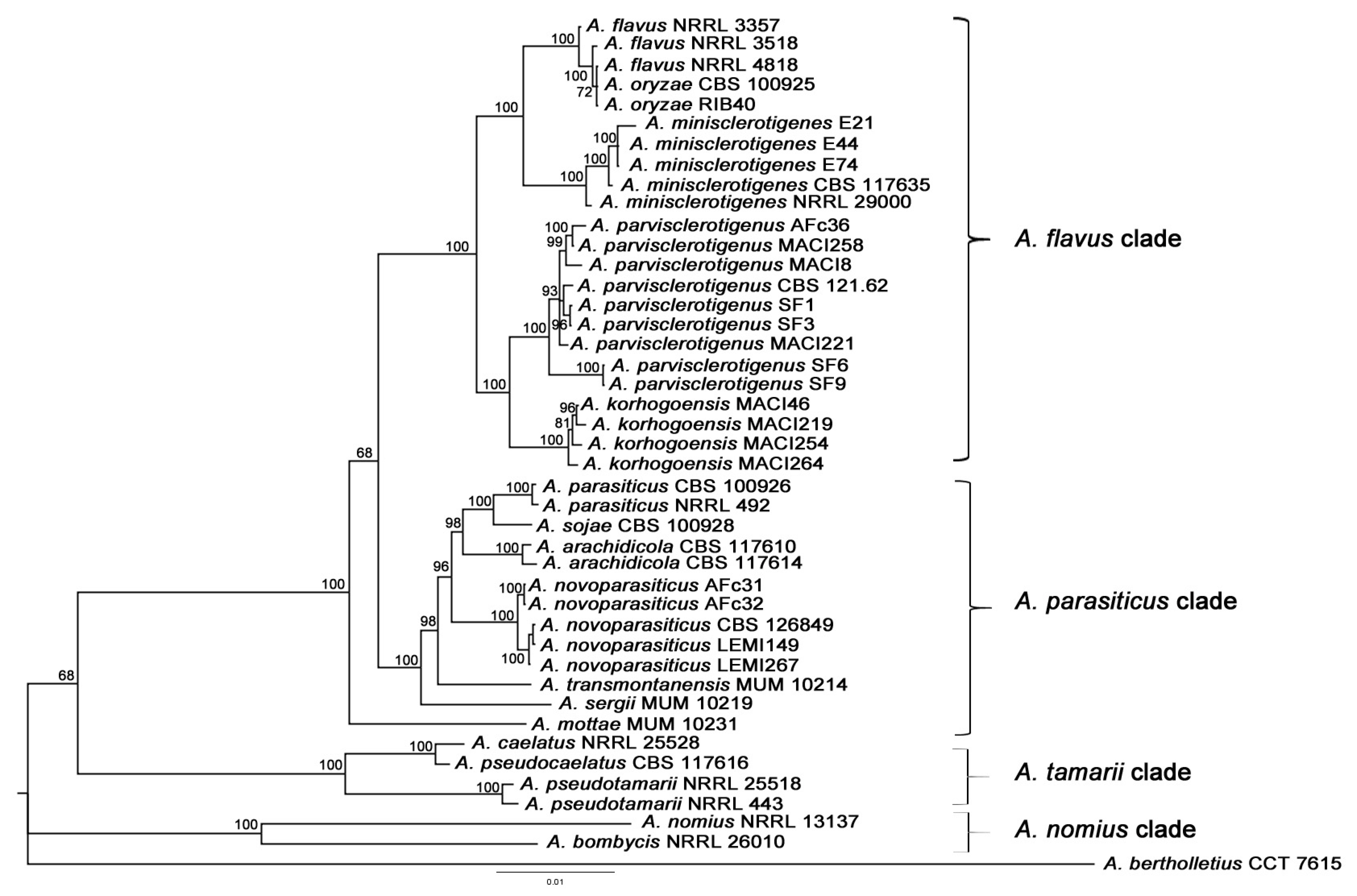
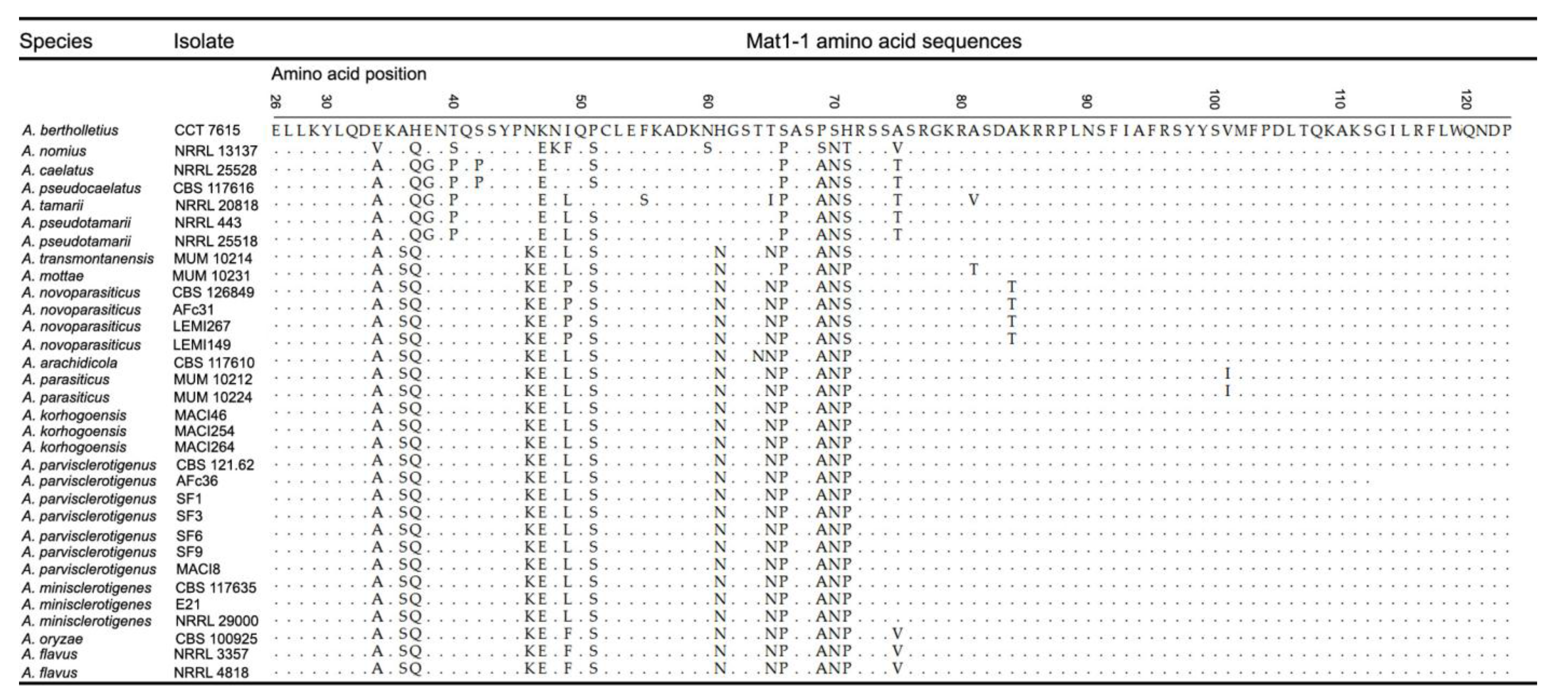
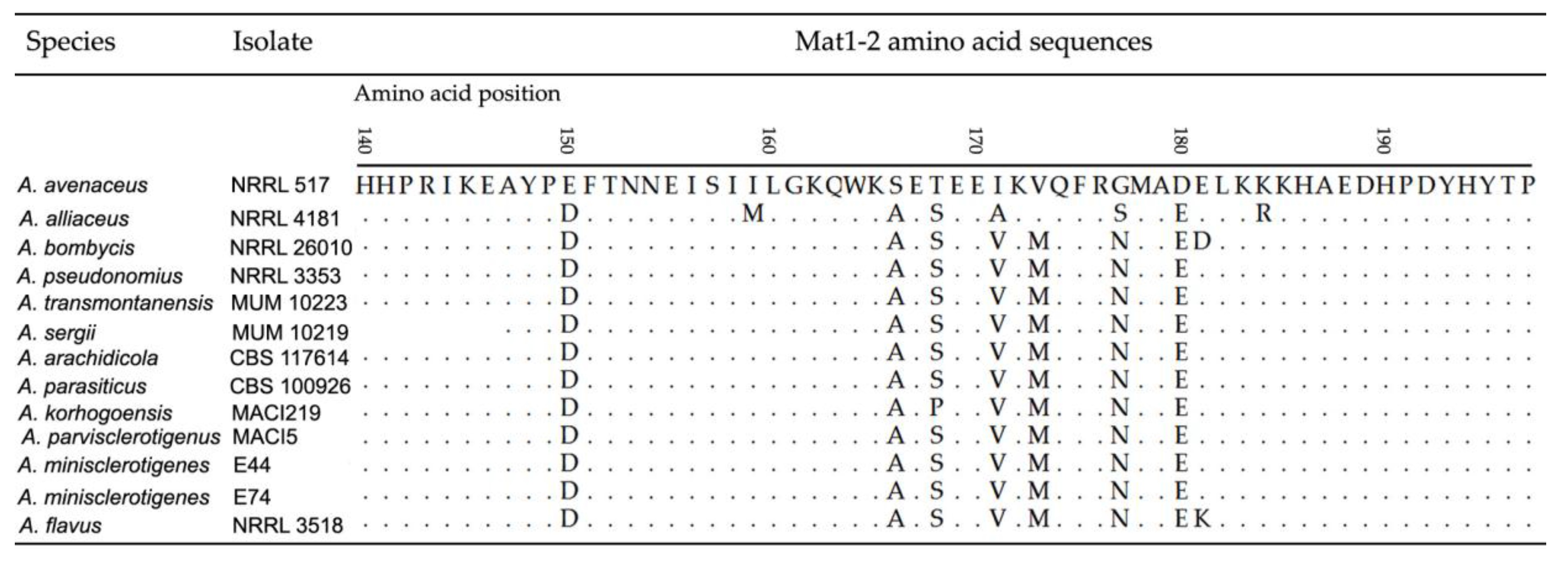
2.1. Molecular Analyses
2.1.1. Multilocus Phylogenetic Analysis
2.1.2. Mating Type Analysis
2.2. Secondary Metabolism Characterization
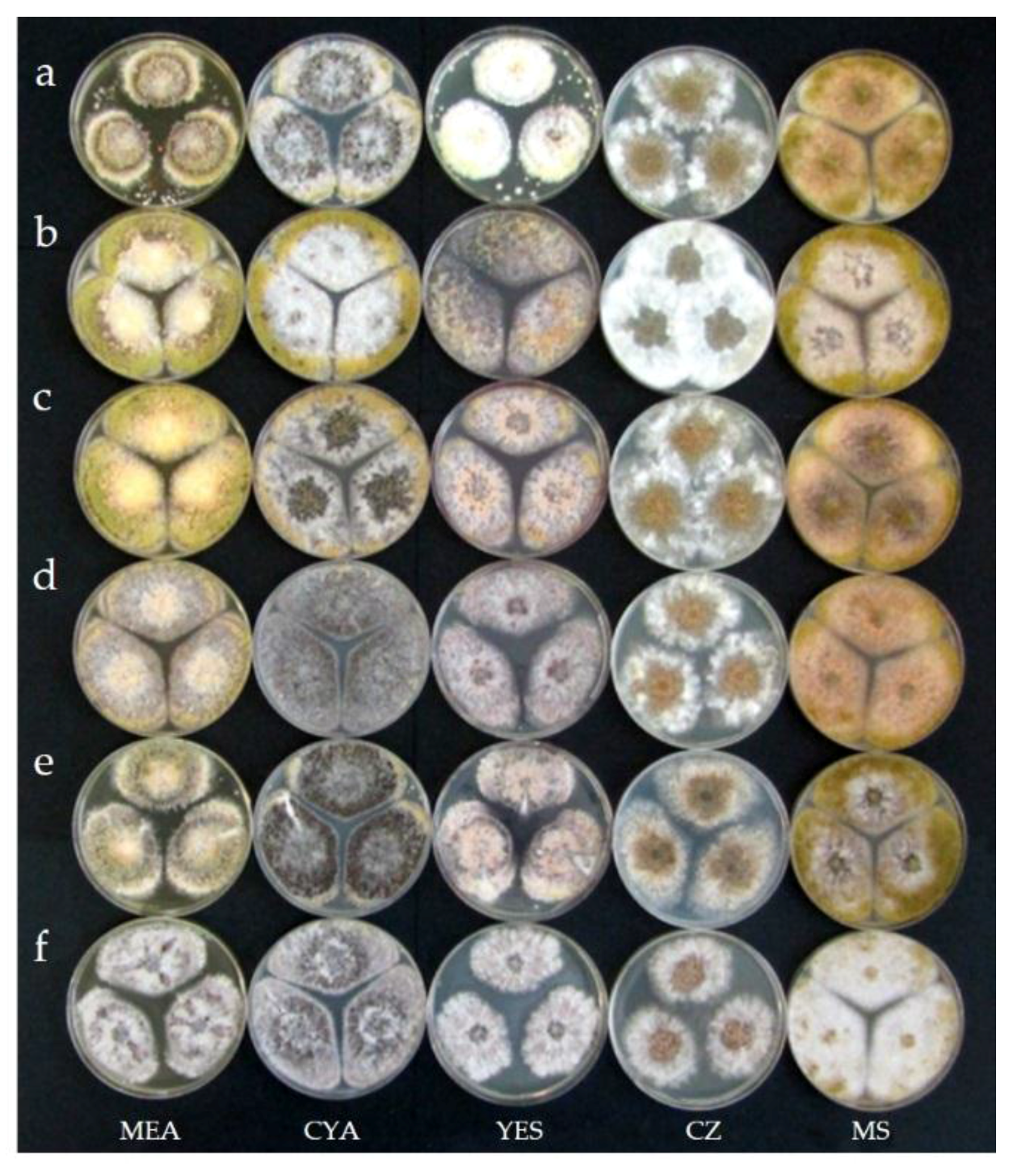
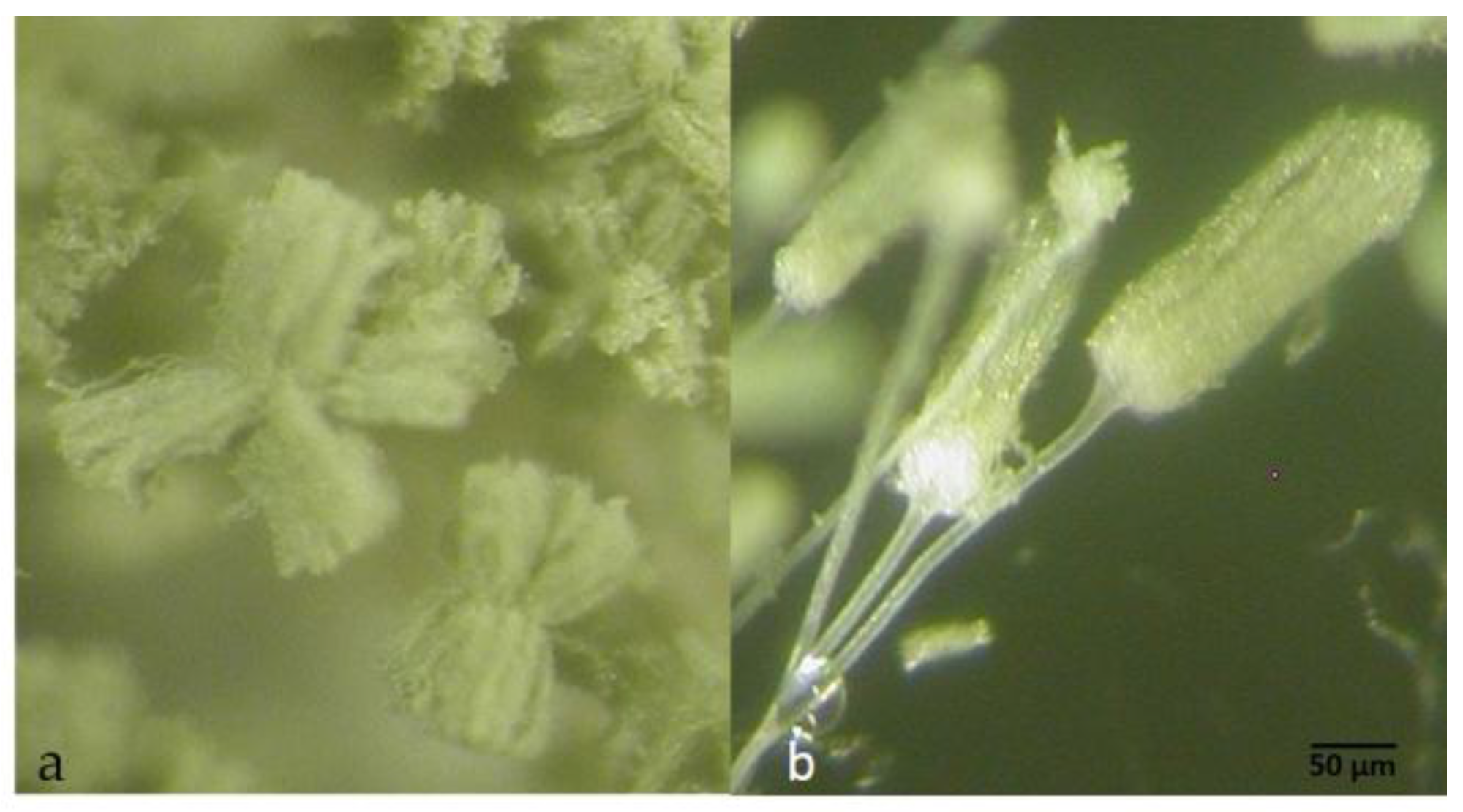
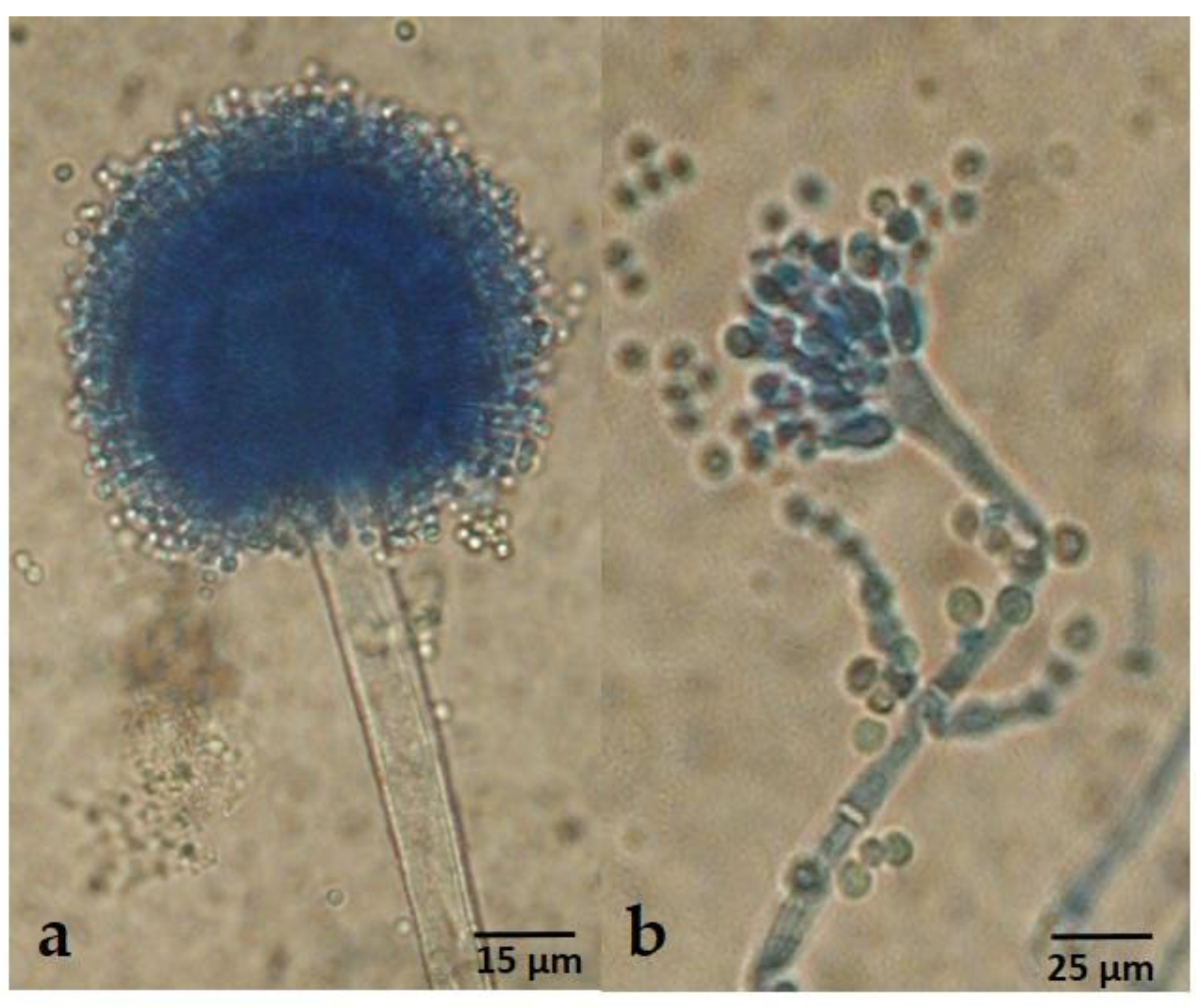
2.3. Taxonomy
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Fungal Isolates and Culture Conditions
4.3. DNA Extraction and Amplification
4.4. Amplification and Sequencing of Genomic Loci
4.5. Alignment, Model Selection, and Molecular Analyses
4.6. Morphological and Physiological Studies
4.7. LC/MS Secondary Metabolic Characterization
4.7.1. Secondary Metabolic Characterization of Whole Fungal Culture
4.7.2. Secondary Metabolic Characterization of Sclerotia
4.7.3. Secondary Metabolic Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Horn, B.W. Ecology and Population Biology of Aflatoxigenic Fungi in Soil. Toxin Rev. 2003, 22, 351–379. [Google Scholar] [CrossRef]
- Mitchell, N.J.; Bowers, E.; Hurburgh, C.; Wu, F. Potential economic losses to the US corn industry from aflatoxin contamination. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2016, 33, 540–550. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Groopman, J.D.; Pestka, J.J. Public health impacts of foodborne mycotoxins. Annu. Rev. Food Sci. Technol. 2014, 5, 351–372. [Google Scholar] [CrossRef] [PubMed]
- Azziz-Baumgartner, E.; Lindblade, K.; Gieseker, K.; Rogers, H.S.; Kieszak, S.; Njapau, H.; Schleicher, R.; McCoy, L.F.; Misore, A.; DeCock, K.; et al. Aflatoxin Investigative Group. Case-control study of an acute aflatoxicosis outbreak, Kenya, 2004. Environ. Health Perspect. 2005, 113, 1779–1783. [Google Scholar] [CrossRef] [PubMed]
- Cano, P.; Puel, O.; Oswald, I.P. Mycotoxins: Fungal Secondary Metabolites with Toxic Properties. In Fungi: Applications and Management Strategies; Deshmukh, S.K., Misra, J.K., Tewari, J.P., Papp, T., Eds.; CRC Press: Boca Raton, FL, USA, 2016; pp. 318–371. ISBN 9781498724913. [Google Scholar]
- Bennett, J.W.; Klich, M. Mycotoxins. Clin. Microbiol. Rev. 2003, 16, 497–516. [Google Scholar] [CrossRef] [PubMed]
- De Ruyck, K.; De Boevre, M.; Huybrechts, I.; De Saeger, S. Dietary mycotoxins, co-exposure, and carcinogenesis in humans: Short review. Mutat. Res. Rev. Mutat. Res. 2015, 766, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Bartoli, A.; Maggi, O. Four new species of Aspergillus from Ivory Coast soil. Trans. Br. Mycol. Soc. 1978, 71, 383–394. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Skouboe, P.; Samson, R.A. Taxonomic comparison of three different groups of aflatoxin producers and a new efficient producer of aflatoxin B1, sterigmatocystin and 3-O-methylsterigmatocystin, Aspergillus rambellii sp. nov. Syst. Appl. Microbiol. 2005, 28, 442–453. [Google Scholar] [CrossRef] [PubMed]
- Abe, K.; Gomi, K.; Hasegawa, F.; Machida, M. Impact of Aspergillus oryzae genomics on industrial production of metabolites. Mycopathologia 2006, 162, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Horn, B.W.; Ramirez-Prado, J.H.; Carbone, I. The sexual state of Aspergillus parasiticus. Mycologia 2009, 101, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Samson, R.A.; Varga, J. What is a species in Aspergillus? Med. Mycol. 2009, 47, S13–S20. [Google Scholar] [CrossRef] [PubMed]
- Cotty, P.J.; Bhatnagar, D. Variability among atoxigenic Aspergillus flavus strains in ability to prevent aflatoxin contamination and production of aflatoxin biosynthetic pathway enzymes. Appl. Environ. Microbiol. 1994, 60, 2248–2251. [Google Scholar] [PubMed]
- Adjovi, Y.C.S.; Bailly, S.; Gnonlonfin, B.J.G.; Tadrist, S.; Querin, A.; Sanni, A.; Oswald, I.P.; Puel, O.; Bailly, J.D. Analysis of the contrast between natural occurence of toxigenic Aspergilli of the Flavi section and aflatoxin B1 in cassava. Food Microbiol. 2014, 38, 151–159. [Google Scholar] [CrossRef] [PubMed]
- El Mahgubi, A.E.; Puel, O.; Bailly, S.; Tadrist, S.; Querin, A.; Ouadia, A.; Oswald, I.P.; Bailly, J.D. Distribution and toxigenicity of Aspergillus section Flavi in spices marketed in Morocco. Food Control 2013, 32, 143–148. [Google Scholar] [CrossRef]
- Perrone, G.; Gallo, A.; Logrieco, A.F. Biodiversity of Aspergillus section Flavi in Europe in relation to the management of aflatoxin risk. Front. Microbiol. 2014, 5, 377. [Google Scholar] [CrossRef] [PubMed]
- Varga, J.; Frisvad, J.C.; Samson, R.A. Two new aflatoxin producing species, and an overview of Aspergillus section Flavi. Stud. Mycol. 2011, 69, 57–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soares, C.; Rodrigues, P.; Peterson, S.W.; Lima, N.; Venâncio, A. Three new species of Aspergillus section Flavi isolated from almonds and maize in Portugal. Mycologia 2012, 104, 682–697. [Google Scholar] [CrossRef] [PubMed]
- Taniwaki, M.H.; Pitt, J.I.; Iamanaka, B.T.; Sartori, D.; Copetti, M.V.; Balajee, A.; Fungaro, M.H.P.; Frisvad, J.C. Aspergillus bertholletius sp. nov. from Brazil Nuts. PLoS ONE 2012, 7, e42480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pildain, M.B.; Frisvad, J.C.; Vaamonde, G.; Cabral, D.; Varga, J.; Samson, R.A. Two novel aflatoxin-producing Aspergillus species from Argentinean peanuts. Int. J. Syst. Evol. Microbiol. 2008, 58, 725–735. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, P.; Santos, C.; Venâncio, A.; Lima, N. Species identification of Aspergillus section Flavi isolates from Portuguese almonds using phenotypic, including MALDI-TOF ICMS, and molecular approaches. J. Appl. Microbiol. 2011, 111, 877–892. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, K.C.; Chang, P.-K.; Yu, J.; Cotty, P.J. Aflatoxin biosynthesis cluster gene cypA is required for G aflatoxin formation. Appl. Environ. Microbiol. 2004, 70, 6518–6524. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, S.S.; Stchigel, A.M.; Cano, J.F.; Godoy-Martinez, P.C.; Colombo, A.L.; Guarro, J. Aspergillus novoparasiticus: A new clinical species of the section Flavi. Med. Mycol. 2012, 50, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Arroyo-Manzanares, N.; Di Mavungu, J.D.; Uka, V.; Malysheva, S.V.; Cary, J.W.; Ehrlich, K.C.; Vanhaecke, L.; Bhatnagar, D.; De Saeger, S. Use of UHPLC high-resolution Orbitrap mass spectrometry to investigate the genes involved in the production of secondary metabolites in Aspergillus flavus. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2015, 32, 1656–1673. [Google Scholar] [CrossRef] [PubMed]
- Paguigan, N.D.; El-Elimat, T.; Kao, D.; Raja, H.A.; Pearce, C.J.; Oberlies, N.H. Enhanced dereplication of fungal cultures via use of mass defect filtering. J. Antibiot. 2017, 70, 553–561. [Google Scholar] [CrossRef] [PubMed]
- Jakšić, D.; Puel, O.; Canlet, C.; Kopjar, N.; Kosalec, I.; Klarić, M.Š. Cytotoxicity and genotoxicity of versicolorins and 5-methoxysterigmatocystin in A549 cells. Arch. Toxicol. 2012, 86, 1583–1591. [Google Scholar] [CrossRef] [PubMed]
- Uka, V.; Moore, G.G.; Arroyo-Manzanares, N.; Nebija, D.; De Saeger, S.; Di Mavungu, J.D. Unravelling the diversity of the cyclopiazonic acid family of mycotoxins in Aspergillus flavus by UHPLC Triple-TOF HRMS. Toxins 2017, 9, 35. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Liu, Y.; Ding, T.; Zhang, X.; Chen, H.; Shen, C.; Wu, B.; Niu, W. Determination of kojic acid in foods using high performance liquid chromatography-tandem mass spectrometry. Chin. J. Chromatogr. 2012, 30, 578–583. [Google Scholar] [CrossRef]
- Uhlig, S.; Egge-Jacobsen, W.; Vrålstad, T.; Miles, C.O. Indole–diterpenoid profiles of Claviceps paspali and Claviceps purpurea from high-resolution Fourier transform Orbitrap mass spectrometry. Rapid Commun. Mass Spectrom. 2014, 28, 1621–1634. [Google Scholar] [CrossRef] [PubMed]
- Malysheva, S.V.; Arroyo-Manzanares, N.; Cary, J.W.; Ehrlich, K.C.; Vanden Bussche, J.; Vanhaecke, L.; Bhatnagar, D.; Di Mavungu, J.D.; De Saeger, S. Identification of novel metabolites from Aspergillus flavus by high resolution and multiple stage mass spectrometry. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2014, 31, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Cary, J.W.; Han, Z.; Yin, Y.; Lohmar, J.M.; Shantappa, S.; Harris-Coward, P.Y.; Mack, B.; Ehrlich, K.C.; Wei, Q.; Arroyo-Manzanares, N.; et al. Transcriptome analysis of Aspergillus flavus reveals veA-dependent regulation of secondary metabolite gene clusters, including the novel aflavarin cluster. Eukaryot. Cell 2015, 14, 983–997. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, M.J.; Koulman, A.; Monahan, B.J.; Pritchard, B.L.; Payne, G.A.; Scott, B. Identification of two aflatrem biosynthesis gene loci in Aspergillus flavus and metabolic engineering of Penicillium paxilli to elucidate their function. Appl. Environ. Microbiol. 2009, 75, 7469–7481. [Google Scholar] [CrossRef] [PubMed]
- Sobolev, V.S.; Cole, R.J.; Dorner, J.W.; Horn, B.W.; Harrigan, G.G.; Gloer, J.B. Isolation and Structure Elucidation of a New Metabolite Produced by Aspergillus parasiticus. J. Nat. Prod. 1997, 60, 847–850. [Google Scholar] [CrossRef]
- TePaske, M.R.; Gloer, J.B.; Wicklow, D.T.; Dowd, P.F. Aflavarin and β-Aflatrem: New Anti-Insectan Metabolites from the Sclerotia of Aspergillus flavus. J. Nat. Prod. 1992, 55, 1080–1086. [Google Scholar] [CrossRef]
- Gloer, J.B.; TePaske, M.R.; Sima, J.S.; Wicklow, D.T.; Dowd, P.F. Antiinsectan aflavinine derivative from sclerotia of Aspergillus flavus. J. Org. Chem. 1988, 53, 5457–5460. [Google Scholar] [CrossRef]
- TePaske, M.R.; Gloer, J.B.; Wicklow, D.T.; Dowd, P.F. Three new aflavinines from the sclerotia of Aspergillus tubingensis. Tetrahedron 1989, 45, 4961–4968. [Google Scholar] [CrossRef]
- Pitt, J.I.; Lange, L.; Lacey, A.E.; Vuong, D.; Midgley, D.J.; Greenfield, P.; Bradbury, M.I.; Lacey, E.; Busk, P.K.; Pilgaard, B.; et al. Aspergillus hancockii sp. nov., a biosynthetically talented fungus endemic to southeastern Australian soils. PLoS ONE 2017, 12, e0170254. [Google Scholar] [CrossRef] [PubMed]
- De Queiroz, K. Species concepts and species delimitation. Syst. Biol. 2007, 56, 879–886. [Google Scholar] [CrossRef] [PubMed]
- Giraud, T.; Refrégier, G.; Le Gac, M.; de Vienne, D.M.; Hood, M.E. Speciation in fungi. Fungal Genet. Biol. 2008, 45, 791–802. [Google Scholar] [CrossRef] [PubMed]
- Bosso, L.; Lacatena, F.; Varlese, R.; Nocerino, S.; Cristinzio, G.; Russo, D. Plant pathogen but not antagonists change in soil fungal communities across a land abandonment gradient in a Mediterranean landscape. Acta Oecol. (Montrouge) 2017, 78, 1–6. [Google Scholar] [CrossRef]
- Dyer, P.S.; Kück, U. Sex and the Imperfect Fungi. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef] [PubMed]
- Dyer, P.S.; Paoletti, M.; Archer, D.B. Genomics reveals sexual secrets of Aspergillus. Microbiology 2003, 149, 2301–2303. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.W.; Jacobson, D.J.; Kroken, S.; Kasuga, T.; Geiser, D.M.; Hibbett, D.S.; Fisher, M.C. Phylogenetic Species Recognition and Species Concepts in Fungi. Fungal Genet. Biol. 2000, 31, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Dupuis, J.R.; Roe, A.D.; Sperling, F.A.H. Multi-locus species delimitation in closely related animals and fungi: one marker is not enough. Mol. Ecol. 2012, 21, 4422–4436. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Prado, J.H.; Moore, G.G.; Horn, B.W.; Carbone, I. Characterization and population analysis of the mating-type genes in Aspergillus flavus and Aspergillus parasiticus. Fungal Genet. Biol. 2008, 45, 1292–1299. [Google Scholar] [CrossRef] [PubMed]
- Dyer, P.S.; O’Gorman, C.M. Sexual development and cryptic sexuality in fungi: Insights from Aspergillus species. FEMS Microbiol. Rev. 2012, 36, 165–192. [Google Scholar] [CrossRef] [PubMed]
- Moore, G.G.; Elliott, J.L.; Singh, R.; Horn, B.W.; Dorner, J.W.; Stone, E.A.; Chulze, S.N.; Barros, G.G.; Naik, M.K.; Wright, G.C.; et al. Sexuality generates diversity in the aflatoxin gene cluster: Evidence on a global scale. PLoS Pathog. 2013, 9, e1003574. [Google Scholar] [CrossRef] [PubMed]
- Horn, B.W.; Moore, G.G.; Carbone, I. Sexual reproduction in Aspergillus flavus. Mycologia 2009, 101, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Horn, B.W.; Moore, G.G.; Carbone, I. Sexual reproduction in aflatoxin-producing Aspergillus nomius. Mycologia 2011, 103, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Olarte, R.A.; Worthington, C.J.; Horn, B.W.; Moore, G.G.; Singh, R.; Monacell, J.T.; Dorner, J.W.; Stone, E.A.; Xie, D.; Carbone, I. Enhanced diversity and aflatoxigenicity in interspecific hybrids of Aspergillus flavus and Aspergillus parasiticus. Mol. Ecol. 2015, 24, 1889–1909. [Google Scholar] [CrossRef] [PubMed]
- Sweany, R.R.; Damann, K.E.J.; Kaller, M.D. Comparison of soil and corn kernel Aspergillus flavus populations: Evidence for niche specialization. Phytopathology 2011, 101, 952–959. [Google Scholar] [CrossRef] [PubMed]
- Carbone, I.; Jakobek, J.L.; Ramirez-Prado, J.H.; Horn, B.W. Recombination, balancing selection and adaptive evolution in the aflatoxin gene cluster of Aspergillus parasiticus. Mol. Ecol. 2007, 16, 4401–4417. [Google Scholar] [CrossRef] [PubMed]
- Moore, G.G.; Singh, R.; Horn, B.W.; Carbone, I. Recombination and lineage-specific gene loss in the aflatoxin gene cluster of Aspergillus flavus. Mol. Ecol. 2009, 18, 4870–4887. [Google Scholar] [CrossRef] [PubMed]
- Varga, J.; Baranyi, N.; Chandrasekaran, M.; Vágvölgyi, C.; Kocsubé, S. Mycotoxin producers in the Aspergillus genus: An update. Acta Biol. Szeged. 2015, 59, 151–167. [Google Scholar]
- Probst, C.; Bandyopadhyay, R.; Cotty, P.J. Diversity of aflatoxin-producing fungi and their impact on food safety in sub-Saharan Africa. Int. J. Food Microbiol. 2014, 174, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Cotty, P.J.; Cardwell, K.F. Divergence of West African and North American Communities of Aspergillus Section Flavi. Appl. Environ. Microbiol. 1999, 65, 2264–2265. [Google Scholar] [PubMed]
- Cotty, P.J.; Jaime-Garcia, R. Influences of climate on aflatoxin producing fungi and aflatoxin contamination. Int. J. Food Microbiol. 2007, 119, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Cardwell, K.F.; Cotty, P.J. Distribution of Aspergillus section Flavi among field soils from the four agroecological zones of the Republic of Bénin, West Africa. Plant Dis. 2002, 86, 434–439. [Google Scholar] [CrossRef]
- Donner, M.; Atehnkeng, J.; Sikora, A.R.; Bandyopadhyay, R.; Cotty, P.J. Distribution of Aspergillus section Flavi in soils of maize fields in three agroecological zones of Nigeria. Soil Biol. Biochem. 2009, 41, 37–41. [Google Scholar] [CrossRef]
- Moore, G.G. Sex and recombination in aflatoxigenic Aspergilli: Global implications. Front. Microbiol. 2014, 5, 32. [Google Scholar] [CrossRef] [PubMed]
- Moore, G.G.; Mack, B.M.; Beltz, S.B. Genomic sequence of the aflatoxigenic filamentous fungus Aspergillus nomius. BMC Genom. 2015, 16, 551. [Google Scholar] [CrossRef] [PubMed]
- Moore, G.G.; Mack, B.M.; Beltz, S.B. Draft genome sequences of two closely related aflatoxigenic Aspergillus species obtained from the Ivory Coast. Genome Biol. Evol. 2015, 8, 729–732. [Google Scholar] [CrossRef] [PubMed]
- Moore, G.G.; Mack, B.M.; Beltz, S.B.; Gilbert, M.K. Draft genome sequence of an aflatoxigenic Aspergillus species, A. bombycis. Genome Biol. Evol. 2016, 8, 3297–3300. [Google Scholar] [CrossRef] [PubMed]
- Fapohunda, S.O.; Moore, G.G.; Ganiyu, O.T.; Beltz, S.B. Toxigenic Aspergillus flavus and other fungi of public health concern in food and organic matter in southwest Nigeria. Mycology 2012, 3, 210–219. [Google Scholar]
- Peterson, S.W.; Ito, Y.; Horn, B.W.; Goto, T. Aspergillus bombycis, a New Aflatoxigenic Species and Genetic Variation in Its Sibling Species, A. nomius. Mycologia 2001, 93, 689–703. [Google Scholar] [CrossRef]
- Nierman, W.C.; Yu, J.; Fedorova-Abrams, N.D.; Losada, L.; Cleveland, T.E.; Bhatnagar, D.; Bennett, J.W.; Dean, R.; Payne, G.A. Genome sequence of Aspergillus flavus NRRL3357, a strain that causes aflatoxin contamination of food and feed. Genome Announc. 2015, 3, e00168-15. [Google Scholar] [CrossRef] [PubMed]
- Machida, M.; Asai, K.; Sano, M.; Tanaka, T.; Kumagai, T.; Terai, G.; Kusumoto, K.; Arima, T.; Akita, O.; Kashiwagi, Y.; et al. Genome sequencing and analysis of Aspergillus oryzae. Nature 2005, 438, 1157–1161. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Peterson, S.W.; Wicklow, D.T.; Goto, T. Aspergillus pseudotamarii, a new aflatoxin producing species in Aspergillus section Flavi. Mycol. Res. 2001, 105, 233–239. [Google Scholar] [CrossRef]
- Goto, T.; Wicklow, D.T.; Ito, Y. Aflatoxin and cyclopiazonic acid production by a sclerotium-producing Aspergillus tamarii strain. Appl. Environ. Microbiol. 1996, 62, 4036–4038. [Google Scholar] [PubMed]
- Girardin, H.; Latgé, J.P.; Srikantha, T.; Morrow, B.; Soll, D.R. Development of DNA probes for fingerprinting Aspergillus fumigatus. J. Clin. Microbiol. 1993, 31, 1547–1554. [Google Scholar] [PubMed]
- Maddison, W.P.; Maddison, D.R. Mesquite: A Modular System for Evolutionary Analysis. Version 3.2. 2017. Available online: http://mesquiteproject.org (accessed on 7 March 2017).
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Suchard, M.A.; Xie, D.; Drummond, A.J. Tracer v1.6. 2014. Available online: http://tree.bio.ed.ac.uk/software/tracer (accessed on 1 December 2014).
- Rambaut, A. FigTree v1.4.2, A Graphical Viewer of Phylogenetic Trees. 2014. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 1 December 2014).
- Frisvad, J.C.; Samson, R.A. Polyphasic taxonomy of Penicillium subgenus Penicillium. A guide to identification of food and air-borne terverticilliate Penicillia and their mycotoxins. Stud. Mycol. 2004, 49, 1–174. [Google Scholar]
- Pitt, J.I.; Hocking, A.D.; Glenn, D.R. An improved medium for detection of Aspergillus flavus and A. parasiticus. J. Appl. Bacteriol. 1983, 54, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Raper, K.B.; Farnell, D.I. The Genus Aspergillus; The Williams & Wilkins Company: Baltimore, MD, USA, 1965; pp. 1–686. [Google Scholar]
- Pitt, J.I.; Hocking, A.D. Fungi and Food Spoilage, 3rd ed.; Springer: New York, NY, USA, 2009; pp. 1–519. ISBN 978-0-387-92206-5. [Google Scholar]
- Cano, P.M.; Jamin, E.L.; Tadrist, S.; Bourdaud’hui, P.; Péan, M.; Debrauwer, L.; Oswald, I.P.; Delaforge, D.; Puel, O. New Untargeted Metabolic Profiling Combining Mass Spectrometry and Isotopic Labeling: Application on Aspergillus fumigatus Grown on Wheat. Anal. Chem. 2013, 85, 8412–8420. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.K.; Scharfenstein, L.L.; Li, R.B.; Arroyo-Manzanares, N.; De Saeger, S.; Di Mavungu, J.D. Aspergillus flavus aswA, a gene homolog of Aspergillus nidulans oefC, regulates sclerotial development and biosynthesis of sclerotium-associated secondary metabolites. Fungal Genet. Biol. 2017, 104, 29–37. [Google Scholar] [CrossRef] [PubMed]

| Metabolite | Elemental Composition | m/z | Ion | Retention Time (min) | MS/MS | Error (ppm) | ID Level * | References |
| AFLATOXIN BIOSYNTHESIS PATHWAY | ||||||||
| Aflatoxin B1 | C17H12O6 | 313.07 | [M + H]+ | 17.37 | 285 (100), 298, 284, 270, 257, 243, 229 | −0.398 | 1, 2 | [25] |
| Aflatoxin B2 | C17H14O6 | 315.07 | [M + H]+ | 14.95 | 297, 287 (100), 259, 269, 273 | −5.920 | 1, 2 | [25] |
| Aflatoxin G1 | C17H12O7 | 329.08 | [M + H]+ | 15.25 | 311 (100), 301, 300, 283, 243 | −0.119 | 1, 2 | [25] |
| Aflatoxin G2 | C17H14O7 | 331.08 | [M + H]+ | 12.84 | 313 (100), 303, 285, 275, 257, 245 | −0.511 | 1, 2 | [25] |
| O-methyl-sterigmatocystin | C19H14O6 | 339.08 | [M + H]+ | 24.21 | 324 (100), 311, 306, 295 | 2.817 | 1 | |
| Sterigmatocystin | C18H12O6 | 325.07 | [M + H]+ | 33.59 | 310 (100), 297, 282 | 0.570 | 1, 2 | [26] |
| Versicolorin A | C18H10O7 | 337.03 | [M − H]− | 35.95 | 309 (100), 319, 308, 293, 265, 253 | −2.094 | 1, 2 | [27] |
| Versicolorin B | C18H12O7 | 339.05 | [M − H]− | 34.40 | 311 (100) 310, 309, 295, 297, 283 | −0.578 | 1, 2 | [27] |
| Norsolorinic acid | C20H18O7 | 369.10 | [M − H]− | 42.07 | 351 (100), 341, 325, 308, 297, 270 | 1.528 | 1 | |
| CYCLOPIAZONIC ACID BIOSYNTHETIC PATHWAY | ||||||||
| α-cyclopiazonic acid | C20H20N2O3 | 337.15 | [M + H]+ | 36.77 | 182 (100), 196, 154, 140 | 0.561 | 1, 2 | [28] |
| β-cyclopiazonic acid | C20H22N2O3 | 339.17 | [M + H]+ | 37.58 | 198 (100), 324, 283, 183, 144, 130 | −1.289 | 2 | [28] |
| 2′-oxo-cyclopiazonic acid | C20H20N2O4 | 353.15 | [M + H]+ | 36.20 | 335 (100), 311, 293, 252, 224, 212 | −1.174 | 2 | [28] |
| 3′-hydroxy-speradine A | C21H22 N2O5 | 383.16 | [M + H]+ | 21.19 | 355 (100), 365, 182, 184, 226, 254, 323, 347, 337 | −1.144 | 2 | [28] |
| Speradine C | C20H22 N2O5 | 371.16 | [M + H]+ | 18.19 | 353 (100), 287, 269, 259, 226, 184 | 2.780 | 2 | [28] |
| Speradine D | C20H22 N2O6 | 387.16 | [M + H]+ | 20.80 | 369 (100), 269, 226, 184 | 2.679 | 2 | [28] |
| Speradine F | C21H22 N2O7 | 415.15 | [M + H]+ | 18.99 | 397 (100), 379, 369, 355, 353, 337, 311, 297, 281, 269, 253, 226, 184 | −0.644 | 2 | [28] |
| Cyclopiamide J | C22H24N2O7 | 429.17 | [M + H]+ | 23.96 | 287 (100), 411, 497, 379, 369, 337, 269, 259, 226, 184 | −0.693 | 2 | [28] |
| KOJIC ACID BIOSYNTHETIC PATHWAY | ||||||||
| Kojic acid | C6H6O4 | 143.03 | [M + H]+ | 1.87 | 143 (100) 125, 113, 97 | 1.432 | 1, 2 | [29] |
| AFLATREM BIOSYNTHETIC PATHWAY | ||||||||
| α-aflatrem | C32H39NO4 | 502.29 | [M + H]+ | 41.45 | 444 (100), 484, 426, 412, 376, 198 | 1.144 | ||
| Paspalinine | C27H31NO4 | 434.23 | [M + H]+ | 39.22 | 376 (100), 416, 419, 362, 358, 344, 130 | 0.726 | 2 | [30] |
| Paspaline | C28H39NO2 | 422.31 | [M + H]+ | 43.96 | 130 (100), 404, 407 | −0.583 | 2 | [30] |
| Hydroxyaflatrem | C32H39NO5 | 518.29 | [M + H]+ | 38.22 | 460 (100), 500, 482, 442, 446, 428 | −0.347 | ||
| Paxilline | C27H33NO4 | 436.25 | [M + H]+ | 38.64 | 418 (100), 421, 400, 378, 360, 346, 130 | −2.762 | 1, 2 | [30] |
| 13′-desoxypaxilline | C27H33NO3 | 420.25 | [M + H]+ | 40.31 | 402 (100), 405, 362, 130 | −0.320 | 2 | [30] |
| ASPARASONE BIOSYNTHESIS PATHWAY | ||||||||
| Asparasone A | C18H14O8 | 357.06 | [M − H]− | 22.13 | 339 (100) 299 | 1.315 | 2 | [31] |
| 1,3,4,6,8 pentahydroxy-2-(1′-hydroxy-3′-oxobuty)anthraquinone | C18H14O9 | 373.04 | [M − H]− | 9.36 | 355 (100) 315 | 0.629 | 2 | [31] |
| 1,3,6,8 tetrahydroxy-2-(1′-hydroxyethyl) anthraquinone | C16H1207 | 315.05 | [M − H]− | 27.98 | 297 (100) | 0.775 | 2 | [31] |
| 1,3,6,8 tetrahydroxy-2-(3′ oxobut 1′-en-1′-yl) anthraquinone | C18H1207 | 339.05 | [M − H]− | 29.77 | 297 (100) 321, 296, 295, 311, 306 | 1.428 | 2 | [31] |
| LEPORINS BIOSYNTHESIS PATHWAY | ||||||||
| Leporin B | C22H25NO3 | 352.19 | [M + H]+ | 40.78 | 216 (100), 230, 244, 258, 270, 282, 296, 306 | −1.505 | 2 | [25] |
| Leporin B precursor | C22H25NO2 | 336.20 | [M + H]+ | 37.97 | 200 (100), 214, 228, 242, 254, 266, 280 | 0.102 | 2 | [25] |
| AFLAVARIN BIOSYNTHESIS PATHWAY | ||||||||
| Aflavarin | C24H22O9 | 455.13 | [M + H]+ | 18.22 | 413 (100), 425, 437, 395, 379, 364, 348, 303 | −3.732 | 1, 2 | [32] |
| 7′-demethyl-siderin | C11H10O4 | 207.07 | [M + H]+ | 13.58 | 163 (100), 177, 175, 148, 147, 135, 133, 131, 115, 107 | 0.312 | 2 | [32] |
| Aflavarin precursor 6 | C22H18O8 | 411.11 | [M + H]+ | 20.69 | 369 (100), 381, 379, 352, 343, 337, 279, 207, 177, 147 | −0.569 | 2 | [32] |
| Aflavarin precursor 5 | C23H20O8 | 425.12 | [M + H]+ | 26.75 | 383 (100), 393, 369, 363, 357, 349 | 0.484 | 2 | [32] |
| Aflavarin precursor 4 | C24H22O8 | 439.14 | [M + H]+ | 30.52 | 397 (100), 383, 371, 367, 365, 351, 341, 321 | −0.624 | 2 | [32] |
| AFLAVININE BIOSYNTHESIS PATHWAY | ||||||||
| 20′-hydroxyaflavinine | C28H39O2N | 404.29 | [M − H2O + H]+ | 37.53 | 386 (100), 287, 269, 243, 144, 130 | 0.071 | 1 | |
| Unknown aflavanine | C28H39O2N | 404.29 | [M − H2O + H]+ | 38.14 | 386 (100), 287, 269, 224 | 0.170 | ||
| Strain | Sampling Data | Reference | |
| Substrate | Country | ||
| A. arachidicola | |||
| CBS 117610T = IBT 25020 | Arachis glabatra leaf | Argentina | [20] |
| CBS 117614 = IBT 27183 | Arachis glabatra leaf | Argentina | [20] |
| A. bertholletius | |||
| CCT 7615T | Soil near Bertholletia excelsa trees | Brazil | [19] |
| A. bombycis | |||
| NRRL 26010T = CBS 117187 | Frass, silkworm rearing house | Japan | [66] |
| A. caelatus | |||
| NRRL 25528T = ATCC 201128 = CBS 763.97 = JCM 10151 | Peanut field soil | Georgia, USA | Horn B.W., National Peanut Lab, Dawson, GA (in NRRL database) |
| A. flavus | |||
| NRRL 3518 | Wheat flour | Illinois, USA | Graves NRRL isolate (in NRRL database) |
| NRRL 4818 = CBS 16870 | Food, butter | USA | Fennell D.I., University of Wisconsin, Madison, Wisconsin (in NRRL database) |
| NRRL 3357 = CBS 128202 | Peanut cotyledons | USA | [67] |
| A. minisclerotigenes | |||
| CBS 117635T | Arachis hypogaea seed | Argentina | [20] |
| NRRL 29000 | Peanut soil | Australia | Geiser D., Pennsylvania State University (in [21]) |
| E21 | Cumin | Morocco | [15] |
| E44 | White pepper | Morocco | [15] |
| E74 | Paprika | Morocco | [15] |
| A. mottae | |||
| MUM 10.231T = CBS 130016 | Maize seed | Portugal | [18] |
| A. nomius | |||
| NRRL 13137T = CBS 260.88 | Wheat | Illinois, USA | Schindler A.F., FDA, Washington D.C. (in NRRL database) |
| A. novoparasiticus | |||
| CBS 126849T = LEMI 250 | Sputum, leukemic patient | São Paulo, Brazil | [23] |
| LEMI 149 | Hospital air | São Paulo, Brazil | [23] |
| LEMI 267 | Sputum, leukemic patient | São Paulo, Brazil | [23] |
| AFc31 = NRRL 62794 | Cassava | Benin | [14] |
| AFc32 = NRRL 62795 | Cassava | Benin | [14] |
| A. oryzae | |||
| CBS 100925T = IMI 16266 = NRRL 447 | Unknown source | Japan | [17] |
| RIB40 | Cereal (broad bean) | Kyoto, Japan | [68] |
| A. parasiticus | |||
| CBS 100926T | Pseudococcus calceolariae, sugar cane mealy bug | Hawaii, USA | [17] |
| NRRL 492 | Unknown source | China | [23] |
| A. parvisclerotigenus | |||
| CBS 121.62T | Arachis hypogea | Nigeria | [9] |
| AFc36 = NRRL 62796 | Cassava | Benin | [14] |
| MACI8 | Peanuts | Côte d’Ivoire | This study |
| MACI221 | Peanuts | Côte d’Ivoire | This study |
| MACI258 | Peanuts | Côte d’Ivoire | This study |
| SF1 | Rain forest soil | Nigeria | [65] |
| SF3 | Rain forest soil | Nigeria | [65] |
| SF6 | Rain forest soil | Nigeria | [65] |
| SF9 | Food item | Nigeria | [65] |
| A. pseudocaelatus | |||
| CBS 117616T | Arachis burkartii leaf | Argentina | [17] |
| A. pseudotamarii | |||
| NRRL 443 | Soil | Brazil | [69] |
| NRRL 25518 | Tea field soil | Miyazaki, Japan | [70] |
| A. sergii | |||
| MUM 10.219T = CBS 130017 | Almond shell | Portugal | [18] |
| A. sojae | |||
| CBS 100928T | Soy sauce | Japan | [17] |
| A. transmontanensis | |||
| MUM 10.214T = CBS 130015 | Almond shell | Portugal | [18] |
| A. korhogoensis sp. nov. | |||
| MACI254T | Peanuts | Côte d’Ivoire | This study |
| MACI46 | Peanuts | Côte d’Ivoire | This study |
| MACI219 | Peanuts | Côte d’Ivoire | This study |
| MACI264 | Peanuts | Côte d’Ivoire | This study |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carvajal-Campos, A.; Manizan, A.L.; Tadrist, S.; Akaki, D.K.; Koffi-Nevry, R.; Moore, G.G.; Fapohunda, S.O.; Bailly, S.; Montet, D.; Oswald, I.P.; et al. Aspergillus korhogoensis, a Novel Aflatoxin Producing Species from the Côte d’Ivoire. Toxins 2017, 9, 353. https://doi.org/10.3390/toxins9110353
Carvajal-Campos A, Manizan AL, Tadrist S, Akaki DK, Koffi-Nevry R, Moore GG, Fapohunda SO, Bailly S, Montet D, Oswald IP, et al. Aspergillus korhogoensis, a Novel Aflatoxin Producing Species from the Côte d’Ivoire. Toxins. 2017; 9(11):353. https://doi.org/10.3390/toxins9110353
Chicago/Turabian StyleCarvajal-Campos, Amaranta, Ama Lethicia Manizan, Souria Tadrist, David Koffi Akaki, Rose Koffi-Nevry, Geromy G. Moore, Stephen O. Fapohunda, Sylviane Bailly, Didier Montet, Isabelle P. Oswald, and et al. 2017. "Aspergillus korhogoensis, a Novel Aflatoxin Producing Species from the Côte d’Ivoire" Toxins 9, no. 11: 353. https://doi.org/10.3390/toxins9110353